Lung Cancer Susceptibility and hOGG1 Ser326Cys Polymorphism: A Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Eligibility of Relevant Studies
2.2. Data Extraction and Assessment of Study Quality
2.3. Meta-Analysis
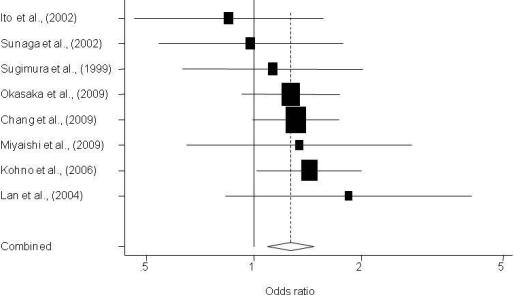
3. Results
3.1. Description of Individual Studies
| Author, published year [reference no.] | Ethnicity | No. of Cases/ Controls | Source of controls | OR (95% CI)* | Prevalence of the Cys allele in controls | Hardy-Weinberg test, P | Quality control of genotyping | |
|---|---|---|---|---|---|---|---|---|
| Ser/Cys | Cys/Cys | |||||||
| Sugimura et al., 1999 [13] | Asian | 241/197 | Hospital | 0.80 (0.52–1.21) | 1.13 (0.63–2.02) | 0.409 | 0.08 | Sequencing |
| Wirkman et al., 2000 [14] | Caucasian | 105/105 | Hospital | 0.66 (0.37–1.17) | 2.20 (0.41–11.8) | 0.224 | 0.07 | Sequencing |
| Ito et al., 2002 [15] | Asian | 138/240 | Hospital | 1.02 (0.63–1.67) | 0.85 (0.46–1.56) | 0.471 | 0.84 | None |
| Sunaga et al., 2002 [16] | Asian | 198/152 | Hospital | 1.49 (0.91–2.43) | 0.98 (0.54–1.77) | 0.454 | 0.13 | None |
| Le Marchand et al., 2002 [28] | Admixture | 298/405 | Population | 0.90 (0.65–1.26) | 1.76 (1.15–2.71) | 0.347 | 0.35 | Sequencing |
| Lan et al., 2004 [17] | Asian | 118/109 | Population | 1.96 (1.10–3.48) | 1.84 (0.83–4.06) | 0.335 | 0.23 | None |
| Park et al., 2004 [29] | Caucasian | 179/350 | Population | 1.89 (1.27–2.80) | 4.10 (1.65–10.2) | 0.147 | 0.86 | Sequencing |
| Vogel et al., 2004 [18] | Caucasian | 256/269 | Population | 1.09 (0.75–1.60) | 0.78 (0.35–1.72) | 0.240 | 0.24 | Replication** |
| Liang et al., 2005 [19] ‡ | Asian | 227/227 | Hospital | 0.94 (0.63–1.41) | 0.98 (0.33–2.87) | 0.606 | 0.04 | Sequencing |
| Hung et al., 2005 [20] | Caucasian | 2,155/2,163 | Hospital | 0.90 (0.79–1.03) | 1.15 (0.84–1.57) | 0.202 | 0.22 | Replication** |
| Zienolddiny et al., 2006 [21] ‡ | Caucasian | 326/386 | Population | 0.91 (0.64–1.29) | 0.63 (0.40–0.97) | 0.346 | 0.00015 | Replication† |
| Matullo et al., 2006 [22] | Caucasian | 116/1,094 | Population | 1.26 (0.83–1.91) | 0.82 (0.21–2.33) | 0.215 | 0.90 | Replication** |
| Kohno et al., 2006 [23] | Asian | 1097/394 | Hospital | 1.24 (0.94–1.63) | 1.43 (1.02–2.01) | 0.447 | 0.63 | None |
| Sorensen et al., 2006 [24] | Caucasian | 431/796 | Population | 1.04 (0.80–1.35) | 1.18 (0.63–2.21) | 0.220 | 0.25 | Replication** |
| De Ruyck et al., 2007 [30] | Caucasian | 110/110 | Hospital | 0.58 (0.33–1.02) | 0.61 (0.13–2.82) | 0.245 | 0.18 | None |
| Karahlil et al., 2008 [25] | Turkish | 165/250 | Hospital | 0.82 (0.54–1.24) | 0.65 (0.32–1.29) | 0.328 | 0.55 | None |
| Miyaishi et al., 2009 [26] | Asian | 208/121 | Hospital | 1.47 (0.79–2.73) | 1.34 (0.65–2.77) | 0.455 | 0.27 | None |
| Chang et al., 2009 [27] | Latino | 112/296 | Population | 0.91 (0.56–1.47) | 1.05 (0.45–2.32) | 0.321 | 0.52 | Replication** |
| Chang et al., 2009 [27] | African-American | 254/280 | Population | 1.32 (0.89 –1.98) | 0.89 (0.25–3.00) | 0.154 | 0.69 | Replication** |
| Chang et al., 2009 [31] | Asian | 1,096/997 | Population | 1.17 (0.89–1.52) | 1.31 (0.99–1.73) | 0.604 | 0.74 | Replication ** |
| Okasaka et al., 2009 [32] | Asian | 515/1,030 | Hospital | 1.00 (0.77–1.33) | 1.27 (0.93–1.75) | 0.493 | 0.08 | None |
3.2. Quantitative Synthesis
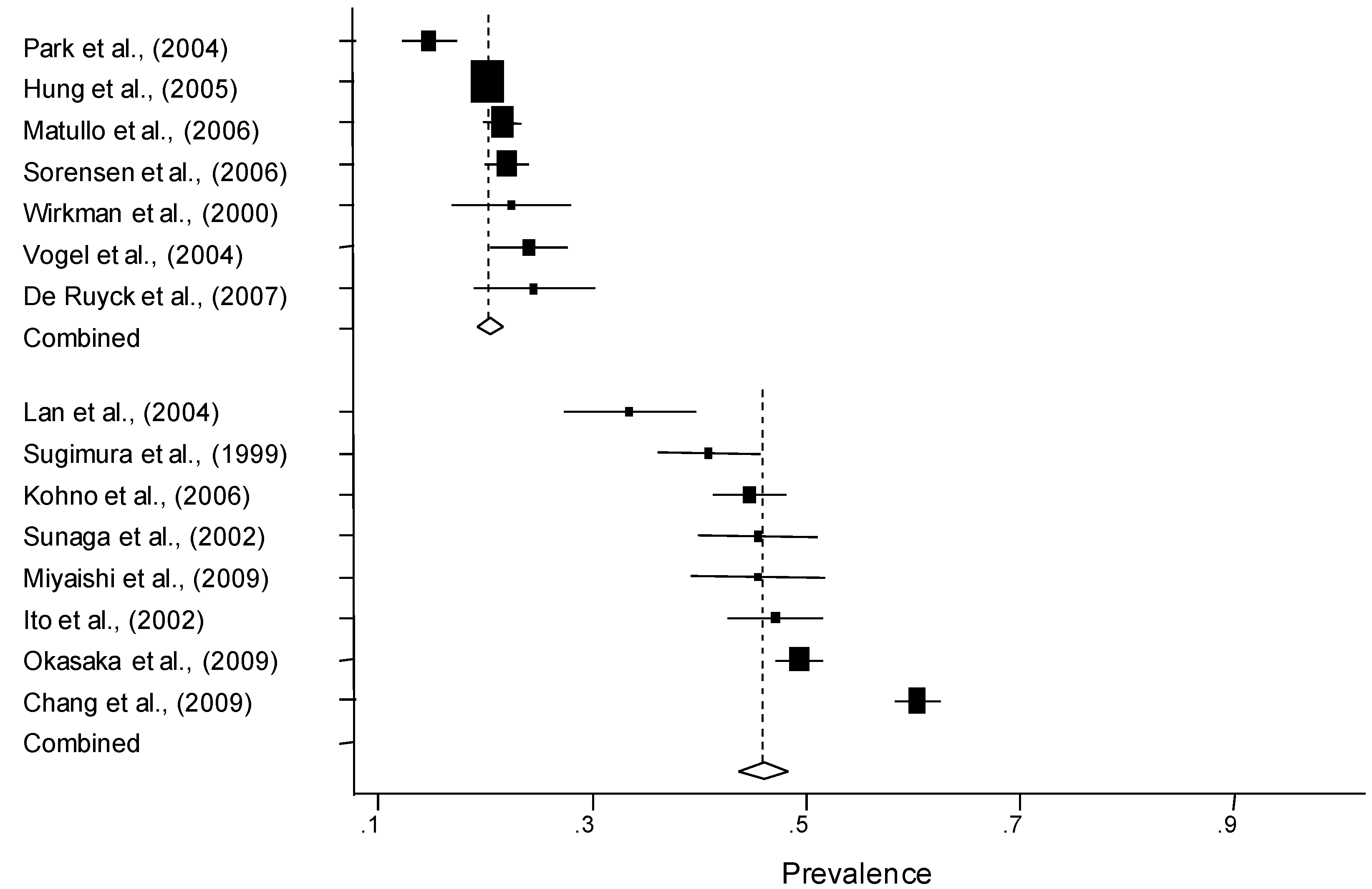
| Subgroup | No. of populations | Number of controls | Prevalence of the Cys allele (%)* | |
|---|---|---|---|---|
| Ethnicity | ||||
| Caucasian | 7 | 4,887 | 20.9 (18.9–22.9) | |
| Asian | 8 | 3,240 | 46.1 (40.2–52.0) | |
| Design | ||||
| Population-based | Overall | 9 | 4,596 | 28.7 (17.9–39.4) |
| Caucasian | 4 | 2,509 | 20.5 (17.1–23.9) | |
| Asian | 2 | 2,212 | 47.1 (20.7–73.5) | |
| Hospital-based | Overall | 10 | 4,762 | 37.3 (27.6–47.0) |
| Caucasian | 3 | 2,178 | 21.1 (18.8–23.3) | |
| Asian | 6 | 2,134 | 45.8 (43.1–48.5) | |
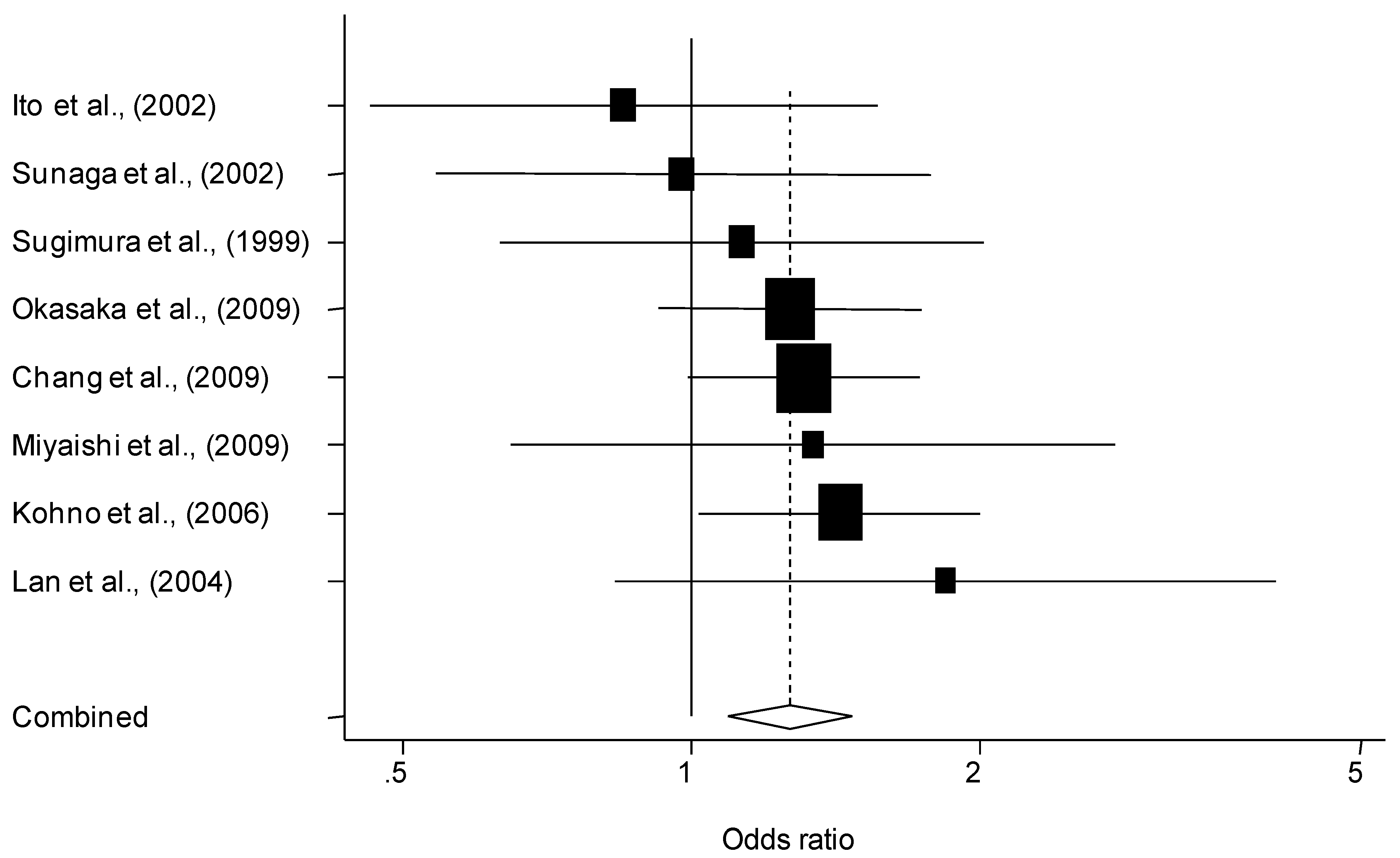
| Subgroup | No. of populations | No. of cases / controls | Ser/Cys vs. Ser/Ser | Cys/Cys vs. Ser/Ser | Cochran Q test for heterogeneity, P | ||
|---|---|---|---|---|---|---|---|
| Ser/Cys | Cys/Cys | ||||||
| Ethnicity | |||||||
| Caucasian | 7 | 3,352/4,887 | 1.02 (0.81–1.29) | 1.24 (0.84–1.83) | 0.004 | 0.13 | |
| Asian | 8 | 3,611/3,240 | 1.16 (1.00–1.36)* | 1.27 (1.09–1.48) | 0.23 | 0.79 | |
| Design | |||||||
| Population-based | Overall | 9 | 2,860/4,596 | 1.21 (1.04–1.40) | 1.38 (1.10–1.74) | 0.12 | 0.26 |
| Caucasian | 4 | 982/2,509 | 1.26 (0.97–1.63) | 1.33 (0.65–2.72) | 0.09 | 0.04 | |
| Asian | 2 | 1,214/2,212 | 1.42 (0.87–2.31) | 1.36 (1.05–1.77) | 0.11 | 0.43 | |
| v | |||||||
| Hospital-based | Overall | 10 | 4,932/4,762 | 0.97 (0.83–1.12) | 1.17 (1.00–1.36)* | 0.09 | 0.63 |
| Caucasian | 3 | 4,932/4,762 | 0.79 (0.60–1.04) | 1.15 (0.85–1.55) | 0.21 | 0.54 | |
| Asian | 6 | 2,370/2,378 | 1.11 (0.94–1.31) | 1.23 (1.02–1.48) | 0.33 | 0.72 | |
| Histology | |||||||
| Adenocarcinoma | Overall | 8 | 2,707/4,479 | 1.14 (0.90–1.45) | 1.43 (1.19–1.72) | 0.02 | 0.68 |
| Caucasian | 3 | 612/2,618 | 1.05 (0.56–1.97) | 1.90 (0.99–3.63)** | 0.01 | 0.30 | |
| Asian | 5 | 2,095/1,861 | 1.24 (1.04–1.48) | 1.38 (1.13–1.69) | 0.82 | 0.79 | |
| Squamous cell carcinoma | Overall | 6 | 1,390/3,933 | 1.02 (0.79–1.33) | 1.07 (0.81–1.40) | 0.13 | 0.43 |
| Caucasian | 3 | 1,008/2,618 | 1.08 (0.60–1.95) | 1.56 (0.54–4.48) | 0.03 | 0.15 | |
| Asian | 3 | 382/1,315 | 0.96 (0.70–1.31) | 1.07 (0.74–1.53) | 0.54 | 0.58 | |
| Small cell carcinoma | Asian | 2 | 127/1,194 | 0.92 (0.55–1.54) | 0.99 (0.51–1.93) | 0.68 | 0.52 |
| Overall | 19 | 7,792/9,358 | 1.07 (0.95–1.21) | 1.24 (1.09–1.42) | 0.007 | 0.32 | |
4. Discussion
5. Conclusions
Acknowledgements
References
- Pharoah, P.D.; Dunning, A.M.; Ponder, B.A.; Easton, D.F. Association studies for finding cancer-susceptibility genetic variants. Nat. Rev. Cancer 2004, 4, 850–860. [Google Scholar] [CrossRef]
- Hecht, S.S. Tobacco smoke carcinogens and lung cancer. J. Natl. Cancer Inst. 1999, 91, 1194–1210. [Google Scholar] [CrossRef]
- Livneh, Z. DNA damage control by novel DNA polymerases: Translesion replication and mutagenesis. J. Biol. Chem. 2001, 276, 25639–25642. [Google Scholar] [CrossRef]
- Berwick, M.; Vineis, P. Markers of DNA repair and susceptibility to cancer in humans: An epidemiologic review. J. Natl. Cancer Inst. 2000, 92, 874–897. [Google Scholar] [CrossRef]
- Wei, Q.; Cheng, L.; Amos, C.I.; Wang, L.E.; Guo, Z.; Hong, W.K.; Spitz, M.R. Repair of tobacco carcinogen-induced DNA adducts and lung cancer risk: A molecular epidemiologic study. J. Natl. Cancer Inst. 2000, 92, 1764–1772. [Google Scholar] [CrossRef]
- Wei, Q.; Cheng, L.; Hong, W.K.; Spitz, M.R. Reduced DNA repair capacity in lung cancer patients. Cancer Res. 1996, 56, 4103–4107. [Google Scholar]
- Rajaee-Behbahani, N.; Schmezer, P.; Risch, A.; Rittgen, W.; Kayser, K.W.; Dienemann, H.; Schulz, V.; Drings, P.; Thiel, S.; Bartsch, H. Altered DNA repair capacity and bleomycin sensitivity as risk markers for non-small cell lung cancer. Int. J. Cancer 2001, 95, 86–91. [Google Scholar] [CrossRef]
- Tomkinson, A.E.; Mackey, Z.B. Structure and function of mammalian DNA ligases. Mutat. Res. 1998, 407, 1–9. [Google Scholar] [CrossRef]
- Rosenquist, T.A.; Zharkov, D.O.; Grollman, A.P. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. USA 1997, 94, 7429–7434. [Google Scholar] [CrossRef]
- Nishioka, K.; Ohtsubo, T.; Oda, H.; Fujiwara, T.; Kang, D.; Sugimachi, K.; Nakabeppu, Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced ogg1 mrnas. Mol. Biol. Cell 1999, 10, 1637–1652. [Google Scholar] [CrossRef]
- Weiss, J.M.; Goode, E.L.; Ladiges, W.C.; Ulrich, C.M. Polymorphic variation in hogg1 and risk of cancer: A review of the functional and epidemiologic literature. Mol. Carcinog. 2005, 42, 127–141. [Google Scholar] [CrossRef]
- Lee, A.J.; Hodges, N.J.; Chipman, J.K. Interindividual variability in response to sodium dichromate-induced oxidative DNA damage: Role of the ser326cys polymorphism in the DNA-repair protein of 8-oxo-7,8-dihydro-2'-deoxyguanosine DNA glycosylase 1. Cancer Epidemiol. Biomarkers Prev. 2005, 14, 497–505. [Google Scholar] [CrossRef]
- Sugimura, H.; Kohno, T.; Wakai, K.; Nagura, K.; Genka, K.; Igarashi, H.; Morris, B.J.; Baba, S.; Ohno, Y.; Gao, C.; Li, Z.; Wang, J.; Takezaki, T.; Tajima, K.; Varga, T.; Sawaguchi, T.; Lum, J.K.; Martinson, J.J.; Tsugane, S.; Iwamasa, T.; Shinmura, K.; Yokota, J. Hogg1 ser326cys polymorphism and lung cancer susceptibility. Cancer Epidemiol. Biomarkers Prev. 1999, 8, 669–674. [Google Scholar]
- Wikman, H.; Risch, A.; Klimek, F.; Schmezer, P.; Spiegelhalder, B.; Dienemann, H.; Kayser, K.; Schulz, V.; Drings, P.; Bartsch, H. Hogg1 polymorphism and loss of heterozygosity (loh): Significance for lung cancer susceptibility in a caucasian population. Int. J. Cancer 2000, 88, 932–937. [Google Scholar] [CrossRef]
- Ito, H.; Hamajima, N.; Takezaki, T.; Matsuo, K.; Tajima, K.; Hatooka, S.; Mitsudomi, T.; Suyama, M.; Sato, S.; Ueda, R. A limited association of ogg1 ser326cys polymorphism for adenocarcinoma of the lung. J. Epidemiol. 2002, 12, 258–265. [Google Scholar] [CrossRef]
- Sunaga, N.; Kohno, T.; Yanagitani, N.; Sugimura, H.; Kunitoh, H.; Tamura, T.; Takei, Y.; Tsuchiya, S.; Saito, R.; Yokota, J. Contribution of the nqo1 and gstt1 polymorphisms to lung adenocarcinoma susceptibility. Cancer Epidemiol. Biomarkers Prev. 2002, 11, 730–738. [Google Scholar]
- Lan, Q.; Mumford, J.L.; Shen, M.; Demarini, D.M.; Bonner, M.R.; He, X.; Yeager, M.; Welch, R.; Chanock, S.; Tian, L.; Chapman, R.S.; Zheng, T.; Keohavong, P.; Caporaso, N.; Rothman, N. Oxidative damage-related genes akr1c3 and ogg1 modulate risks for lung cancer due to exposure to pah-rich coal combustion emissions. Carcinogenesis 2004, 25, 2177–2181. [Google Scholar] [CrossRef]
- Vogel, U.; Nexo, B.A.; Wallin, H.; Overvad, K.; Tjonneland, A.; Raaschou-Nielsen, O. No association between base excision repair gene polymorphisms and risk of lung cancer. Biochem Genet 2004, 42, 453–460. [Google Scholar] [CrossRef]
- Liang, G.; Pu, Y.; Yin, L. Rapid detection of single nucleotide polymorphisms related with lung cancer susceptibility of chinese population. Cancer Lett. 2005, 223, 265–274. [Google Scholar] [CrossRef]
- Hung, R.J.; Brennan, P.; Canzian, F.; Szeszenia-Dabrowska, N.; Zaridze, D.; Lissowska, J.; Rudnai, P.; Fabianova, E.; Mates, D.; Foretova, L.; Janout, V.; Bencko, V.; Chabrier, A.; Borel, S.; Hall, J.; Boffetta, P. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. J. Natl. Cancer Inst. 2005, 97, 567–576. [Google Scholar] [CrossRef]
- Zienolddiny, S.; Campa, D.; Lind, H.; Ryberg, D.; Skaug, V.; Stangeland, L.; Phillips, D.H.; Canzian, F.; Haugen, A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 2006, 27, 560–567. [Google Scholar]
- Matullo, G.; Dunning, A.M.; Guarrera, S.; Baynes, C.; Polidoro, S.; Garte, S.; Autrup, H.; Malaveille, C.; Peluso, M.; Airoldi, L.; Veglia, F.; Gormally, E.; Hoek, G.; Krzyzanowski, M.; Overvad, K.; Raaschou-Nielsen, O.; Clavel-Chapelon, F.; Linseisen, J.; Boeing, H.; Trichopoulou, A.; Palli, D.; Krogh, V.; Tumino, R.; Panico, S.; Bueno-De-Mesquita, H.B.; Peeters, P.H.; Lund, E.; Pera, G.; Martinez, C.; Dorronsoro, M.; Barricarte, A.; Tormo, M.J.; Quiros, J.R.; Day, N.E.; Key, T.J.; Saracci, R.; Kaaks, R.; Riboli, E.; Vineis, P. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis 2006, 27, 997–1007. [Google Scholar] [CrossRef]
- Kohno, T.; Kunitoh, H.; Toyama, K.; Yamamoto, S.; Kuchiba, A.; Saito, D.; Yanagitani, N.; Ishihara, S.; Saito, R.; Yokota, J. Association of the ogg1-ser326cys polymorphism with lung adenocarcinoma risk. Cancer Sci 2006, 97, 724–728. [Google Scholar] [CrossRef]
- Sorensen, M.; Raaschou-Nielsen, O.; Hansen, R.D.; Tjonneland, A.; Overvad, K.; Vogel, U. Interactions between the ogg1 ser326cys polymorphism and intake of fruit and vegetables in relation to lung cancer. Free Radic. Res. 2006, 40, 885–891. [Google Scholar] [CrossRef]
- Karahalil, B.; Emerce, E.; Kocer, B.; Han, S.; Alkis, N.; Karakaya, A.E. The association of ogg1 ser326cys polymorphism and urinary 8-ohdg levels with lung cancer susceptibility: A hospital-based case-control study in turkey. Arh. Hig. Rada Toksikol. 2008, 59, 241–250. [Google Scholar]
- Miyaishi, A.; Osawa, K.; Osawa, Y.; Inoue, N.; Yoshida, K.; Kasahara, M.; Tsutou, A.; Tabuchi, Y.; Sakamoto, K.; Tsubota, N.; Takahashi, J. Mutyh gln324his gene polymorphism and genetic susceptibility for lung cancer in a japanese population. J. Exp. Clin. Cancer Res. 2009, 28, 10. [Google Scholar] [CrossRef]
- Chang, J.S.; Wrensch, M.R.; Hansen, H.M.; Sison, J.D.; Aldrich, M.C.; Quesenberry, C.P., Jr.; Seldin, M.F.; Kelsey, K.T.; Wiencke, J.K. Base excision repair genes and risk of lung cancer among san francisco bay area latinos and african-americans. Carcinogenesis 2009, 30, 78–87. [Google Scholar]
- Le Marchand, L.; Donlon, T.; Lum-Jones, A.; Seifried, A.; Wilkens, L.R. Association of the hogg1 ser326cys polymorphism with lung cancer risk. Cancer Epidemiol. Biomarkers Prev. 2002, 11, 409–412. [Google Scholar]
- Park, J.; Chen, L.; Tockman, M.S.; Elahi, A.; Lazarus, P. The human 8-oxoguanine DNA n-glycosylase 1 (hogg1) DNA repair enzyme and its association with lung cancer risk. Pharmacogenetics 2004, 14, 103–109. [Google Scholar] [CrossRef]
- De Ruyck, K.; Szaumkessel, M.; De Rudder, I.; Dehoorne, A.; Vral, A.; Claes, K.; Velghe, A.; Van Meerbeeck, J.; Thierens, H. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat. Res. 2007, 631, 101–110. [Google Scholar] [CrossRef]
- Chang, C.H.; Hsiao, C.F.; Chang, G.C.; Tsai, Y.H.; Chen, Y.M.; Huang, M.S.; Su, W.C.; Hsieh, W.S.; Yang, P.C.; Chen, C.J.; Hsiung, C.A. Interactive effect of cigarette smoking with human 8-oxoguanine DNA n-glycosylase 1 (hogg1) polymorphisms on the risk of lung cancer: A case-control study in taiwan. Am. J. Epidemiol. 2009, 170, 695–702. [Google Scholar] [CrossRef]
- Okasaka, T.; Matsuo, K.; Suzuki, T.; Ito, H.; Hosono, S.; Kawase, T.; Watanabe, M.; Yatabe, Y.; Hida, T.; Mitsudomi, T.; Tanaka, H.; Yokoi, K.; Tajima, K. Hogg1 ser326cys polymorphism and risk of lung cancer by histological type. J. Hum. Genet. 2009, 54, 739–745. [Google Scholar] [CrossRef]
- Li, H.; Hao, X.; Zhang, W.; Wei, Q.; Chen, K. The hogg1 ser326cys polymorphism and lung cancer risk: A meta-analysis. Cancer Epidemiol. Biomarkers Prev. 2008, 17, 1739–1745. [Google Scholar]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials. Control. Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef]
- Cochran, W. The combination of estimates from different experiments. Biometrics 1954, 10, 101–129. [Google Scholar] [CrossRef]
- Whitehead, A.; Whitehead, J. A general parametric approach to the meta-analysis of randomized clinical trials. Stat. Med. 1991, 10, 1665–1677. [Google Scholar] [CrossRef]
- Begg, C.B.; Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994, 50, 1088–1101. [Google Scholar] [CrossRef]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef]
- Duval, S.; Tweedie, R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000, 56, 455–463. [Google Scholar] [CrossRef]
- Kohno, T.; Shinmura, K.; Tosaka, M.; Tani, M.; Kim, S.R.; Sugimura, H.; Nohmi, T.; Kasai, H.; Yokota, J. Genetic polymorphisms and alternative splicing of the hogg1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene 1998, 16, 3219–3225. [Google Scholar]
- Dhenaut, A.; Boiteux, S.; Radicella, J.P. Characterization of the hogg1 promoter and its expression during the cell cycle. Mutat. Res. 2000, 461, 109–118. [Google Scholar] [CrossRef]
- Hung, R.J.; Hall, J.; Brennan, P.; Boffetta, P. Genetic polymorphisms in the base excision repair pathway and cancer risk: A huge review. Am. J. Epidemiol. 2005, 162, 925–942. [Google Scholar] [CrossRef]
- Vogel, U.; Overvad, K.; Wallin, H.; Tjonneland, A.; Nexo, B.A.; Raaschou-Nielsen, O. Combinations of polymorphisms in xpd, xpc and xpa in relation to risk of lung cancer. Cancer Lett. 2005, 222, 67–74. [Google Scholar] [CrossRef]
- Loft, S.; Svoboda, P.; Kasai, H.; Tjonneland, A.; Vogel, U.; Moller, P.; Overvad, K.; Raaschou-Nielsen, O. Prospective study of 8-oxo-7,8-dihydro-2'-deoxyguanosine excretion and the risk of lung cancer. Carcinogenesis 2006, 27, 1245–1250. [Google Scholar] [CrossRef]
- Hecht, S.S.; Morse, M.A.; Amin, S.; Stoner, G.D.; Jordan, K.G.; Choi, C.I.; Chung, F.L. Rapid single-dose model for lung tumor induction in a/j mice by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and the effect of diet. Carcinogenesis 1989, 10, 1901–1904. [Google Scholar] [CrossRef]
- Hecht, S.S. DNA adduct formation from tobacco-specific n-nitrosamines. Mutat. Res. 1999, 424, 127–142. [Google Scholar] [CrossRef]
- Hoffmann, D.; Brunnemann, K.D.; Prokopczyk, B.; Djordjevic, M.V. Tobacco-specific n-nitrosamines and areca-derived n-nitrosamines: Chemistry, biochemistry, carcinogenicity, and relevance to humans. J. Toxicol. Environ. Health 1994, 41, 1–52. [Google Scholar] [CrossRef]
- Arimoto-Kobayashi, S.; Sakata, H.; Mitsu, K.; Tanoue, H. A possible photosensitizer: Tobacco-specific nitrosamine, 4-(n-methylnitrosamino)-1-(3-pyridyl)-1-butanone (nnk), induced mutations, DNA strand breaks and oxidative and methylative damage with uva. Mutat. Res. 2007, 632, 111–120. [Google Scholar] [CrossRef]
- Igarashi, M.; Watanabe, M.; Yoshida, M.; Sugaya, K.; Endo, Y.; Miyajima, N.; Abe, M.; Sugano, S.; Nakae, D. Enhancement of lung carcinogenesis initiated with 4-(n-hydroxymethylnitrosamino)-1-(3-pyridyl)-1-butanone by ogg1 gene deficiency in female, but not male, mice. J. Toxicol. Sci. 2009, 34, 163–174. [Google Scholar] [CrossRef]
- Kiyohara, C.; Otsu, A.; Shirakawa, T.; Fukuda, S.; Hopkin, J.M. Genetic polymorphisms and lung cancer susceptibility: A review. Lung Cancer 2002, 37, 241–256. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kiyohara, C.; Takayama, K.; Nakanishi, Y. Lung Cancer Susceptibility and hOGG1 Ser326Cys Polymorphism: A Meta-Analysis. Cancers 2010, 2, 1813-1829. https://doi.org/10.3390/cancers2041813
Kiyohara C, Takayama K, Nakanishi Y. Lung Cancer Susceptibility and hOGG1 Ser326Cys Polymorphism: A Meta-Analysis. Cancers. 2010; 2(4):1813-1829. https://doi.org/10.3390/cancers2041813
Chicago/Turabian StyleKiyohara, Chikako, Koichi Takayama, and Yoichi Nakanishi. 2010. "Lung Cancer Susceptibility and hOGG1 Ser326Cys Polymorphism: A Meta-Analysis" Cancers 2, no. 4: 1813-1829. https://doi.org/10.3390/cancers2041813
APA StyleKiyohara, C., Takayama, K., & Nakanishi, Y. (2010). Lung Cancer Susceptibility and hOGG1 Ser326Cys Polymorphism: A Meta-Analysis. Cancers, 2(4), 1813-1829. https://doi.org/10.3390/cancers2041813