Abstract
Isolation and studies of novel, crude oil biodegrading thermophilic strains may provide a wider knowledge in understanding their role in petroleum degradation. In this study, the screening of ten new thermophilic strains revealed that all strains were alkane hydroxylase producers and seven of them produced lipase concurrently. Three best strains were characterized and identified through 16S rRNA sequence analysis as Geobacillus sp. D4, Geobacillus sp. D7, and Anoxybacillus geothermalis D9 with GenBank accession numbers MK615934.1, MK615935.1, and MK615936.1, respectively. Gas chromatography (GC) analysis showed that all three strains were able to breakdown various compounds in crude oil such as alkanes, toxic poly-aromatic hydrocarbons (PAHs), organosulfur, carboxylic acids, alkene, resins, organosilicon, alcohol, organochlorine, and ester. For the first time, alkane hydroxylase and lipase activity as well as crude oil degradation by A. geothermalis species were reported. Geobacillus sp. D7 is the best alkane degrader followed by A. geothermalis D9 and Geobacillus sp. D4 with 17.3%, 13.1%, and 12.1% biodegradation efficiency (BE%), respectively. The potential of thermophiles isolated can be explored further for bioremediation of sites polluted by petroleum and oil spills.
1. Introduction
Crude oil hydrocarbons are considered as one of the organic pollutants of high concern due to their wide distribution and persistence in nature [1,2,3]. The presence of complex compounds in crude oil such as alkanes, monocyclic, and polycyclic aromatic hydrocarbons causes toxicity to the environment and human health [2,4,5]. Crude oil biodegradation has been widely studied for the past few decades [6,7,8,9]. These studies have led to numerous microorganisms and their enzymes to be isolated and characterized. Despite its complexity and inertness, alkane, which made up the major components of crude oil, can be broken down aerobically by groups of cooperative enzymes such as alkane hydroxylase, alcohol dehydrogenase, aldehyde dehydrogenase, esterase, and lipase [10,11].
Alkane hydroxylase is known to be the first enzyme that initiates n-alkane degradation. Many researches were focused on the degradation of mainly the saturated portion of alkane in crude oil. In addition, only a few studies on alkane hydroxylase with a wide substrates range can be found. Alkane hydroxylase from Pseudomonas oleovorans GPol was reported to oxidize multiple forms of alkane and aromatic compounds [10]. Another similar observation was demonstrated by Alcanivorax dieselolei B-5 with multiple alkane hydroxylase systems [3]. On the other hand, alkane was proven to induce different protein expressions in Geobacillus thermoleovans B23 that could contribute to a wide range of crude oil components degradation [12].
Furthermore, most of the studies and current available commercial enzymes were done on various mesophilic microorganisms [3,11,13,14]. Thermophiles and their enzymes are more robust and stable as compared to mesophiles and psychrophiles. The presence of abundant disulfide bonds and positively charged amino acids in thermostable protein was responsible for their stability [12]. These proteins are often more tolerant of high pressure and organic solvents [14]. Moreover, the pressure resistance exhibits by β-glucosidases resulted from their thermal-resistance properties [15]. Thus, a search for special alkane hydroxylase in terms of thermostability and substrate range for bioremediation and industrial purposes is of interest. The isolations of thermophilic bacteria that produced alkane hydroxylase are necessary to accommodate these demands. In this study, the screening of thermophilic alkane hydroxylase producers was done on the east coast and the central region of peninsular Malaysia. The best three alkane hydroxylase producers—Geobacillus sp. D4, Geobacillus sp. D7, and Anoxybacillus geothermalis D9—were characterized and the degradation of various complex compounds in crude oil was recorded.
2. Results
2.1. Isolation, Screening, and Selection of Thermophilic Bacteria for Crude oil Degradation
In this study, 23 thermophilic bacteria that are able to utilize crude oil as a sole carbon source were isolated from seawater and soil. Ten out of 23 were chosen based on their thermostability to screen for potential crude oil degraders. Figure 1 shows OD600 and changes of pH culture medium every 24 h after the inoculation of isolates in enriched media supplemented with 1% of crude oil (w/v) for an incubation period of three days. Isolates D2, D5, D6, D8, and D10 grew moderately while exponential growth was observed with D1 and D7 along the incubation period. The highest growth was recorded on the second day of incubation for both D4 and D9 before it started to drop in the following day. However, isolate D3 only grew after two days of incubation (Figure 1a) which indicates that D3 might be a slow-growing bacterium. pH analysis of the culture medium shows an increase of alkalinity from the initial pH of 7.5 for all ten isolates once the bacteria cells proliferate. Likewise, when Pseudomonas synxantha LSH-7 were grown in crude oil, the pH of media became alkaline due to acid and alkaline phosphatase produced during the synthesis of protein and lipid [16]. In Figure 1b, the culture medium of isolate D3 was slightly acidic for the first two days of incubation due to the absence of bacterial growth.
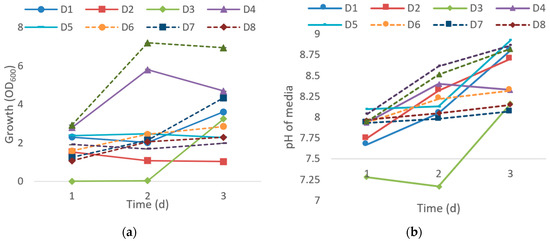
Figure 1.
Effects of modified ocean water with 1% crude oil after inoculation of ten isolates in an incubation period of three days. (a) Optical density of culture growth every 24 h. (b) The changes in pH media from initial pH 7.5 after inoculation every 24 h. The values presented were the averages of three replicates. Error bar represents the standard error while no error bar denotes the standard error values that are too small.
During the three days of incubation, D7 produced the highest alkane hydroxylase activity while D1, D4, D8, D9, and D10 showed more than 50% activity relative to D7 (Figure 2). Whereas for D2, D3, D5, and D6, the alkane hydroxylase relative activity was in a range of 7% to 25%. The production of lipase by all isolates, except D1, D3, and D5, has emphasized the role of lipase in crude oil degradation (Figure 2). In this study, D2 produced the highest lipase activity followed by D4, D9, and D7 with relative activity of 48%, 44%, and 42%, respectively. Contrarily, D6, D8, and D10 only produced a small amount of lipase with not more than 21% of relative activity. Thus, the three isolates with the highest alkane hydroxylase and lipase activity were selected, which were D4, D7, and D9.
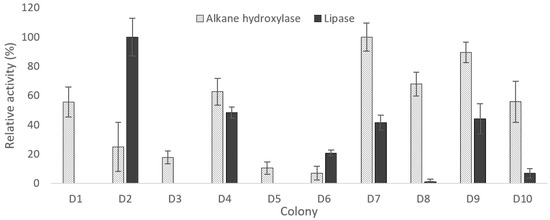
Figure 2.
The average of extracellular alkane and lipase relative activity of each isolates after three days of incubation in modified ocean water media with 1% crude oil. The data shows a significant difference between alkane hydroxylase and lipase activity with a P-value of 0.02. The values presented were the averages of three replicates. Error bar represents the standard error.
2.2. Characterization and identification of strains.
The morphological and biochemical characterization of D4, D7, and D9 were as stated in Table 1. The three colonies are Gram-positive rod and endospore-forming bacteria. The colonies are circular with creamy to whitish pigmentation when streaked on nutrient agar at 50 °C. They are aerobic catalase producing bacteria that could hydrolyze lipid and starch. From the biochemical test, the isolates utilized glucose, lactose, and crude oil as their sole carbon source. Furthermore, D7 and D9 could utilize sucrose, whereas only D9 could hydrolyze casein in 24 h of incubation.

Table 1.
Morphological and biochemical identification of the three isolates.
The 16S rRNA sequence analysis showed that both isolates D4 and D7 belong to the Geobacillus species with 99.9% identity to Geobacillus kaustrophilus NBRC 102445 and 99.3% to the Geobacillus jurassicus NBRC 107829 strain R-35651, respectively (Figure 3). The constructed phylogenetic tree proves that D4 and D7 are from a different species completely. Contrarily, in this study D9 was identified as an A. geothermalis species. Phylogenetic analysis showed 100% confidence that D9 was closely related to Anoxybacillus rupiensis R270. However, the phylogenetic relationship was constructed from a partial 16S rRNA nucleotide sequence through the missing gap deletion process. Therefore, the BLAST result for a complete 16S rRNA nucleotide sequence of D9 determined 99.8% of identity for Anoxybacillus geothermalis ATCC BAA-2555. Nucleotide sequence of 16S rRNA for D4, D7, and D9 were submitted to the NCBI GenBank nucleotide database with accession numbers MK615934.1, MK615935.1, and MK615936.1, respectively.

Figure 3.
Phylogenetic tree of three isolates and the relationship of their 16S rRNA nucleotide sequences with other 24 related bacteria where Bacillus subtilis IAM 12118 served as outgroup.
2.3. Total Protein Content and Alkane Hydroxylase for D4, D7, and D9
During the cell propagation in 1% crude oil medium, the extracellular protein content increased gradually for all isolates in a period of three days. The data shows that the highest extracellular protein content was recorded on the third day with 0.134 mg/mL, 0.175 mg/mL, and 0.065 mg/mL for D4, D7, and D9, respectively. In contrast, the intracellular protein concentrations decreased except for D4 as it dropped on the second day of incubation and continued to rise again on the third day (Figure 4). The protein concentration was proportional to its cell mass, thus the reduction of protein content might be due to a drop of cell mass production on that day (data not shown). Nevertheless, enzyme activity was measured at a standardized protein content (1 ng) in every experiment. The intracellular protein concentration was highest on the first day for D9 with 0.586 mg/mL, second day for D7 with 0.712 mg/mL, and the third day for D4 with 0.719 mg/mL. The alkane hydroxylase secreted on the first day (Figure 4b) was higher as compared with the one produced intracellularly (Figure 4c). Isolates D4, D7, and D9 had the highest extracellular alkane hydroxylase activity on the first day with 0.192 U/mL, 1.120 U/mL, and 0.331 U/mL, respectively, where it gradually decreased over the incubation period (Figure 4b). In the meantime, D7 and D9 showed the highest intracellular alkane hydroxylase activity with 0.163 U/mL and 0.244 U/mL, respectively, whereas for D4, the activity was highest on the third day with 0.140 U/mL (Figure 4c).
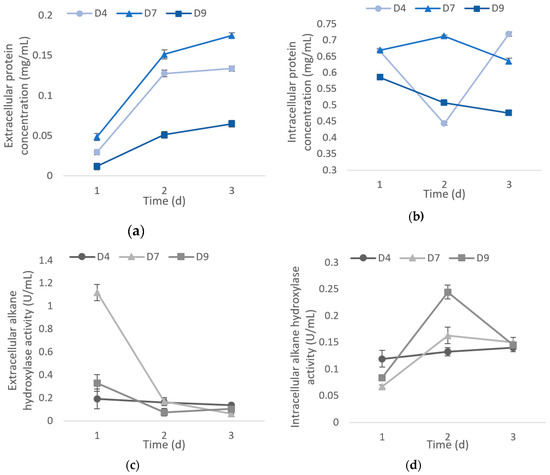
Figure 4.
Analysis of crude enzyme concentration of isolates and their alkane hydroxylase activity every 24 h in three days of incubation period. Extracellular (a) and intracellular (b) total protein concentration. Extracellular (c) and intracellular (d) alkane hydroxylase relative activity at 1 ng of protein. The data shows that there is a significant difference between extracellular and intracellular alkane hydroxylase activity measured at each day (P < 0.05). The values presented were the averages of three replicates. Error bar represents the standard error while no error bar denotes the standard error values that are too small.
2.4. Gas Chromatography (GC) Analysis
GC analysis in Figure 5a visualized n-alkane as the major component of crude oil with 87.2% followed by 6.3% of branched alkane, 1.8% of aromatic compounds, 1.3 % of alkene, and 3.4% of trace compounds which included cyclo-alkane, resins, organosulfur, ester, organochlorine, carboxylic acids, organosilicon, and alcohol. Regardless of having a degradation of only 12.6%, Figure 5b shows that n-alkane still represents the largest fractions of degraded crude oil. Branched alkane, aromatic, and organosilicon, on the other hand, have average degradation values of 25.5%, 34.9%, and 37.8%, respectively (Figure 5b). However, the average degradation percentage of alkene, cyclo-alkane, ester, organochlorine, and resins were all recorded to be less than 10%. All colonies showed more than 50% preference towards alcohol, organosulfur, and carboxylic acids with degradation rates of 94.8%, 72.3%, and 50.8%, respectively.
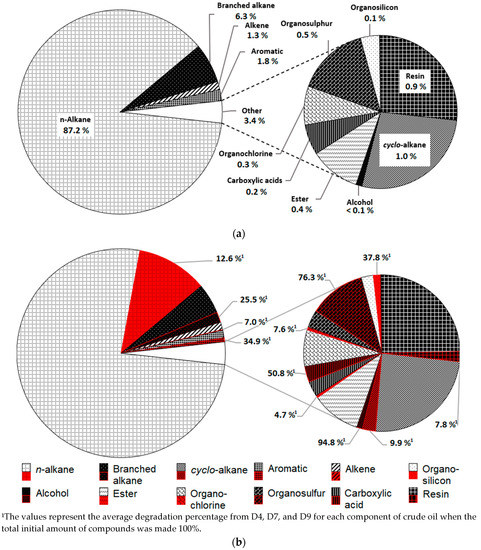
Figure 5.
The fractions of crude oil based on gas chromatography (GC) analysis. (a) Compound groups that made up crude oil components before treatment. (b) Red shading visualized the compound removal after inoculation based on average degradation percentage by the three isolates for each compound type.
In this study, all strains were able to remove n-decane (C10) completely while a moderate breakdown of n-undecane (C11) was observed with 40.8%, 42.4%, and 51% for D4, D7, and D9, respectively (Table 2). Meanwhile, n-tetradecane (C14) was removed by D4, D7, and D9 with 52.8%, 63.5%, and 59.1% degradation, respectively. Degradation of n-tricosane (C23) and n-hexacosane (C26) was recorded to be more than 14% by all isolates. Generally, the isolates prefer to degrade shorter to medium-chain alkane where the degradation percentage for C15 to C17 was observed between 10.9% and 21.6%. In addition, C18 and above showed less than 10% of degradation (Table 2). Moreover, the isolates were also able to oxidize branched alkane molecules with 100% degradation of 3-ethylhexane (C8) for D7 and 46% to 80% degradation of 2,6,10-trimethyldodecane (C15) and 2-methyl-6-propyldodecane (C18) for all isolates. Longer branched alkane from C18 to C20 was being degraded at a slower rate with an average degradation percentage of 19% for all isolates. A small portion of saturated cyclo-alkane molecules such as 1-Nonylcycloheptane (C16) is also degraded with 21%, 37.5%, and 25.2% for D4, D7, and D9, respectively. Contrarily, only D7 and D9 could degrade cyclotetracosane (C24) and 1,1,3,6-tetramethyl-2-(3,6,10,13,14-pentamethyl-3-ethyl-pentadecyl) cyclohexane (C32) with an average of 13.6% removal. However, all isolates show very little affinity towards nonadecylcyclohexane (C25) with less than 10% of degradation. In addition, some alkene fraction in crude oil was degraded where a total breakdown of (2Z,4E)-3,7-Dimethyl-2,4-octadiene (C10) by D7 and D9 was achieved, while about 81.4% were broken down by D4. Slow removal of hexacosene (C26) was also measured with degradation ranging from 2.6% to 10.8% by isolates.

Table 2.
The tabulated data of degradation percentage for each compound type and their RT. The Biodegradation Efficiency BE% for D4, D7, and D9 on overall crude oil degradation was listed.
Furthermore, the isolates have the ability to degrade toxic mono- and polyaromatic compounds with 100% removal of 1,2,3,5-tetramethylbenzene. In addition, 2,3-dimethylnaphthalene, 1,6,7-trimethylnaphthalene, and 2,3,5-trimethylphenanthrene were removed with degradation percentages of 31.6% to 34.3%, 48.5% to 54.6%, and 7.5% to 11.5%, respectively. Furthermore, only isolates from the Geobacillus species could degrade polyaromatic compound 9,10-dimethylanthracene with 3.2% and 20.1% degradation for D4 and D7, respectively. From this study, resin group 28-Nor-17.beta.(H)-hopane (C29) which is one of the most persistent compounds was also removed with less than 10.1% by all isolates. Apart from that, alcohol which is composed of the smallest fraction of crude oil was removed effectively where the degradation of 2-methyl-1-undecanol is 100% for D4 and D9, while the degradation for D7 is 84.5%. The data collected shows that only a small removal of ester and organochloride molecules was recorded, where the degradation percentage is less than 12.6%. The GC results also showed complete removal of carboxylic acid compound, which is 2-tetradecyl ester methoxyacetic acid. On the other hand, some organosilicon compounds were removed at a higher rate where 100% degradation of trichlorodocosylsilane and more than 28% degradation of octamethylcyclotetrasiloxane were observed by all isolates. Degradation of organosulfur compounds was also recorded where a 100% breakdown of butyl octadecyl ester sulfurous acid was demonstrated by both Geobacillus isolates. Moreover, more than 70% of 1-octadecanesulphonyl chloride were also removed by all isolates.
The negative values in Table 2 indicate the presence of new compounds that are originally absent in the control. Increase of n-hexane (C6), n-octadecane (C18), n-pentacosane (C25), n-heptacosane (C27), n-nonacosane (C29), and n-pentatriacontane (C35) in crude oil compositions by some of the isolates have resulted from the breakdown of longer n-alkane and other hydrocarbons. Similarly, the presence of 1-Tricosene in D9 could have resulted from a breakdown of longer alkene molecules and the degradation of the products is slower than their formation. The Biodegradation Efficiency (BE%) calculated in Table 2 proposed that D7 has a better overall efficiency to degrade crude oil followed by D9 and D4 with BE% of 17.3, 13.1, and 12.1, respectively. Total ion chromatogram (TIC) of the crude oil degradation by each isolate conducted in this study can be found in the Supplementary Materials.
3. Discussion
3.1. Growth and Enzyme Analysis of Isolates
Most of the Geobacillus species was known to utilize n-alkanes as energy source such as G. kautophilus TERI and G. jurassicus DS1T [6,17]. Some of the Geobacillus species possess an alkane hydroxylase gene, AlkB, or its homologs as found in G. thermoleovorans B23 and G. toebii B-1024 [12,18]. So far, there is only one reported study on genus Anoxybacillus on alkane hydroxylase involved in crude oil degradation as recorded by Anoxybacillus sp. WJ-4 [8]. Another study demonstrated that organic solvent tolerant Anoxybacillus sp. PGDY12 is able to grow in the presence of n-alkanes for more than five days [19]. However, the degradation of n-alkane and the presence of alkane hydroxylase were not mentioned. Thus, this study demonstrated for the second time crude oil degradation and alkane hydroxylase produced by the Anoxybacillus genus and the first time reported for the A. geothermalis species.
The growth of bacteria in hydrocarbon-rich environments induced the high protein concentration by all isolates (Figure 4a,b). Similarly, the protein content of G. thermoleovorans B23 was increased when the bacteria were cultivated in the presence of alkanes [12]. The increase of extracellular alkane hydroxylase as compared with intracellular enzymes in the first day (Figure 4c,d) corresponds to the new alkane degradation pathway as proposed by Meng (2018). As reported in the study, alkane was degraded outside the cell by extracellular alkane hydroxylase before uptake, thus explaining the high extracellular alkane hydroxylase activity in the first day [20]. As the degradation goes by, the degraded alkanes entered the cells, thus reducing the extracellular alkane hydroxylase produced. This is because small alkane molecules that could possibly diffuse into the cells caused the intracellular alkane hydroxylase to increase along the incubation period.
Moreover, the majority of thermophiles in this study produced lipase apart from the main alkane oxidizing enzyme. This result highlights the importance of lipase in crude oil degradation. Research states that lipase is involved in inducing the lipolytic reactions of emulsified hydrocarbon at the lipid–water interface, thus assisting the hydrocarbon uptake [7]. Furthermore, many studies showed the presence of lipase intracellularly or extracellularly produced by hydrocarbon degraders from Bacillus and Pseudomonas species [7,9,16]. Nevertheless, in this study, lipase production does not affect the pH media (Figure 1b and Figure 2). Although lipase activity resulted in the production of acidic products, however, fatty acid produced in three days crude oil degradation is not enough to change the medium pH. The alkalinity of the pH medium was instead caused by nutrients metabolism during bacteria growth [16].
3.2. Biodegradation of Crude Oil Compositions
Despite being saturated hydrocarbons, n-alkane is the most degraded compound in this study. The degradation summary (Figure 5) shows that the order of the most degraded compounds in relative comparison is n-alkane > branched alkane > aromatic compounds > organosulfur > carboxylic acids > cyclo-alkane > alkene > resins > organosilicon > alcohol > organochlorine > ester. A similar order of alkane biodegradation preference was observed as linear alkane being the most favorable followed by branched and cyclic alkanes, respectively [1].
In this study, all isolates prefer short-chain n-alkane as comparison to longer n-alkane. Lower molecular weight alkanes have a higher cell surface hydrophobicity which will increase the chances of uptake and small-sized n-alkane can pass through lipoproteins present in the membrane [7,20]. Therefore, longer chain n-alkane have a lower degradation rate as they either need to form emulsion by the help of surfactants or they need to be degraded extracellularly prior to uptake [20]. Branched alkanes, which have bulkier structures as compared to linear alkanes, are more difficult to be degraded as the alkyl branch hinders them from initial oxidation [21].
On the other hand, aromatics with high molecular weights are considered to be persistent and recalcitrant in nature. Nevertheless, poly-aromatic hydrocarbons (PAHs) were reported to be one of the most common compounds to be degraded in crude oil as the alkyl substituent or double bond of the benzene ring are more susceptible to enzymatic attack as compared to other complex hydrocarbons [22]. Thus, the data collected in this study shows that PAHs were being the third most degraded compounds where their removal was recorded to be more than 30% of the total amount by the isolates (Figure 5b). However, phenanthrene and anthracene in the study were being degraded at a much slower rate and there is an increase for anthracene compounds in crude oil treated with D9. Phenanthrene and anthracene were reported to be the most resistant PAH compounds [2].
Some researches state that it is thermodynamically less favorable to oxidize hopanes to their corresponding carboxylic acids [23,24]. This is because alcohols are reported to be readily degraded into its corresponding aldehydes [25]. Moreover, various toxic compounds other than aromatics such as organochlorine, organosilicon, and organosulfur found in crude oil can also be found in the environment as pollutants from industrial waste and in pharmaceutical products. Organochlorine compounds which are commonly used as pesticides were degraded by Pseudomonas, Flavimonas, and Morganella species [26]. The findings indicate both Geobacilus species and A. geothermalis isolates also have similar degradation capabilities towards these chloroalkane compounds. Despite being undesirable products [27], siloxane compounds were reported to be removed by Methylibium sp. and P. aeruginosa [28]. Likewise, organosulfur was widely reported as one of the common compounds to be biodegraded through alkanesulfonates monooxygenases [29,30].
A study shows that there is an increase of isoprenanes, steranes, and herpanes by G. jurassicus DST when grown in crude oil [17]. Similarly, the increase of acidic components are common intermediates produced in biodegraded oils [31,32]. Data also show an increasing amount of ester (Table 2.) such as 4-dodecyl dimethyl ester 1,2,4-benzenetricarboxylic acid might result from the degradation of 1,2,3,5-Tetramethylbenzene that reacted with other hydrocarbons. Nocardia and Vocardia species were reported to convert 1,2,3,4-Tetramethylbenzene (prehnitene) to 2,3-Dihydroxy-4,5,6-trimethyl benzoic acid during the degradation process [33]. Therefore, the accumulation of new compounds might result from the intermediates produced by the bacteria during crude oil metabolism.
4. Materials and Methods
4.1. Chemicals and Samples
The crude oil used in this study was sourced from PETRONAS Research Sdn Bhd, Malaysia, and stored at room temperature. Unless specified, chemical reagents used in this study were purchased from Sigma-Aldrich, St. Louis, MO, USA, with a purity of ≥99.0%. Culture media were purchased from Merck, Darmstadt, HE., Germany, and Fisher Scientific, Hampton, NH, USA. Seawater sample and oil contaminated soil were collected from Cherating beach, 26080 Balok, Pahang (4°7′15.80″ N, 103°23′20.20″ E) and a car workshop in Batu Caves, Selangor, Malaysia (3°15′28.09″ N, 101°40′38.65″ E), respectively.
4.2. Isolation and Screening of Crude Oil-Degrading Bacteria
Seawater and soil samples were cultivated in Bushnell and Haas minimal medium with the following composition (g/L): 1 g of KHPO4, 1 g of K2HPO4, 1 g of NH4NO3, 0.2 g MgSO4.7H2O, 0.05 g of FeCl3, 0.02 g of CaCl2.2H2O, and 1% of crude oil pH 7.0 [34]. Cultures were incubated at 50 °C and 70 °C with 100 rpm agitation for seven days. The harvested bacteria present in the culture media were isolated every one, two, three, and seven days on nutrient agar and incubated at 50 °C and 70 °C for 24 h.
Enzymes production was obtained from growing the bacteria at 60 °C with 100 rpm agitation for three days in modified ocean water media [35] pH 7.5 with the following components (g/L): 5 g of yeast extract, 5 g of peptone, 3 g of beef extract, 19.9 g of NaCl, 0.6 g of KCl, 4.4 g of MgCl2.6H2O, 5.8 g of MgSO4.7H2O, 0.08 g of CaCl2, and 1% of crude oil.
4.3. Alkane Hydroxylase Activity
Alkane hydroxylase activity was measured according to the standard NADH assay method with modification by measuring absorbance change at 340 nm (ε = 6,220 M−1 cm−1) [10,29,36]. The reaction mixture containing a final concentration of 500 µM of NADH, 1 mM of n-hexadecane, 0.02% of DMSO, and 1 ng of enzyme was added in 100 mM PBS buffer with pH 8.0. The reaction was initiated by the addition of NADH to the reaction mixture and incubated at 50 °C for 5 min. The reaction mixture without enzyme acts as the control. Activity was expressed as unit/mL (U/mL) enzyme where one unit of alkane hydroxylase activity is defined as the amount of enzymes required for consumption of 1 μM of NADH per minute at 50 °C.
4.4. Lipase Activity
Measurement of lipase activity was performed by a modified Winkler and Stuckmann method by using p-nitrophenol as a substrate [37]. The reaction mixture contained 25 µL of 5 mM PNP palmitate solution in isopropanol, 1 µL of crude enzyme, and 224 µL of 100 mM PBS buffer, pH 8.0. The mixture was incubated at 70 °C with shaking for 10 min and the absorbance was recorded at 410 nm. One unit of lipase activity was defined as the amount of enzymes releasing 1 µmol of p-nitrophenol per minute at 70 °C.
4.5. Protein Quantitation
The modified Bradford method [38] was used in the detection and quantitation of the total protein content in the crude enzyme. The reaction consisted of 200 μL of Bradford reagent (Sigma-Aldrich, St. Louis, MO, USA), 10 μL of 0.15 M NaCl, and 10 μL of sample, and mixed in a 96-well microplate. The reaction was incubated for 10 min at room temperature and read the absorbance at 595 nm. Bovine serum albumin (BSA) was used standard.
4.6. Characterization and Identification of Strains
Bacterial isolates were characterized and identified by physiological analysis and biochemical tests according to Bergey’s Manual of Determinative Bacteriology [39]. Genomic DNA was extracted using the Qiagen Dneasy Blood and Tissue Kit (Qiagen, Hilden, NW., Germany). The 16S rRNA sequence of strain was amplified using Taq DNA polymerase (ABM Inc., Richmond, BC, Canada) with a universal forward primer (8F): 5′AGAGTTTGATCCTGGCTCAG3′ and a reverse primer (1492R): 5′ACGGCTACCTTGGTTACGACTT3′ [40] under standard conditions. Sequence similarity was performed using the BLASTn program at the National Centre for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST.html).
4.7. Construction of the Phylogenetic tree
The 16S rRNA of bacterial strains was aligned with nucleotide sequences of related bacteria obtained from NCBI GenBank using the MUSCLE program. A phylogenetic tree was constructed by the Neighbour-Joining method [41] using Jukes and Cantor’s model [42] with 1000 resampling repeats of bootstrapping analysis to evaluate the tree algorithms data. The tree and evolutionary analyses were conducted in the MEGA X software [43]. The evolutionary distances were computed using the Maximum Composite Likelihood and are in the units of the number of base substitutions per site. This analysis involved 28 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated (complete deletion option). There were a total of 1171 positions in the final dataset.
4.8. Gas Chromatography (GC) Analysis
Characterization of total hydrocarbon fractions during biodegradation was analyzed by the gas chromatographic–mass spectrometry (GC–MS) model Agilent 7890A, HP-5MS (30 m × 0.25 µm × 0.25 µm, Santa Clara, CA, USA). The operating conditions were as follows: helium was used as the carrier gas; MS source and quad temperature were 230 °C and 150°C, respectively; the oven temperature was kept at 35 °C for 1 min then raised to 160 °C and 315 °C at a rate of 15 °C/minute and 5 °C/minute, respectively.
4.9. Statistical Analysis
All experiments were carried out in triplicates. Statistical analyses were performed using a two-way analysis of variance (ANOVA) and t-Test to determine a significant level (P-value) between the independent variables. The analysis was done using Microsoft Excel data analysis tools with standard protocols.
5. Conclusions
The presence of lipase in almost all screened thermophiles indicates that lipase plays an important role in hydrocarbons metabolism. The best three isolates were characterized as Gram-positive rod and endospore-forming bacteria. They are catalase and amylase positive and utilized crude oil as their sole carbon source. The 16S rRNA sequences of Geobacillus sp. D4, Geobacillus sp. D7, and A. geothermalis D9 described here are available via GenBank accession numbers MK615934.1, MK615935.1, and MK615936.1, respectively. The extracellular and intracellular alkane hydroxylase analysis has shown that some of the alkanes degraded outside the cells into their corresponding acids before the uptake. The breakdown of various alkylated compounds might be promoted by alkane hydroxylase produced and the involvement of other enzymes that have yet to be discovered. In this study, crude oil degradation by A. geothermalis was reported for the first time. Therefore, to have a complete knowledge of these isolates and their bioremediation potential, further characterization and studies on the pathways, enzymes produced, and gene expressions are needed to be carried out in the future.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4344/10/8/851/s1, Figure S1: Head to tail comparisons of total ion chromatogram (TIC) exhibits the relative abundance of crude oil at different RT between the control (black) and treated (blue) with D4 (a), D7 (b) and D9 (c). The TIC of all the three isolates shows the relative abundance of lighter end compounds were notably reduced.
Author Contributions
Conceptualization, D.F.Y., R.N.Z.R.A.R., and M.M.; methodology, D.F.Y., M.M., and R.N.Z.R.A.R.; validation, D.F.Y. and M.M.; formal analysis, D.F.Y. and M.M.; investigation, D.F.Y.; resources, R.N.Z.R.A.R., M.S.M.A., and T.C.L.; data curation, D.F.Y. and R.N.Z.R.A.R.; writing—original draft preparation, D.F.Y.; writing—review and editing, M.M., R.N.Z.R.A.R., and M.S.M.A.; visualization, D.F.Y.; supervision, R.N.Z.R.A.R., M.S.M.A., and T.C.L.; project administration, R.N.Z.R.A.R.; funding acquisition, R.N.Z.R.A.R., M.S.M.A., and T.C.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
D.F.Y. is supported by the Graduate Research Fellowship (GRF) program from Universiti Putra Malaysia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chaineau, C.H.; Rougeux, G.; Yepremian, C.; Oudot, J. Effects of nutrient concentration on the biodegradation of crude oil and associated microbial populations in the soil. Soil Biol. Biochem. 2005, 37, 1490–1497. [Google Scholar] [CrossRef]
- Fernández-Álvarez, P.; Vila, J.; Garrido-Fernández, J.M.; Grifoll, M.; Lema, J.M. Trials of bioremediation on a beach affected by the heavy oil spill of the Prestige. J. Hazard. Mater. 2006, 137, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.W.G.; Chang, T.C.; Whang, L.M.; Kao, C.H.; Pan, P.T.; Cheng, S.S. Bioremediation of petroleum hydrocarbon contaminated soil: Effects of strategies and microbial community shift. Int. Biodeter. Biodegrad. 2011, 65, 1119–1127. [Google Scholar]
- Hamdoun, A.M.; Griffin, F.J.; Cherr, G.N. Tolerance to biodegraded crude oil in marine invertebrate embryos and larvae is associated with expression of a multixenobiotic resistance transporter. Aquat. Toxicol. 2002, 61, 127–140. [Google Scholar] [CrossRef]
- Zhang, Z.; Hou, Z.; Yang, C.; Ma, C.; Tao, F.; Xu, P. Degradation of n-alkanes and polycyclic aromatic hydrocarbons in petroleum by a newly isolated Pseudomonas aeruginosa DQ8. Bioresour. Technol. 2011, 102, 4111–4116. [Google Scholar] [CrossRef]
- Sood, N.; Lal, B. Isolation and characterization of a potential paraffin-wax degrading thermophilic bacterial strain Geobacillus kaustophilus TERI NSM for application in oil wells with paraffin deposition problems. Chemosphere 2008, 70, 1445–1451. [Google Scholar] [CrossRef]
- Sakthipriya, N.; Doble, M.; Sangwai, J.S. Fast degradation and viscosity reduction of waxy crude oil and model waxy crude oil using Bacillus subtilis. J. Petrol. Sci. Eng. 2015, 134, 158–166. [Google Scholar] [CrossRef]
- Xia, W.; Dong, H.; Zheng, C.; Cui, Q.; He, P.; Tang, Y. Hydrocarbon degradation by a newly isolated thermophilic Anoxybacillus sp. with bioemulsifier production and new alkB genes. RSC Adv. 2015, 5, 102367–102377. [Google Scholar] [CrossRef]
- Mulani, N.; Fulke, A.B.; D’souza, E.; Ram, A.; Maloo, A.; Sayed, F.; Gajbhiye, S.N. Biodegradation of crude oil using marine Bacillus species from Vadinar coast, Gujarat, India. Curr. Sci. 2017, 112, 569–576. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Kingma, J.; Witholt, B. Substrate specificity of the alkane hydroxylase system of Pseudomonas oleovorans GPo1. Enzyme Microb. Technol. 1994, 16, 904–911. [Google Scholar] [CrossRef]
- Park, C.; Park, W. Survival and Energy Producing Strategies of Alkane Degraders under Extreme Conditions and Their Biotechnological Potential. Front. Microbiol. 2018, 9, 1081. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Miyanaga, A.; Kanaya, S.; Morikawa, M. Alkane inducible proteins in Geobacillus thermoleovorans B23. BMC Microbiol. 2009, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, A.; Nercessian, O.; Fayolle, F.; Blanchet, D.; Jeanthon, C. Microbial diversity in production waters of a low-temperature biodegraded oil reservoir. FEMS Microbiol. Ecol. 2005, 54, 427–443. [Google Scholar] [CrossRef]
- Atalah, J.; Cáceres-Moreno, P.; Espina, G.; Blamey, J.M. Thermophiles and the applications of their enzymes as new biocatalysts. Bioresour. Technol. 2019, 280, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Hamon, V.; Dallet, S.; Legoy, M.D. The pressure-dependence of two β-glucosidases with respect to their thermostability. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1996, 1294, 195–203. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Bao, M.; Sun, P. Metabolic pathway for a new strain Pseudomonas synxantha LSH-7′: From chemotaxis to uptake of n-hexadecane. Sci. Rep. 2017, 7, 39068. [Google Scholar] [CrossRef]
- Gordadze, G.N.; Poshibaeva, A.R.; Giruts, M.V.; Gayanova, A.A.; Semenova, E.M.; Koshelev, V.N. Formation of Petroleum Hydrocarbons from Prokaryote Biomass: 2. Formation of Petroleum Hydrocarbon Biomarkers from Biomass of Geobacillus jurassicus Bacteria Isolated from Crude Oil. Pet. Chem. 2018, 58, 1005–1012. [Google Scholar]
- Tourova, T.P.; Sokolova, D.S.; Semenova, E.M.; Shumkova, E.S.; Korshunova, A.V.; Babich, T.L.; Poltaraus, A.B.; Nazina, T.N. Detection of n-alkane biodegradation genes alkB and ladA in thermophilic hydrocarbon-oxidizing bacteria of the genera Aeribacillus and Geobacillus. Microbiology 2016, 85, 693–707. [Google Scholar] [CrossRef]
- Gao, Y.; Dai, J.; Peng, H.; Liu, Y.; Xu, T. Isolation and characterization of a novel organic solvent-tolerant Anoxybacillus sp. PGDY12, a thermophilic Gram-positive bacterium. J. Appl. Microbiol. 2011, 110, 472–478. [Google Scholar]
- Meng, L.; Li, W.; Bao, M.; Sun, P. Promoting the treatment of crude oil alkane pollution through the study of enzyme activity. Int. J. Biol. Macromol. 2018, 119, 708–716. [Google Scholar] [CrossRef]
- Atlas, R.M.; Bartha, R. Hydrocarbon biodegradation and oil spill bioremediation. In Advances in Microbial Ecology, 1st ed.; Marshall, K.C., Ed.; Springer: Boston, MA, USA, 1992; Volume 12, pp. 287–338. [Google Scholar]
- Gibson, D.T.; Subramanian, V.; Gibson, D.T. Microbial degradation of aromatic hydrocarbons. In Microbial Degradation of Organic Compounds, 1st ed.; Gibson, D.T., Ed.; Marcel Dekker, Inc.: New York, NY, USA, 1984; Volume 13, pp. 181–242. [Google Scholar]
- Prince, R.C.; Elmendorf, D.L.; Lute, J.R.; Hsu, C.S.; Haith, C.E.; Senius, J.D.; Butler, E.L. 17. alpha.(H)-21. beta.(H)-hopane as a conserved internal marker for estimating the biodegradation of crude oil. Environ. Sci. Technol. 1994, 28, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.S.; Jones, D.M.; Swannell, R.P.J.; Van Duin, A.C.T. Formation of carboxylic acids during aerobic biodegradation of crude oil and evidence of microbial oxidation of hopanes. Org. Geochem. 2002, 33, 1153–1169. [Google Scholar] [CrossRef]
- Schaefer, C.E.; Yang, X.; Pelz, O.; Tsao, D.T.; Streger, S.H.; Steffan, R.J. Aerobic biodegradation of iso-butanol and ethanol and their relative effects on BTEX biodegradation in aquifer materials. Chemosphere 2010, 81, 1104–1110. [Google Scholar] [CrossRef]
- Barragan-Huerta, B.E.; Costa-Pérez, C.; Peralta-Cruz, J.; Barrera-Cortés, J.; Esparza-García, F.; Rodríguez-Vázquez, R. Biodegradation of organochlorine pesticides by bacteria grown in microniches of the porous structure of green bean coffee. Int. Biodeter. Biodegr. 2007, 59, 239–244. [Google Scholar] [CrossRef]
- Hirner, A.V.; Flassbeck, D.; Gruemping, R. Organosilicon compounds in the environment. In Oganometallic Compounds in the Environment, 2nd ed.; Peter, J.C., Ed.; John Wiley and Sons: West Sussex, UK, 2003; pp. 305–351. [Google Scholar]
- Boada, E.; Santos-Clotas, E.; Bertran, S.; Cabrera-Codony, A.; Martín, M.J.; Bañeras, L.; Gich, F. Potential use of Methylibium sp. as a biodegradation tool in organosilicon and volatile compounds removal for biogas upgrading. Chemosphere 2020, 240, 124908. [Google Scholar] [CrossRef]
- Eichhorn, E.; van der Ploeg, J.R.; Leisinger, T. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 1999, 274, 26639–26646. [Google Scholar] [CrossRef]
- Leidner, H.; Gloor, R.; Wüest, D.; Wuhrmann, K. The influence of the sulphonic group on the biodegradability of n-alkylbenzene sulphonates. Xenobiotica 1980, 10, 47–56. [Google Scholar] [CrossRef]
- Behar, F.H.; Albrecht, P. Correlations between carboxylic acids and hydrocarbons in several crude oils. Alteration by biodegradation. Org. Geochem. 1984, 6, 597–604. [Google Scholar] [CrossRef]
- Langbehn, A.; Steinhart, H. Biodegradation studies of hydrocarbons in soils by analyzing metabolites formed. Chemosphere 1995, 30, 855–868. [Google Scholar] [CrossRef]
- Abbott, B.J.; Gledhill, W.E. The extracellular accumulation of metabolic products by hydrocarbon-degrading microorganisms. In Advances in Applied Microbiology, 1st ed.; Perlman, D., Ed.; Academic Press, Inc.: New York, NY, USA, 1971; Volume 14, pp. 249–388. [Google Scholar]
- Bushnell, L.D.; Haas, H.F. The Utilization of Certain Hydrocarbons by Microorganisms. J. Bacteriol. 1941, 41, 653–673. [Google Scholar] [CrossRef]
- Kester, D.R.; Duedall, I.W.; Connors, D.N.; Pytkowicz, R.M. Preparation of artificial seawater 1. Limnol. Oceanogr. 1967, 12, 176–179. [Google Scholar] [CrossRef]
- Li, P.; Wang, L.; Feng, L. Characterization of a novel Rieske-type alkane monooxygenase system in Pusillimonas sp. strain T7-7. J. Bacteriol. 2013, 195, 1892–1901. [Google Scholar] [CrossRef] [PubMed]
- Winkler, U.K.; Stuckmann, M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J. Bacteriol. 1979, 138, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Holt, J.G.; Krieg, N.R.; Sneath, P.H.; Staley, J.T.; Williams, S.T. Bergey’s Manual of Determinative Bacteriology, 9th ed.; William & Wilkins: Baltimor, MD, USA, 1994. [Google Scholar]
- Magray, M.S.U.D.; Kumar, A.; Rawat, A.K.; Srivastava, S. Identification of Escherichia coli through analysis of 16S rRNA and 16S-23S rRNA internal transcribed spacer region sequences. Bioinformation 2011, 6, 370. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism, 3rd ed.; Munro, H.N., Ed.; Academic Press, Inc.: New York, NY, USA, 1969; Volume 3, pp. 22–126. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).