Abstract
Noncommunicable chronic diseases (NCDs) affect a large part of the population. With the emergence of COVID-19, its most severe cases impact people with NCDs, increasing the mortality rate. For this reason, it is necessary to develop personalized solutions to support healthcare considering the specific characteristics of individuals. This paper proposes an ontology to represent the knowledge of educational assistance in NCDs. The purpose of ontology is to support educational practices and systems oriented towards preventing and monitoring these diseases. The ontology is implemented under Protégé 5.5.0 in Ontology Web Language (OWL) format, and defined competency questions, SWRL rules, and SPARQL queries. The current version of ontology includes 138 classes, 31 relations, 6 semantic rules, and 575 axioms. The ontology serves as a NCDs knowledge base and supports automatic reasoning. Evaluations performed through a demo dataset demonstrated the effectiveness of the ontology. SWRL rules were used to define accurate axioms, improving the correct classification and inference of six instantiated individuals. As a scientific contribution, this study presents the first ontology for educational assistance in NCDs.
1. Introduction
In 2019, the World Health Organization (WHO) estimated that 41 million people died due to noncommunicable chronic diseases (NCDs), equivalent to 74% of a total of 55.4 million people who died worldwide [1]. Hypertension, obesity, physical inactivity, smoking, harmful use of alcohol, and an unhealthy diet are the main modifiable risk factors that can be controlled to prevent these diseases [2].
COVID-19 (Coronavirus Disease 2019), an infectious disease caused by the novel coronavirus (SARS-CoV-2), was declared by WHO [2] as a global pandemic on 11 March 2020. This disease, responsible for more than 3.2 million deaths around the world by 1 May 2021, has generated an additional alert for NCDs care since people with these diseases and their comorbidities have a higher risk of developing severe cases of COVID-19 [2].
Recent studies have shown that the pandemic of COVID-19 has affected lifestyles and compromised people’s healthcare, especially considering increased consumption of alcohol, tobacco [3], and ultra-processed foods [4], as well as reduced physical activity [5]. The study by Malta et al. [6] indicated that 58% of adults with NCDs reduced the practice of physical activity, and 53.7% increased the consumption of frozen food during the pandemic. In turn, in adults without NCDs the values are 60% and 43.6%, respectively. Given the current situation and the social isolation and remoteness of millions of people, telemedicine and health information services have gained space, especially regarding health promotion and prevention actions [7]. These services and information require adequate strategies to produce and reinforce people’s self-care habits.
An ontology refers to the representation of a given knowledge domain, which must be formal, shareable, and composed of well-defined concepts and rules. The scope of the ontology can be defined using a list of competency questions (CQ). These questions need to specify the requirements that the ontology should be able to answer. The finding for the answers to questions can be possible through reasoners [8,9,10].
According to Gruber [8], an ontology is an explicit specification of a conceptualization, so when defining a domain of interest with ontologies the objective is sharing knowledge, specifying a representative vocabulary in a domain, and understanding the semantics of the domain data. Therefore, the author understands that generic ontology can help to reason about a specific knowledge domain by mapping the most general concepts and relations. Ontology can be represented by Ontology Web Language (OWL), a language standard of the World Wide Web Consortium (W3C) (https://www.w3.org/, accessed on 10 October 2021).
An OWL ontology contains four basic elements: concepts, instances, properties, and restrictions. Concepts are used to identify the objects in the ontology definition, described through classes. Instances define the individuals of the classes. Individuals can be related to other individuals through properties. Properties are binary relationships used to define the relationships among the individuals of a class or among itself. Constraints are used to define boundaries for the individuals that belong to a class. The contents of an ontology can be checked using the SPARQL query language. Inferences about classes and individuals can be made using Semantic Web Rule Language (SWRL). Thus, ontologies are used to represent domains and infer knowledge, from construction methodologies [11] in several areas, such as the Web of Things (WoT) [12] and Decision Support Systems (DSS) [13], among others. This work is interested in an ontology to represent the knowledge of educational assistance in NCDs.
In this context, the proposed ontology allows representing the relationships between NCDs and their risk factors, as well as information about the person’s health profile. Thus, it becomes possible to propose personalized assistance for health promotion and prevention actions. Moreover, the ontology supports the development of systems to care for people in their health and quality of life, whether they are subjects with or without NCDs. Autonomy in learning strengthens the subject’s ability to learn and develops awareness of the need to maintain a constant learning process toward a healthy life.
The scientific contribution of this work is the proposal of the ontology, which represents the knowledge needed to assist individuals in improving their health conditions. Individuals with the manifested disease can follow the evolution of the NCD and minimize its aggravating factors. Through rules, a system can indicate content about the disease, places for medical assistance, notifications containing alerts, and reminders. In addition, the system can indicate groups and networks of people who share similar characteristics. Based on the related works, it can be stated that the proposed ontology is the first for educational assistance on NCDs.
This paper is structured in six sections. Section 2 approaches topics related to this work, providing a background to understanding the proposal and the related scientific works. The methodology for implementing the ontology is presented in Section 3. Section 4 approaches the results of the evaluation, and Section 5 discusses the results and addresses some limitations of the proposed approach. Finally, Section 6 summarizes the main contribution of this work and indicates future works.
2. Background
This section introduces the fundamentals of diseases and concepts of ubiquitous learning related to the importance of educational assistance in diseases. Section 2.1 describes the contextualization of diseases and their risk factors. Section 2.2 introduces concepts of ubiquitous learning and its importance for the field of the study, and Section 2.3 presents the related scientific works.
2.1. Noncommunicable Diseases and Risk Factors
Chronic lung diseases, cardiovascular diseases, cancers, and diabetes affect people of all age groups in all regions and countries of the world and are characterized as one of the major health challenges of the 21st century [14]. In 2019, data indicated that 55% of the 55.4 million deaths worldwide were associated with ten leading causes, being: ischemic heart disease, stroke, chronic obstructive pulmonary disease, lower respiratory infections, neonatal conditions, trachea, bronchus, and lung cancers, Alzheimer’s disease and other dementias, diarrhoeal diseases, diabetes mellitus, and kidney diseases. Of these, seven are associated with NCDs, which is 44% of all deaths. However, all NCDs accounted for 74% of deaths worldwide in 2019 [1]. According to the WHO [1], monitoring people’s leading causes of death is vital for evaluating the effectiveness of health systems and directing resources to the causes most needed. Although several other NCDs exist, in this study we limited the work to the NCDs defined by the WHO. We will consider other NCDs in future work.
NCDs have four common risk factors, which are considered the most frequent and modifiable: tobacco use, harmful use of alcohol, physical inactivity, and unhealthy diet. In turn, these behaviors result in metabolic/physiological changes, such as hypertension, being overweight, obesity, increased blood glucose, and increased lipids (fat) [14,15]. As well as these mentioned factors, air pollution is also considered a significant risk factor [14]. Prevention and control of NCDs can be done by combating these risk factors, which requires a comprehensive care approach, including different institutions in society [14]. The pandemic of COVID-19 has reinforced the importance of prevention and control, not only of NCDs, but of diseases in general. Investment in health systems, and systems with the capacity to manage and analyze vital statistical data was created to allow the control of cases and direct prevention and treatment actions [1].
Tobacco use is one of the main risk factors for the development of NCDs, including cancer, respiratory diseases, cardiovascular diseases, and diabetes, representing an enormous threat to public health. The reports from the WHO indicated that smoking kills more than 8 million people each year, disproportionately affecting low- and middle-income countries [16]. Of these, more than 7 million are the result of direct tobacco use, while about 1 million are non-smokers exposed to secondhand smoke. Of the world’s estimated 1 billion smokers, 80% live in low- and middle-income countries. The pandemic of COVID-19 has shown that smokers are more prone to hospitalization and death due to COVID-19, mainly because of its relationship with respiratory diseases that directly affect the lungs [16].
Harmful use of alcohol is also considered a major risk factor for premature death and disability worldwide. It can cause cardiovascular disease, cancer, liver disease, mental, and behavioral disorders and other chronic diseases. Data from the WHO [17] estimate that in 2016, harmful use of alcoholic beverages resulted in about 3 million deaths (5.3% of all deaths) worldwide. In 2019, the estimated level of alcohol consumption was 5.8 L of pure alcohol per person aged 15 years or older [7].
Lack of physical activity, physical inactivity, or sedentarism, can lead to the development of diseases such as diabetes, hypertension, cancer, respiratory diseases, and cardiovascular diseases. Like smoking, lack of physical activity is a significant risk factor for NCDs and is treated as a global epidemic by the WHO [14]. When individuals are insufficiently physically active, their mortality risk increases compared to those who engage in at least 30 min of moderate physical activity five times a week. In 2016, 28% of adults aged 18 years and older were insufficiently physically active. Physical activity has benefits for the health of the heart, body, and mind, being an ally in reducing the risks of cardiovascular disease, diabetes, hypertension, and depression [14].
A diet high in salt has contributed to increased hypertension and the risk of developing cardiovascular disease and stroke. The recommended daily intake of sodium should be less than 2 g or 5 g of salt [14]. The intake of foods that contain high fat and sugar has contributed to the development of obesity and being overweight. Obesity is related to increased hypertension and diseases such as diabetes, cancers, and cardiovascular diseases [14]. According to the WHO [14], in 2016, more than 1.9 billion people over the age of 18 were overweight, with more than 650 million considered obese. Children and adolescents aged 5 to 19 years totaled 340 million cases of being overweight or obese, and in children under 5 years, it was 40 million [14]. A balanced and healthy diet coupled with regular physical activity contributed to weight maintenance, prevention, and treatment of NCDs [7].
Like other risk factors, hypertension or raised blood pressure contributes to cardiovascular morbidity and mortality [7]. Data from 2015 indicated that 22% of all adults aged 18 years and older were hypertensive [14]. Once uncontrolled, hypertension can pose risks mainly for heart failure, peripheral vascular disease, renal failure, retinal hemorrhage, visual impairment, stroke, and dementia [14].
Diabetes mellitus (DM) is considered a significant and growing health problem in all countries of the world. In 2019, the International Diabetes Federation (IDF) estimated that 463 million people aged 20 to 79 years were living with diabetes [18]. By the year 2045, these rates are estimated to reach 700 million cases. This increase is mainly due to the growing and aging population, the increasing number of obese and sedentary people, greater urbanization, and the increasingly prolonged survival of patients with DM. There are three types of diabetes: type 1 diabetes, type 2 diabetes, and gestational diabetes. The first occurs because the pancreas does not produce enough insulin, and the second because the body cannot use the insulin it produces effectively. On the other hand, the third type is a characteristic of high blood glucose levels during pregnancy. In all types, blood glucose increases, causing severe health problems, including heart attack, stroke, renal failure, lower limb amputation, blindness, and peripheral neuropathy [14].
2.2. Ubiquitous Learning in Health
Information and communication technologies (ICTs) play an essential role in developing actions that enable self-care and self-management of health conditions. Electronic health (e-Health), ubiquitous health (u-Health), or telehealth refers to “health services and information delivered or enhanced through the Internet and related technologies” [19]. ICTs enable access to information through different digital resources at any place and time. In this scenario, ubiquitous computing emerges as a computational model that is always present to meet the needs of the individual naturally and imperceptibly [20,21,22,23]. This approach is applied in different areas, such as education [24,25,26,27], health [28,29,30,31,32,33], well-being [34], accessibility [35,36], agriculture [37,38,39], and data privacy [40,41].
In healthcare education, developing educational systems to support learning activities involving ubiquitous computing technologies has been an emerging research topic [42,43]. Healthcare for the individual goes beyond offices and hospitals. Public places and people’s homes are also spaces where health and education services can be offered. In turn, ubiquitous learning [44,45] has shown itself as a possibility developing of educational systems that support ubiquitous activities, taking into account the context of the individual [46,47]. From the capture and storage of context data, it is possible to create context histories [29,35,48,49,50], and make them available for future analysis [51,52,53,54].
A ubiquitous learning system must be able to understand the individual’s behavior and real-world parameters. The parameters are profile information of individuals, the situation they are in (stationary or moving), their preferences, the activities performed in the context, the notion of time and location, and the hardware and software devices used. In this way, learning must occur in a continuous and contextualized way [44,45]. A study conducted by Larentis et al. [55] identified research opportunities related to the absence of ubiquitous computing-based systems focused on personalized educational assistance in NCDs. The use of these technologies corroborates to expand the possibilities for developing systems that can be used to assist in the prevention of risk factors and treatment of NCDs and, not limited to that, all diseases.
2.3. Related Work
The use of ontologies to support healthcare systems is an emerging field of research. Some works involve collaborative efforts in the development of ontology and semantic web systems involving International Classification of Diseases (ICD-11) defined by the WHO [56,57]. Other works involve ontologies for smart home [58,59] focused on assisted living solutions. These solutions provide support to the residents and also consider issues such as disabilities and/or impairments related to the ageing of the person. In the area of technology applied to healthcare in NCDs, several pieces of research have also focused on the creation of ontologies. This section presents a review of ontologies that address the representation of knowledge in the healthcare of individuals with NCDs, whether in the prevention, monitoring, or treatment of the disease.
Alian et al. [10] developed a recommendation system for diabetic patients. The system recommended diet, exercise, and medication through rules created in the ontology. The ontology constitutes a knowledge base of the patient including biological, cultural, socioeconomic, and environmental information that affects the patient’s well-being. This base was built by using already consolidated wellness guidelines, including four dimensions: diabetes management general guidelines, food and nutrition guidelines, physical exercise guidelines, and American Indians (AI) related healthcare guidelines. The authors reused other existing ontologies about food, nutrition, and workouts. Recommendation rules were created and tested through case studies.
Bravo et al. [60] implemented a model to manage the profile of diabetic patients with ontology. The ontology considered personal and clinical information of the patient, physical activities, diet, and medications. The authors considered reusing existing ontologies for knowledge representation with the patient profile, such as National Drug File - Reference Terminology (NDF-RT), Human Disease Ontology (DOID), Symptom Ontology (SYMP), and Logical Observation Identifiers Names and Codes (LOINC). Fourteen competency questions were classified into three groups: patient profile, diabetes diagnosis, and diabetes clinical diagnosis. Three case studies were defined and instances created to check consistency through reasoner.
Chen et al. [61] presented a model for diabetes risk prediction, diagnosis, and treatment with ontology. The ontology describes personal and clinical information of the patient, exercise, diet, and medication. In building the ontology the authors considered reusing some existing standards such as: content of disease was based on DOID; some content of drugs was based on RxNorm Ontology; symptom content was based on SYMP; and gene content was based on Gene Ontology, Physical Ontology, Food Ontology, and Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT). Two top-level ontologies were merged in the proposed ontology: Basic Formal Ontology (BFO) and Ontology for General Medical Science (OGMS). Semantic rules were created and tested with 766 medical records of patients from the First Affiliated Hospital, Sun Yat-sem University, 269 of whom were diagnosed with diabetes. The ontology was implemented in Protégé software.
El-Sappagh and Ali [62] developed an ontology for type 2 diabetes diagnosis called Diabetes Mellitus Diagnosis Ontology (DDO). The ontology was built following the principles of Open Biomedical Ontologies (OBO) Foundry. Two top-level ontologies were merged in the proposed ontology: BFO and OGMS definitions. Some information makes up the ontology such as laboratory test results, demographic data, existing complications and symptoms, physical exam results, and medication intake. The ontology does not consider treatment plans, medications, and the patient’s family history. The authors considered the reuse of ontologies such as DOID, SYMP, Units of Measurement Ontology (UO), OBO Relation Ontology (RO), and RxNorm Ontology. The ontology was implemented in Protégé software and a consistency test was performed using reasoner.
El-Sappagh et al. [63] described an ontology for type 2 diabetes treatment called Diabetes Mellitus Treatment Ontology (DMTO). The ontology was built following the principles of OBO Foundry. Two top-level ontologies were merged in the proposed ontology: BFO and OGMS definitions. The authors incorporated the concepts previously defined in DDO Ontology [62] and reused other ontologies such as RxNorm Ontology, NDF-RT, SNOMED CT, OWL Time, Drug Interactions Ontology (DINTO), and OntoFood Ontology (FO). The ontology was implemented in Protégé software. Competency questions were created. A case study was designed and created manually in Protégé software for testing inferences and answers to the competency questions.
Madhusanka et al. [64] developed an ontology model for use in clinical decision support systems called Clinical Decision Support Systems (CDSS). The ontology was created through a conceptual model. This model considered clinical guidelines for patients with type 1 diabetes, type 2 diabetes, and gestational diabetes from the Ceylon College of Physicians, the Sri Lanka College of Obstetricians and Gynecologists, and the Sri Lanka Diabetes Federation. Two competency questions were created for evaluation. The competency questions were answered through SPARQL queries. The decision support system used the ontology as a knowledge base to provide the recommendations considering information such as age, Body Mass Index (BMI), fasting plasma glucose, postprandial blood sugar, and glycated hemoglobin.
Somodevilla et al. [65] proposed a system that integrates six ontologies (person, physical activity, NCDs, nutritional information, geographic regions, and chronic disease symptoms). Semantic rules were created to answer four competency questions: (1) What are the main risk factors for having diabetes? (2) Is the risk of having diabetes related to a high fat diet? (3) How is an average level lifestyle pattern defined? and (4) What are the characteristics of the person who has a high fat diet? The authors defined rules to perform the ontology tests; for example, if the person is a teenager and if the person is sedentary. Inference tests considered 85 individuals instantiated in the Protégé software.
Vianna et al. [66] proposed an ontology to detect the influence of social relationships on the spread of risk factors for NCDs. Information such as gender, whether smoking, whether obese, whether happy, and the person’s social relations (friend, relative, or acquaintance) are described by the ontology. The concepts modeled in the ontology were extracted from three studies that analyzed the influence of social networks on weight gain, the spread of happiness, and also smoking cessation. Semantic rules were created to answer 12 competency questions. A social network containing six individuals was created to demonstrate the solution to one of the questions.
Zhang et al. [67] created an ontology to specify the variables used in cancer research called Ontology for Cancer Research Variables (OCRV). The ontology constitutes a knowledge base considering individual risk factors including race, gender, tumor stage, and tumor type, as well as contextual factors such as socioeconomic status, minority status, and language, adult mental and physical health status, smoking rate, and alcohol consumption rate. The ontology construction used the following definitions from other ontologies: BFO, NCI Thesaurus (NCIt), and Time Event Ontology (TEO). In addition to these, the ontology was created through the semantic integration of five databases that supported the definition of individual and contextual risk factors of cancer patients. Queries were created to demonstrate the consistency between the ontology structures and the integration between the databases. The ontology was implemented in Protégé software.
Table 1 presents the comparison between this study and related works. The proposed ontology stands out in the modeling of knowledge directed to the healthcare of the individual throughout life, either in the prevention or in the monitoring of NCDs. This differential is not explored in the analyzed works, since they consider the person with the diagnosis of the disease. None of the related works focus on educational assistance in NCDs.

Table 1.
Comparison of related works.
3. Modeling and Implementation
In general, there is no methodology or method that should be followed in the construction of ontologies. According to Noy and McGuinness [68], there are definitions for ontology projects that can help researchers in choosing a methodology. These definitions include the fact that there is no single way to model a domain, i.e., alternatives must be considered. Furthermore, the development of an ontology is always an iterative process, and the concepts defined for the ontology must be related to the objects and relationships in its domain.
Methodologies were studied through a literature search such as Ontology Development 101 [68], Toronto Virtual Enterprise (TOVE) [69], Methontology [70], NeOn Methodology [71], On-To-Knowledge Methodology [72], UPON Methodology [73], and Enterprise Ontology [74,75]. These methodologies are already consolidated and used in knowledge modeling, language formalization, and ontology implementation. They show similarities between the main steps in ontology construction, however, differences exist with respect to the application domain of the ontology, for example, the TOVE [69] and Enterprise [74,75] methodologies are directed to the business application domain.
The ontology construction was based on the Ontology Development 101 methodology [68]. The use of this methodology is due mainly to the balance between the knowledge representations, use of restrictions, and the possibility of extending the ontology, allowing future definitions. This methodology consists of an iterative process divided into seven steps: (1) Determine the domain, scope, and competence issues of the ontology, (2) Consider reusing existing ontologies, (3) List important terms, (4) Define the classes and hierarchy, (5) Define relationships and class properties, (6) Define the semantic rules, and (7) Create the instances. The use of the technique of competency questions proposed by Grüninger and Fox [69] was also used in the ontology definition.
3.1. Determine the Domain, Scope, and Competency Issues
The knowledge domain of ontology is educational assistance in NCDs. The scope of the ontology is defined within the scope of prevention and follow-up of these diseases. The ontology can be used by information systems that support people in their health and quality of life process, whether an individual with or without NCDs. The goal is to provide a mapping of concepts about education and NCDs to assist the person. The competency questions are questions that the ontology must be able to answer [69]. Such questions, besides justifying the existence of the ontology, serve for its evaluation. The competency questions are:
(CQ1) “Which people have high risk for developing cardiovascular disease?”
(CQ2) “Which people have moderate risk for developing cardiovascular disease?”
(CQ3) “What content is suitable for people with diabetes?”
(CQ4) “What content is suitable for people who smoke?”
(CQ5) “Which people are at risk of developing hypertension?”
(CQ6) “Which people have a family history of NCDs?”
(CQ7) “What content is suitable for people with hypertension?”
3.2. Consider Reusing Existing Ontologies
In this step the reuse of other ontologies was considered. The ontology was built based on the CAMeOnto ontology developed by Aguilar et al. [9] and the ontology of Skillen et al. [76]. Both represent context concepts, i.e., where the person is and activities performed. The reuse included the classes “Context”, “Activity”, “Time”, “Location”, and “Device”. In representing the concepts of NCDs, the ontologies OntoMex proposed by Somodevilla et al. [65] and Alian et al. [10], and the Disease Ontology (DO) by Schriml et al. [77], were used. In addition, the DOID ontology was selected because it represents disease terms correlated with the domain, and SNOMED CT that consists of a collection of medical vocabularies aiming at supporting the clinical data.
3.3. List Important Ontology Terms
The ontology terms were defined from exploratory and descriptive research about the researched theme. A concept map [78] was used to organize and represent the knowledge related to the domain.
The concept map is an instrument to graphically represent the knowledge, or part of the knowledge, acquired in the definition of the domain of a conceptual field. This map aims to represent meaningful relationships between concepts through propositions. A proposition constitutes one or more conceptual terms interconnected through links [79]. As an example, “Concept A” relates to “Concept B”, where “Concept A” and “Concept B” refer to the conceptual term and “relates to” refers to the proposition, that is, the relationship between the two conceptual terms.
The construction of the concept map was performed from the definition of the focal theme, being: “Education assistance in NCDs”. First, questions were defined to assist in the construction of knowledge. Additionally, the questions were answered through bibliographic research. After the elaboration of the questions and answers, words were chosen among them to compose the concepts. Afterwards, propositions were created to represent the relationship between two concepts. Two questions and their respective answers are presented below:
Question 1: “What are the NCDs defined by the WHO?” Answer 1: “Cancer, diabetes, cardiovascular diseases and chronic respiratory diseases [80].”
Question 2: “What are the categories of risk factors associated with NCDs?” Answer 2: “Nonmodifiable, intermediates and modifiable [80].”
The CmapTools https://cmap.ihmc.us/cmaptools/, accessed on 10 October 2021 tool was used to organize the content and demonstrate the concepts. Figure 1 shows the conceptual model of ontology composed of the concepts and their relationships highlighted by propositions.
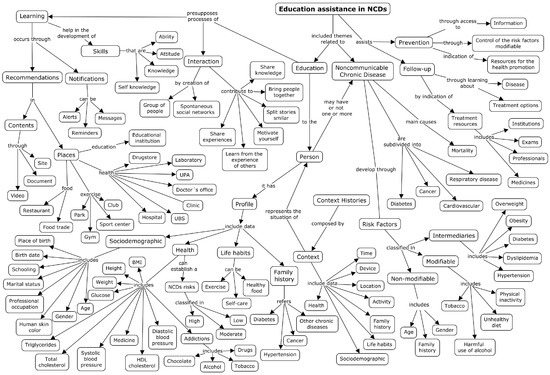
Figure 1.
Conceptual map on educational assistance in NCDs.
3.4. Define the Classes and Hierarchy
Figure 2 presents the main classes. The ontology is composed of 14 main classes and their relationships. In addition to these, other classes were created and represented through subclasses. The ontology was developed in OWL language in Protégé software version 5.5.0 [81].
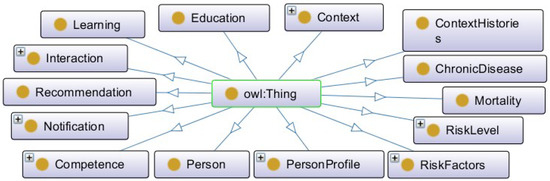
Figure 2.
Hierarchical view of ontology classes.
The classes have been defined in the singular. Each concept can represent one or more instances. The English language was used because it allows greater visibility and reusability of the ontology. Table 2 describes the main classes.

Table 2.
Core classes of ontology.
3.5. Define Relationships and Class Properties
The fifth step defined the relationships and properties of the classes. Figure 3 presents a hierarchical view of the ontology classes (class hierarchy) in yellow. The relationships between the classes are highlighted in blue (object property) and the attributes of the classes are highlighted in green (data property). All information was included in Protégé.
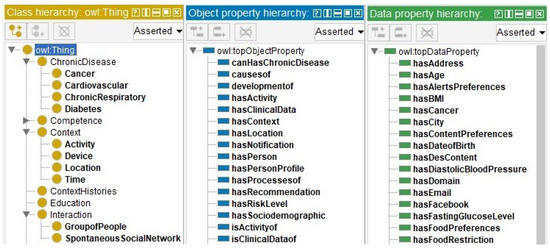
Figure 3.
Hierarchy of classes, relationships, and attributes.
Figure 4 presents the relationships between the classes. Relationships’ names are highlighted in the right frame.
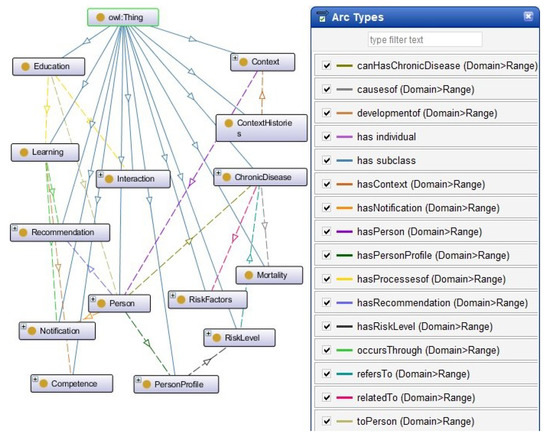
Figure 4.
Hierarchical view of ontology classes and relationships.
3.6. Define the Semantic Rules
The ontology defined restrictions for the class RiskLevelCVD. CQ1 and CQ2 defined in Section 3.1 refer to cardiovascular disease. According to the Framingham Heart Study [82] the risk for a person to develop cardiovascular disease is low if the score is less than 10%, moderate if the score is between 10% and 19%, and high if greater than or equal to 20%. Figure 5 shows three logical expressions created in the equivalence axiom for low, moderate, and high. The first expression in Figure 5 can be read as “If the risk score for cardiovascular disease is smaller than 10 the risk level is low.”

Figure 5.
Logical expression describing the axiom of equivalence for (1) low, (2) moderate, and (3) high risk level for cardiovascular disease.
The second expression can be read as “If the risk score for cardiovascular disease is greater than or equal to 10 and less than 20 the risk level moderate”, and the third can be read as “If the risk score for cardiovascular disease is greater than or equal to 20 the risk level is classified as high”. The expressions were created and linked to the class RiskLevel, which in turn has a subclass RiskLevelCVD and other subclasses of low, moderate, and high. The restrictions may vary according to the use of the ontology for different types of NCDs. New restrictions for other diseases will be detailed in future work.
Rules can also be created using SWRL. SWRL allows creating rules to infer new knowledge for individuals (instances) and has support for clauses in Horn’s format (if a → b, implication sentence), not supported by OWL [83].
The SWRL rules created in the ontology allow content inference considering the main risk factors for NCDs. The contents have keywords that relate to the theme. Through these keywords it is possible to verify the relationship of the content with NCDs. An evaluation, through the tools ©Google Trends and SEMrush https://www.semrush.com/, accessed on 10 October 2021 identified the keywords that relate to the criteria that make up the Framingham Risk Score (FRS) [82]: (a) “colesterol total” and “colesterol HDL” relates to the keywords “colesterol alto” and “hipercolesterolemia”; (b) “se fumante” relates to the keywords “tabagismo” and “parar de fumar”; (c) “pressão arterial” relates to the keywords “hipertensão”, “hipotensão”, “pressão alta”, and “pressão baixa”; (d) “se diabético” the keywords “glicemia” and “glicose alta”, and (e) “idade” relates to the keywords “geriatria”, “velhice”, and “terceira idade”. The criteria were translated to the Portuguese language; in this way, the keywords and content sites kept the same pattern. Additionally, a survey was carried out to identify the most relevant content for the keywords through the SEMrush tool. This tool provides data of a diverse nature including information consumption, use of keywords, and access to internet sites. Each content is related to a URL and is available on an internet domain. The result showed references of 1.179 most accessed contents when a search is performed on an internet search engine, such as ©Google. Extensions for English language are considered for future work.
Table 3 presents the rules for content inference in the ontology. The SWRL expression created for the rule “R4_Hypertension” can be read as “A content whose keyword equals hipertensao will be indicated for the person who has systolic blood pressure greater than 140”. The diagnostic classification for hypertension was taken from the Brazilian Guidelines of Hypertension [84]. For diabetes, three SWRL expressions were created, “R1_Diabetic_diabetes”, “R2_Diabetic_glycemia”, and “R3_Diabetic_glucose”. The expressions can be read, respectively, as “A content whose keyword equals diabetes will be indicated for the person who has diabetes”, “A content whose keyword equals “glicemia” will be indicated for the person who has diabetes”, and “A content whose keyword equals glicose will be indicated for the person who has diabetes”. Similar expressions were defined for people who smoke. It should also be noted that other expressions can be created according to the risk factors associated with NCDs. Others expressions with keywords, such as overweight, obesity, cancer, physical inactivity, unhealthy diet, harmful use of alcohol, and asthma will be considered in future work.

Table 3.
SWRL rules for content inference.
3.7. Create the Instances
The last step consisted of defining instances to perform inference and validate the ontology. Instances were created for the Person, PersonProfile, ClinicalData, Sociodemographic, and FamiliarHistory classes. Figure 6 shows the instances of the classes in Protégé.
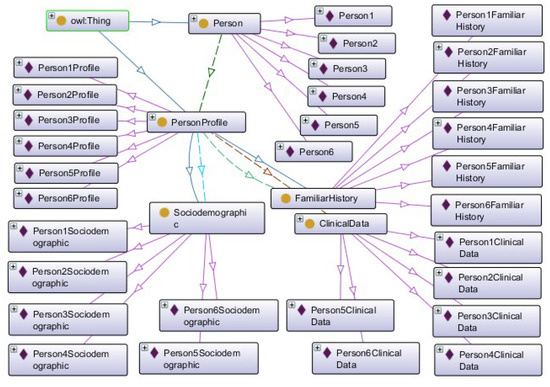
Figure 6.
Instances created in Protégé.
Table 4 presents the ontology metrics extracted from Protégé. The metric “Axiom” represents the quantity of logical expressions that define a concept. The quantity of ontology elements is given by the metrics “Classe count and Subclasses count”. The metric “Object property count” indicates the existing relations between two individuals (instances). In turn, the “Data property count” metric indicates the amount of literal data types, which can be a number, date, or text. The instances created are represented by the “Individual count” metric.

Table 4.
The ontology metrics.
4. Evaluation and Results
First, a coherence test was executed using the reasoning tasks of consistency checking and classification. The ontology was evaluated through an automated reasoning process run by the Pellet plugin version 2.2.0 installed in Protégé. Figure 7 shows the result of executing Pellet reasoning services of class hierarchy, object property hierarchy, data property hierarchy, class assertions, and object property assertions, with the same individuals returning a consistent ontology model.
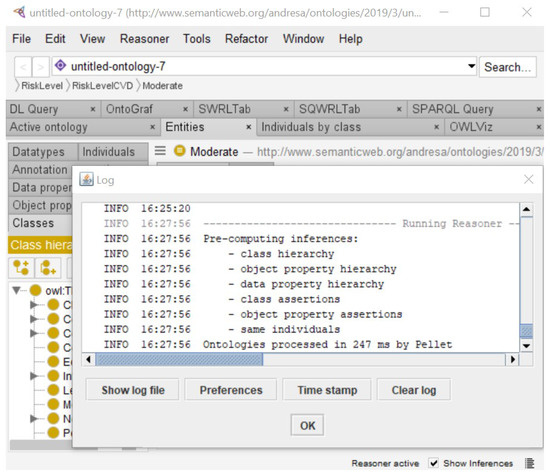
Figure 7.
Execution of Pellet plugin reasoning tasks.
Second, data from a demonstration database of the Medical Information Mart for Intensive Care-III (MIMIC-III) [85] was used in the instances created in Section 3.7. MIMIC-III contains data from patient admissions from Beth Israel Deaconess Medical Center, located in Boston, from 2001 to 2012 [86,87]. We identified six individuals with complete total cholesterol, HDL cholesterol, glucose, and systolic blood pressure data for use in the evaluation. The logical expressions described in Section 3.6 were processed by the reasoner and the results are presented in Figure 8 and Figure 9.
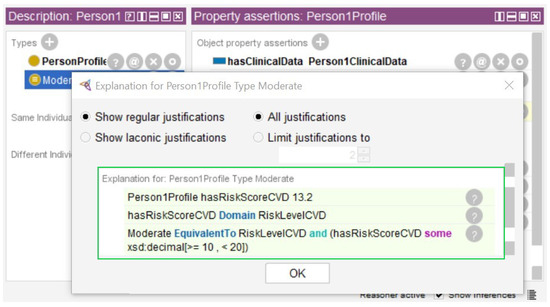
Figure 8.
Outcome of the NCD risk inference process.
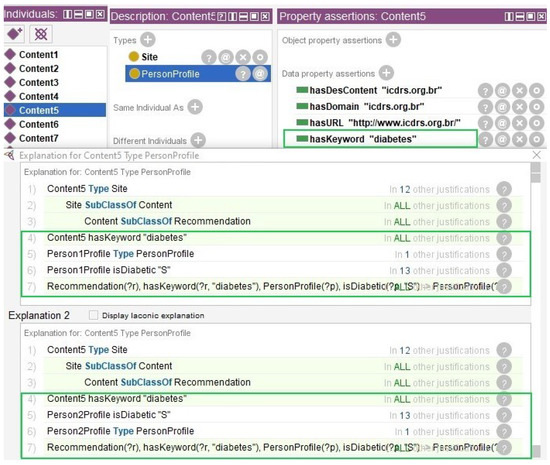
Figure 9.
Result of the content inference process for instances “Person2Profile” and “Person5Profile”.
Figure 8 shows the result of the risk for cardiovascular disease. The Person1Profile instance was created with the data property hasRiskScoreCVD=13.2. After running the reasoner it was inferred that the Person1Profile instance has the moderate risk. In green, the rectangle shows the risk score of 13.2%. The equivalence rule applied was “If the risk score for cardiovascular disease is greater than or equal to 10 and less than 20 it is classified as moderate”.
Figure 9 shows the result of the content inference. Instances were created for the Content class (Content1, Content2, Content3, Content4, Content5, Content6, Content7, Content8, and Content9). Each instance has in the data property hasKeyword: “colesterol_alto”, “parar_fumar”, “tabagismo”, “glicose_alta”, “diabetes”, “glicemia”, “diabetes”, “hipertensao”, and “hipotensao”. The domain and URL data were entered with SEMrush feedback. The Content5 instance has the data property hasKeyword=“diabetes”. The figures show two inferences, one for Person5Profile and one for Person2Profile. Both have the data property isDiabetic=“S”, so the equivalence rule was applied where individuals who have diabetes are classified with content whose keyword is “diabetes”.
Once the ontology is tested, it is possible to perform queries with SPARQL, similar to SQL. Figure 10 shows a query to select all instances that have the relationship Person x PersonProfile x ClinicalData x Sociodemographic.
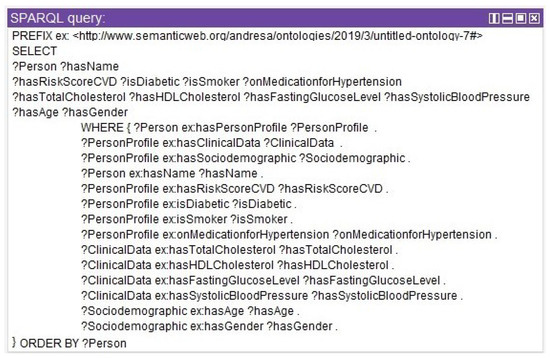
Figure 10.
SPARQL query for profile, clinical, and sociodemographic data.
Figure 11 shows six records returned from the SPARQL query. The result refers to the existing relationships between Person x PersonProfile x ClinicalData x Sociodemographic.
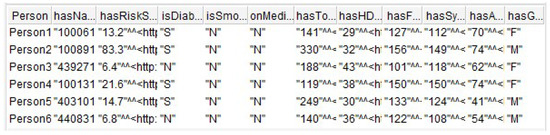
Figure 11.
Result of the SPARQL query.
In addition to this, seven queries were created by adding criteria in the FILTER field. These queries allowed the CQ defined in Section 3.1 to be answered. Table 5 shows the queries created for CQ1, CQ2, CQ3, CQ4, CQ5, CQ6, and CQ7.

Table 5.
SPARQL queries for the competency questions.
Figure 12 shows the results of the CQ1 and CQ2. The first returned two individuals with risk level high for cardiovascular disease. The second returned two individuals with risk level moderate. Figure 13 shows the results of the CQ3 and CQ4. The first returned three results including contents indicated for individuals with diabetes and the second returned two contents indicated for smoking individuals.
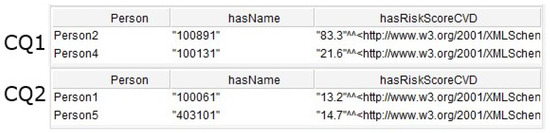
Figure 12.
Result of the SPARQL queries for CQ1 and CQ2.
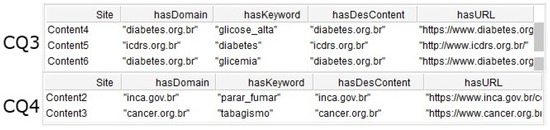
Figure 13.
Result of the SPARQL queries for CQ3 and CQ4.
Figure 14 shows the results of CQ5, CQ6, and CQ7. The first returned two individuals with risk for hypertension. The second returned two contents indicated for individuals with hypertension and hypotension, and the third result showed two individuals with familiar history for NCDs.
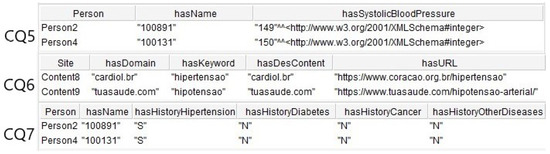
Figure 14.
Results of the SPARQL queries for CQ5, CQ6, and CQ7.
5. Discussion
This work enabled the creation of an ontology to represent the knowledge needed for health promotion through education. Once the individual’s profile is mapped, it is possible to provide information and knowledge to help him take better care of his health, especially in a preventive way. In a current scenario in which everyone is exposed to an immeasurable amount of information through ICTs, it becomes a challenge to transform data of interest into information to contribute significantly to the construction of knowledge in a given domain. This article conducted a literature search on ontology development methodologies. The ontology engineering ranges from methodologies for ontology development to methods dedicated to support models’ modification and evolution through their life cycle. The main activities to development an ontology to be considered in general are domain analysis, conceptualization, and implementation. It is important to stress the knowledge acquisition, reuse of existing ontologies, evaluation of the ontology, and documentation of the development process [88]. The search allowed us to choose Ontology Development 101 [68] due to the balance between knowledge representations, use of constraints, and the possibility of extending the ontology. The theory of concept maps created by Novak et al. [78] served as a technique to map knowledge about educational assistance in NCDs. This technique is known to aid in the teaching and learning process in the classroom, since the visual representation of knowledge about a domain aids in capturing student knowledge, summarization, and knowledge assessments. A questionnaire containing questions assisted in capturing the concepts and defining the relationships. The questions were answered through a literature search conducted on the internet. No meetings with domain experts were conducted at this early stage. After finalizing the concept map, this work extracted a list of initial terms to compose the domain of educational assistance in NCDs.
Regarding the reuse of existing ontologies, the research work reviewed a number of ontologies including the DOID and SNOMED CT ontologies. The first was used to standardize descriptions of human disease terms and the latter to assist in the definition of an individual’s clinical record data. In addition to these, the proposed ontology reused the ontologies defined by Aguilar et al. [9] and Skillen et al. [76] as both define context concepts. In relation to NCDs, other ontologies were also analyzed and considered including the ontologies OntoMex proposed by Somodevilla et al. [65] and Alian et al. [10], and DO by Schriml et al. [77].
Using the Ontology Development 101 methodology [68], which consists of seven steps, it was possible to develop and implement in the Protégé software. The iterative model makes the ontology development flexible, and new elements may need to be included in each iteration. The Protégé software was chosen because it is an open source tool widely used in the definition of ontologies. Once the ontology was implemented, instances were created with the goal of performing inferences and queries on these instances. Seven competency questions were created and validated through SPARQL queries. The research work mapped the knowledge needed to develop the ontology through consolidated knowledge sources such as books, international standards, and scientific articles. Other limitation of this study included the immeasurable amount of information available on the internet and the need to check the content available. We used the SEMrush tool as a content provider, but the result was not checked by experts.
6. Conclusions
This paper proposed an ontology for the domain of educational assistance in NCDs. The ontology works in a multidisciplinary way, joining the fields of education and health. The modeling of knowledge directed to the lifelong healthcare of people allows the interaction between systems and people, whether to prevent or control the diseases. In this sense, for the development of systems aimed at educational assistance, it is necessary that domains are established and that the person’s life context can be considered in order to provide means for this subject to learn and self-regulate. One of the characteristics of ontology is the modeling aimed at preventing and monitoring chronic diseases. In this research, a conceptual model was constructed based on the elaboration of the questions and answers about education assistance in NCDs. The conceptual model is one of the contributions of this research that provides greater understandability of the domain by converting questions and answers from text format to well-structured models. The related work section presented the state-of-the-art on health ontologies applied to NCDs. Furthermore, the related work made it possible to reuse definitions for representing entities in the modeling of knowledge. The ontology modeling considered characteristics of the individual’s context including sociodemographic information, health, family history, life habits, activities, location, time, and devices used. All this information composes the individual’s profile and is stored through context histories.
The ontology structure was designed with the guidelines in the Noy and Mcguinness methodology [68]. Next, the designed ontology structure was implemented using OWL and the Protégé software. The ontology includes 138 classes, 31 relations, 6 semantic rules, and 575 axioms. The definition of SWRL rules allowed automatic inference. Finally, the ontology was internally evaluated using SPARQL queries. These queries made it possible to answer the seven competency questions that the ontology should satisfy. Other questions can be created considering its domain and scope.
Results demonstrated the usefulness of the ontology in risk classification for developing NCDs and indicating content about the diseases. As future work, we suggest the construction of semantic rules for other NCDs, also considering WHO standards for the definition of people’s health conditions [11,13], expanding the evaluation with more real data, and implementing an evaluation with a system for educational assistance in NCDs that uses the inferences made by the ontology. In addition, we plan to work with individuals and domain experts to increase the validity of the model [89]. Another important future work is to conduct evaluations with experts in the field of health, especially considering the content provider. In the future, we will require medical professionals to check and maintain the content base and we will test other plug-ins for Prótége, such as Snap-SPARQL, to improve the results.
Author Contributions
Conceptualization, A.V.L., D.N.F.B. and J.L.V.B.; investigation, D.N.F.B., J.L.V.B. and A.V.L.; methodology, A.V.L. and J.L.V.B.; software, A.V.L.; project administration, J.L.V.B.; supervision, D.N.F.B. and J.L.V.B.; validation, A.V.L. and J.L.V.B.; writing—original draft, A.V.L., E.G.d.A.N. and J.L.V.B.; writing—review and editing, A.V.L., E.G.d.A.N., J.L.V.B., D.N.F.B., V.R.Q.L. and S.D.C.; financial, V.R.Q.L. and S.D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by national funds through the Fundação para a Ciência e a Tecnologia, I.P. (Portuguese Foundation for Science and Technology) by the project UIDB/05064/2020 (VALORIZA—Research Centre for Endogenous Resource Valorization) and it was partially supported by Fundação para a Ciência e a Tecnologia under Project UIDB/04111/2020, and ILIND–Instituto Lusófono de Investigação e Desenvolvimento, under projects COFAC/ILIND/COPELABS/1/2020 and COFAC/ILIND/COPELABS/3/2020.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank the University of Vale do Rio dos Sinos (Unisinos), the Applied Computing Graduate Program (PPGCA), the Mobile Computing Laboratory (Mobilab), the Research Support Foundation of the State of Rio Grande do Sul (FAPERGS), the National Development Council Scientific and Technological (CNPq), and the Coordination for the Improvement of Higher Education Personne-Brazil (CAPES)-Code Funding 001.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | American Indians |
| CQ | Competency Questions |
| BFO | The Basic Formal Ontology |
| CDSS | Clinical Decision Support Systems |
| COVID-19 | Coronavirus Disease 2019 |
| DDO | Diabetes Mellitus Diagnosis Ontology |
| DINTO | Drug Interactions Ontology |
| DM | Diabetes Mellitus |
| DMTO | Diabetes Mellitus Treatment Ontology |
| DOID | Human Disease Ontology |
| e-Health | Electronic Health |
| FO | OntoFood Ontology |
| IDF | International Diabetes Federation |
| ICTs | Information and communication technologies |
| LOINC | Logical Observation Identifiers Names and Codes |
| MIMIC-III | Medical Information Mart for Intensive Care-III |
| NCDs | Noncommunicable chronic diseases |
| NCIt | NCI Thesaurus |
| NDF-RT | National Drug File-Reference Terminology |
| OBO | Open Biomedical Ontologies |
| OCRV | Cancer Research Variables |
| OGMS | Ontology for General Medical Science |
| OWL | Ontology Web Language |
| RO | OBO Relation Ontology |
| SNOMED CT | Systematized Nomenclature of Medicine-Clinical Terms |
| SWRL | Semantic Web Rule Language |
| SYMP | Symptom Ontology |
| TEO | Time Event Ontology |
| TOVE | Toronto Virtual Enterprise |
| u-Health | Ubiquitous Health |
| UO | Units of Measurement Ontology |
| WHO | World Health Organization |
| W3C | World Wide Web Consortium |
References
- WHO. The Top 10 Causes of Death. Available online: http://who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 9 September 2021).
- WHO. The Global Health Observatory. Available online: http://who.int/data/gho/data/themes/noncommunicable-diseases/GHO/noncommunicable-diseases (accessed on 9 September 2021).
- García-Álvarez, L. Will changes in alcohol and tobacco use be seen during the COVID-19 lockdown? Adicciones 2020, 32, 85–89. Available online: https://digibuo.uniovi.es/dspace/bitstream/handle/10651/56548/1546-4314-1-PB.pdf?sequence=1 (accessed on 9 September 2021). [CrossRef] [PubMed]
- Malta, D.C. The COVID-19 Pandemic and changes in adult Brazilian lifestyles: A cross-sectional study. Epidemiol. Serv. Saude 2020, 29, e2020407. [Google Scholar] [CrossRef] [PubMed]
- Stanton, R.; To, Q.G.; Khalesi, S.; Williams, S.L.; Alley, S.J.; Thwaite, T.L.; Fenning, A.S.; Vandelanotte, C. Depression, Anxiety and Stress during COVID-19: Associations with Changes in Physical Activity, Sleep, Tobacco and Alcohol Use in Australian Adults. Int. J. Environ. Res. Public Health 2020, 17, 4065. [Google Scholar] [CrossRef] [PubMed]
- Malta, D.C.; Gomes, C.S.; Barros, M.; Lima, M.G. Noncommunicable diseases and changes in lifestyles during the COVID-19 pandemic in Brazil. Rev. Bras. Epidemiol. 2021, 24, e210009. [Google Scholar] [CrossRef]
- WHO. World Health Statistics 2021: Monitoring Health for the SDGs, Sustainable Development Goals. Available online: http://who.int/data/gho/publications/world-health-statistics/2021 (accessed on 9 September 2021).
- Gruber, T.R. A translation approach to portable ontology specifications. Knowl. Acquis. 1993, 5, 199–220. [Google Scholar] [CrossRef]
- Aguilar, J.; Jerez, M.; Rodríguez, T. CAMeOnto: Context awareness meta ontology modeling. Appl. Comput. Inform. 2018, 14, 202–213. [Google Scholar] [CrossRef]
- Alian, S.; Li, J.; Pandey, V. A personalized recommendation system to support diabetes self-Management for American Indians. IEEE Access 2018, 6, 73041–73051. [Google Scholar] [CrossRef]
- Fernandez, L.M.; Gômez, P.A. Overview and analysis of methodologies for building ontologies. Knowl. Eng. Rev. 2002, 17, 129–156. [Google Scholar] [CrossRef]
- Blomqvist, E. The Use of Semantic Web Technologies for Decision Support—A Survey. Semant. Web 2014, 5, 177–201. [Google Scholar] [CrossRef]
- De, S.; Zhou, Y.; Moessner, K. Chapter 1—Ontologies and context modeling for the Web of Things. Manag. Web Things 2017, 3–36. [Google Scholar] [CrossRef] [Green Version]
- WHO. Noncommunicable Diseases Country Profiles 2018. Available online: https://apps.who.int/iris/handle/10665/274512 (accessed on 9 September 2021).
- WHO. Global Action Plan for the Prevention and Control of Noncommunicable Diseases 2013–2020. 2013. Available online: https://www.who.int/publications/i/item/9789241506236 (accessed on 9 September 2021).
- WHO. WHO Report on the Global Tobacco Epidemic 2021: Addressing New and Emerging Products. Available online: https://www.who.int/publications/i/item/9789240032095 (accessed on 9 September 2021).
- WHO. Global Status Report on Alcohol and Health 2018. Available online: https://www.who.int/substance_abuse/publications/global_alcohol_report/en/ (accessed on 9 September 2021).
- IDF. International Diabetes Federation. IDF Diabetes Atlas, 9th edition 2019. Available online: https://www.diabetesatlas.org. (accessed on 9 September 2021).
- Eysenbach, G. What is e-health? J. Med. Internet Res. 2001, 3, 2. [Google Scholar] [CrossRef]
- Weiser, M. The Computer for the 21st Century. Sci. Am. 1991, 265, 94–104. Available online: https://www.jstor.org/stable/24938718 (accessed on 9 September 2021).
- Satyanarayanan, M. Pervasive Computing: Vision and Challenges. IEEE Pers. Commun. 2001, 4, 10–17. [Google Scholar] [CrossRef]
- Barbosa, J.L.V. Ubiquitous Computing: Applications and Research Opportunities. In Proceedings of the VI IEEE International Conference on Computational Intelligence and Computing Research (ICCIC), Madurai, India, 10–12 December 2015; pp. 1–8. [Google Scholar] [CrossRef]
- de Souza, R.S.; Lopes, J.L.; Geyer, C.F.R.; Cardozo, A.; Yamin, A.C.; Barbosa, J.L. An Architecture for IoT Management Targeted to Context Awareness of Ubiquitous Applications. J. Univers. Comput. Sci. 2018, 24, 1452–1471. [Google Scholar] [CrossRef]
- da Silva, L.G.; Neto, E.G.D.A.; Francisco, R.; Barbosa, J.L.V.; Silva, L.A.; Leithardt, V.R.Q. ULearnEnglish: An Open Ubiquitous System for Assisting in Learning English Vocabulary. Electronics 2021, 10, 1692. [Google Scholar] [CrossRef]
- Wagner, A.; Barbosa, J.L.V.; Barbosa, D.N.F. A model for profile management applied to ubiquitous learning environments. Expert Syst. Appl. 2014, 41, 2023–2034. [Google Scholar] [CrossRef]
- Barbosa, J.L.V.; Barbosa, D.N.F.; de Oliveira, J.M.; Andrade, R.S., Jr. A decentralized infrastructure for ubiquitous learning environments. J. Univers. Comput. Sci. 2014, 20, 1649–1669. [Google Scholar]
- Barbosa, J.L.V.; Hahn, R.M.; Barbosa, D.N.F.; Costa, Z.S.A.I.D. A ubiquitous learning model focused on learner interaction. Int. J. Learn. Technol. 2011, 6, 62–83. [Google Scholar] [CrossRef]
- Vianna, H.D.; Barbosa, J.L.V. A scalable model for building context-aware applications for noncommunicable diseases prevention. Inf. Process. 2019, 148, 1–6. [Google Scholar] [CrossRef]
- Machado, S.D.; Tavares, J.E.d.R.; Martins, M.G.; Barbosa, J.L.V.; Gonzalez, G.V.; Leithardt, V.R.Q. Ambient Intelligence Based on IoT for Assisting People with Alzheimer’s Disease Through Context Histories. Electronics 2021, 10, 1260. [Google Scholar] [CrossRef]
- Dias, L.P.S.; Barbosa, J.L.V.; Feijó, L.P.; Vianna, H.D. Development and testing of iAware model for ubiquitous care of patients with symptoms of stress, anxiety and depression. Comput. Methods Progr. Biomed. 2020, 187, 105113. [Google Scholar] [CrossRef] [PubMed]
- Petry, M.M.; Barbosa, J.L.V.; José, R.; Sandro, D.; Lucas, P.S.; Büttenbender, P. Toward a ubiquitous model to assist the treatment of people with depression. Univers. Access Inf. Soc. 2019, 19, 841–854. [Google Scholar] [CrossRef]
- Bavaresco, R.; Barbosa, J.; Vianna, H.; Büttenbender, P.; Dias, L. Design and evaluation of a context-aware model based on psychophysiology. Comput. Methods Progr. Biomed. 2020, 189, 105299. [Google Scholar] [CrossRef] [PubMed]
- Vianna, H.D.; Barbosa, J.L.V. A model for ubiquitous care of noncommunicable diseases. IEEE J. Biomed. Health Inform. 2014, 18, 1597–1606. [Google Scholar] [CrossRef]
- Vianna, H.D.; Barbosa, J.L.V.; Pittoli, F. In the Pursuit of Hygge Software. IEEE Softw. 2017, 34, 48–52. [Google Scholar] [CrossRef]
- Barbosa, J.; Tavares, J.; Cardoso, I.; Alves, B.; Martini, B. TrailCare: An Indoor and Outdoor Context-aware System to Assist Wheelchair Users. Int. J.-Hum.-Comput. Stud. 2018, 116, 1–14. [Google Scholar] [CrossRef]
- Tavares, J.; Barbosa, J.; Cardoso, I.; Costa, C.; Yamin, A.; Real, R. Hefestos: An intelligent system applied to ubiquitous accessibility. Univ. Access Inf. Soc. 2016, 15, 589–607. [Google Scholar] [CrossRef]
- Oliveira, J.L.; Souza, R.; Geyer, C.F.R.; Costa, C.A.; Barbosa, J.L.V.; Pernas, A.; Yamin, A.C. A Middleware Architecture for Dynamic Adaptation in Ubiquitous Computing. J. Univers. Comput. Sci. 2014, 20, 1327–1351. [Google Scholar]
- De Souza, R.S.; Afonso, C.; Anderson, Y.; Adenauer, G.; Ingrid, G.; Lopes, J.; Geyer, C.; João, S.; da Rosa, L.; Barbosa, J. Continuous monitoring seed testing equipments using internet of things. Comput. Electron. Agric. 2019, 158, 122–132. [Google Scholar] [CrossRef]
- Helfer, G.A.; Barbosa, J.L.V.; Santos, R.D.; Costa, A.B. A computational model for soil fertility prediction in ubiquitous agriculture. Comput. Electron. Agric. 2020, 175, 105602. [Google Scholar] [CrossRef]
- Lopes, H.; Pires, I.M.; Sánchez San Blas, H.; García-Ovejero, R.; Leithardt, V. PriADA: Management and Adaptation of Information Based on Data Privacy in Public Environments. Computers 2020, 9, 77. [Google Scholar] [CrossRef]
- Lucca, A.V.; Silva, L.A.; Luchtenberg, R.; Garcez, L.; Mao, X.; Ovejero, R.G.; Pires, I.M.; Barbosa, J.L.V.; Leithardt, V.R.Q. A Case Study on the Development of a Data Privacy Management Solution Based on Patient Information. Sensors 2020, 20, 6030. [Google Scholar] [CrossRef]
- Quinde, M.; Augusto, J.C.; Khan, N.; van Wyk, A. ADAPT: Approach to Develop context-Aware solutions for Personalised asthma management. J. Biomed. Inform. 2020, 111, 103586. [Google Scholar] [CrossRef]
- Seys, S.F.; de Bont, S.; Fokkens, W.J. Real-life assessment of chronic rhinosinusitis patients using mobile technology: The mySinusitisCoach project by EUFOREA. Allergy 2020, 75, 2867–2878. [Google Scholar] [CrossRef]
- Cárdenas-Robledo, L.A.; Peña-Ayala, A. Ubiquitous learning: A systematic review. Telemat. Inform. 2018, 35, 1097–1132. [Google Scholar] [CrossRef]
- Mota, F.; Tolêdo, F.; Kwecko, V.; Devicenzi, S.; Nunez, P.; Botelho, S. Ubiquitous Learning: ASystematic Review. In Proceedings of the IEEE Frontiers in Education Conference (FIE), Covington, KY, USA, 16–19 October2019. [Google Scholar]
- Dey, A.K. Understanding and Using Context. J. Pers. Ubiquitous Comput. 2001, 5, 4–7. [Google Scholar] [CrossRef]
- Dey, A.K.; Abowd, G.; Salber, D. A Conceptual Framework and a Toolkit for Supporting the Rapid Prototyping of Context-Aware Applications. Hum.-Comput. Interact. 2001, 16, 97–166. [Google Scholar] [CrossRef]
- Barbosa, J.L.V.; Martins, C.J.; Franco, L.K.; Barbosa, D.N.F. TrailTrade: A model for trail-aware commerce support. Comput. Ind. 2016, 80, 43–53. [Google Scholar] [CrossRef]
- Martini, B.G.; Helfer, G.A.; Barbosa, J.L.V.; Espinosa, M.R.C.; da Silva, M.R.; de Figueiredo, R.M.; Mendes, A.S.; Silva, L.A.; Leithardt, V.R.Q. IndoorPlant: A Model for Intelligent Services in Indoor Agriculture Based on Context Histories. Sensors 2021, 21, 1631. [Google Scholar] [CrossRef]
- Rosa, J.H.; Barbosa, J.L.V.; Kich, M.R.; Brito, L.K. A Multi-Temporal Context-aware System for Competences Management. Int. J. Artif. Intell. Educ. 2015, 25, 455–492. [Google Scholar] [CrossRef] [Green Version]
- Filippetto, A.S.; Lima, R.K.; Barbosa, J.L.V. A Risk Prediction Model for Software Project Management based on Similarity Analysis of Context Histories. Inf. Softw. Technol. 2021, 131, 106497. [Google Scholar] [CrossRef]
- Cardoso, I.G.; Barbosa, J.L.V.; Mota, B.; Dias, L.P.S.; Nesi, L.C. Vulcont: A recommender system based on context history ontology. IET Softw. 2021, 1, 1–13. [Google Scholar] [CrossRef]
- Dupont, D.; Barbosa, J.L.V.; Alves, B.M. CHSPAM: A multi-domain model for sequential pattern discovery and monitoring in contexts histories. Pattern Anal. Appl. 2020, 23, 725–734. [Google Scholar] [CrossRef]
- Da Rosa, J.H.; Barbosa, J.L.V.; Ribeiro, G.D. ORACON: An adaptive model for context prediction. Expert Syst. Appl. 2016, 45, 56–70. [Google Scholar] [CrossRef]
- Larentis, A.V.; Barbosa, D.N.F.; da Silva, C.R.; Barbosa, J.L.V. Applied Computing to Education on Noncommunicable Chronic Diseases: A Systematic Mapping Study. Telemed. e-Health 2019, 26, 147–163. [Google Scholar] [CrossRef]
- Tudorache, T.; Nyulas, C.I.; Noy, N.F.; Musen, M.A. Using Semantic Web in ICD-11: Three Years Down the Road. Comput. Sci. 2013, 8219, 195–211. [Google Scholar] [CrossRef]
- Tudorache, T. Ontology Development for the Masses: Creating ICD-11 in WebProtégé. Comput. Sci. 2010, 6317, 74–89. [Google Scholar] [CrossRef]
- Spoladore, D.; Arlati, S.; Colombo, V.; Modoni, G.; Sacco, M. A Semantic-Enabled Smart Home for AAL and Continuity of Care. In IoT in Healthcare and Ambient Assisted Living; Springer: Singapore, 2021. [Google Scholar] [CrossRef]
- Spoladore, D.; Sacco, M. Semantic and Dweller-Based Decision Support System for the Reconfiguration of Domestic Environments: RecAAL. Electronics 2018, 7, 179. [Google Scholar] [CrossRef] [Green Version]
- Bravo, M. Management of diabetic patient profiles using ontologies. Contaduría y Administración 2020, 65, 218. Available online: https://dialnet.unirioja.es/servlet/articulo?codigo=7731048 (accessed on 9 September 2021). [CrossRef]
- Chen, L. OMDP: An ontology-based model for diagnosis and treatment of diabetes patients in remote healthcare systems. Int. J. Distrib. Sens. Netw. 2019, 15. [Google Scholar] [CrossRef]
- El-Sappagh, S.; Ali, F. DDO: A diabetes mellitus diagnosis ontology. Appl. Inform. 2016, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- El-Sappagh, S.; Kwak, D.; Ali, F.; Kwak, K. DMTO: A realistic ontology for standard diabetes mellitus treatment. J. Biomed. Semant. 2018, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madhusanka, S.; Walisadeera, A.; Dantanarayana, G.; Goonetillake, J.; Ginige, A. An Ontological Clinical Decision Support System Based on Clinical Guidelines for Diabetes Patients in Sri Lanka. Healthcare 2020, 8, 573. [Google Scholar] [CrossRef] [PubMed]
- Somodevilla, M.J. Discovering semantic relationships between NCD and lifestyle patterns using ontologies. Res. Comput. Sci. 2015, 99, 19–28. Available online: https://rcs.cic.ipn.mx/2015_99/RCS_99_2015.pdf#page=19 (accessed on 9 September 2021). [CrossRef]
- Vianna, H.D.; Barbosa, J.L.V.; Gluz, J.C.; dos Santos, R.B.S. Design of an ontology for detecting the social influence on non-communicable diseases risk factors. Int. J. Metadata Semant. Ontol. 2018, 13, 120–130. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, Y.; Li, Q.; George, T.J.; Shenkman, E.; Modave, F.; Bian, J. An ontology-guided semantic data integration framework to support integrative data analysis of cancer survival. BMC Med. Inform. Decis. Mak. 2018, 18, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noy, N.F.; Mcguinness, D.L. Ontology Development 101: A guide to creating your first ontology. Stanford KSL Technical Report KSL-01-05 and Stanford Medical Informatics Technical Report SMI-2001-0880. Knowl. Syst. Lab. 2001, 32, 1–25. [Google Scholar]
- Grüninger, M.; Fox, M.S. Methodology for the design and evaluation of ontologies. In Workshop on Basic Ontological Issues in Knowledge Sharing; IJCAI-95; Montreal, QC, Canada, 1995; Available online: http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.44.8723 (accessed on 10 October 2021).
- Corcho, O.; Fernández-López, M.; Gómez-Pérez, A.; López-Cima, A. Building Legal Ontologies with METHONTOLOGY and WebODE. Lect. Notes Comput. Sci. 2005, 3369, 142–157. [Google Scholar] [CrossRef] [Green Version]
- Villazón-Terrazas, B.; Ramírez, J.; Suárez-Figueroa, M.C.; Gómez-Perez, A. A network of ontology networks for building e-employment advanced systems. Expert Syst. Appl. 2011, 38, 13612–13624. [Google Scholar] [CrossRef] [Green Version]
- Sure, Y.; Staab, S.; Studer, R. On-To-Knowledge Methodology (OTKM). In Handbook on Ontologies; Staab, S., Studer, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; Volume 6, pp. 117–132. [Google Scholar] [CrossRef]
- De Nicola, A.; Missikoff, M.; Navigli, R. A proposal for a unified process for ontology building: UPON. In Database and Expert Systems Applications. DEXA; Andersen, K.V., Debenham, J., Wagner, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 655–664. [Google Scholar]
- Uschold, M.; Gruninger, M. Ontologies: Principles, methods and applications. Knowl. Eng. Rev. 1996, 2, 93–136. [Google Scholar] [CrossRef] [Green Version]
- Uschold, M.; King, M. Towards a methodology for building ontologies. In Workshop on Basic Ontological Issues in Knowledge Sharing, Held in Conjunction with IJCAI-95; Artificial Intelligence Applications Institute, University of Edinburgh: Edinburgh, Scotland, 1995; Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.697.8733&rep=rep1&type=pdf (accessed on 21 September 2021).
- Skillen, K.L. Ontological user profile modeling for context-aware application personalization. In Proceedings of the International Conference on Ubiquitous Computing and Ambient Intelligence (UCAml), Vitoria-Gasteiz, Spain, 3–5 December 2012; pp. 261–268. [Google Scholar] [CrossRef]
- Schriml, L.M. Human Disease Ontology 2018 update: Classification, content and workflow expansion. Nucleic Acids Res. 2019, 47, 955–962. [Google Scholar] [CrossRef] [Green Version]
- Novak, J.D.; Cañas, A.J. Theoretical Origins of Concept Maps, How to Construct Them, and Uses in Education; Reflecting Education: London, UK, 2007; Volume 3, pp. 29–42. Available online: https://www.researchgate.net/profile/Alberto_Canas/publication/228761562_Theoretical_origins_of_concept_maps_how_to_construct_them_and_uses_in_education/links/0fcfd50be1868a1f0c000000/Theoretical-origins-of-concept-maps-how-to-construct-them-and-uses-in-education.pdf (accessed on 9 September 2021).
- Novak, J.; Gowin, D.; Kahle, J. Concept mapping for meaningful learning. In Learning How to Learn; Cambridge University Press: Cambridge, UK, 1984; pp. 15–54. [Google Scholar] [CrossRef]
- WHO. Preventing Chronic Diseases: A Vital Investment: WHO Global Report. Available online: https://apps.who.int/iris/handle/10665/43314 (accessed on 9 September 2021).
- Musen, M.A. The protégé project: A look back and a look forward. AI Matters 2015, 1, 4–12. [Google Scholar] [CrossRef]
- FHS. Framingham Heart Study. History of the Framingham Heart Study. Available online: https://framinghamheartstudy.org/fhs-about/history/ (accessed on 9 September 2021).
- O’Connor, M. Supporting Rule System Interoperability on the semantic web with SWR. In Fourth International Semantic Web Conference; Springer: Berlin/Heidelberg, Germany, 2005; pp. 974–986. [Google Scholar] [CrossRef] [Green Version]
- Barroso, W.K.S.; Rodrigues, C.I.S.; Bortolotto, L.A.; Mota-Gomes, M.A.; Brandão, A.A.; Feitosa, A.D.M. Diretrizes Brasileiras de Hipertensão Arterial—2020. Arq. Bras. Cardiol. 2021, 116, 516–658. [Google Scholar] [CrossRef]
- Johnson, A.; Pollard, T.; Mark, R. MIMIC-III Clinical Database Demo. PhysioNet. Available online: https://physionet.org/content/mimiciii-demo/1.4/ (accessed on 9 September 2021).
- Goldberger, A.L. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.E.W. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [Green Version]
- Simperl, E.P.B.; Tempich, C. Ontology engineering: A reality check. In On the Move to Meaningful Internet Systems, Proceedings of the OTM Confederated International Conferences; Springer: Berlin/Heidelberg, Germany, 2006; pp. 836–854. [Google Scholar]
- Spoladore, D.; Pessot, E. Collaborative Ontology Engineering Methodologies for the Development of Decision Support Systems: Case Studies in the Healthcare Domain. Electronics 2021, 10, 1060. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).