Reduced HLA-I Transcript Levels and Increased Abundance of a CD56dim NK Cell Signature Are Associated with Improved Survival in Lower-Grade Gliomas
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval of LGG Patient Transcriptomes from TCGA and Chinese Glioma Genome Atlas (CGGA) Databases
2.2. Construction of Transcriptional Signatures for CD56bright and CD56dim NK Cells
2.3. Deconvolution of Bulk RNAseq Datasets to Obtain the Relative Abundance of CD56bright and CD56dim NK Cell Subsets
2.4. Estimation of HLA-I Gene Abundance
2.5. LGG Prognostic Association
2.6. Single-Cell RNA-seq Data Analysis
2.7. Statistical Analysis
3. Results
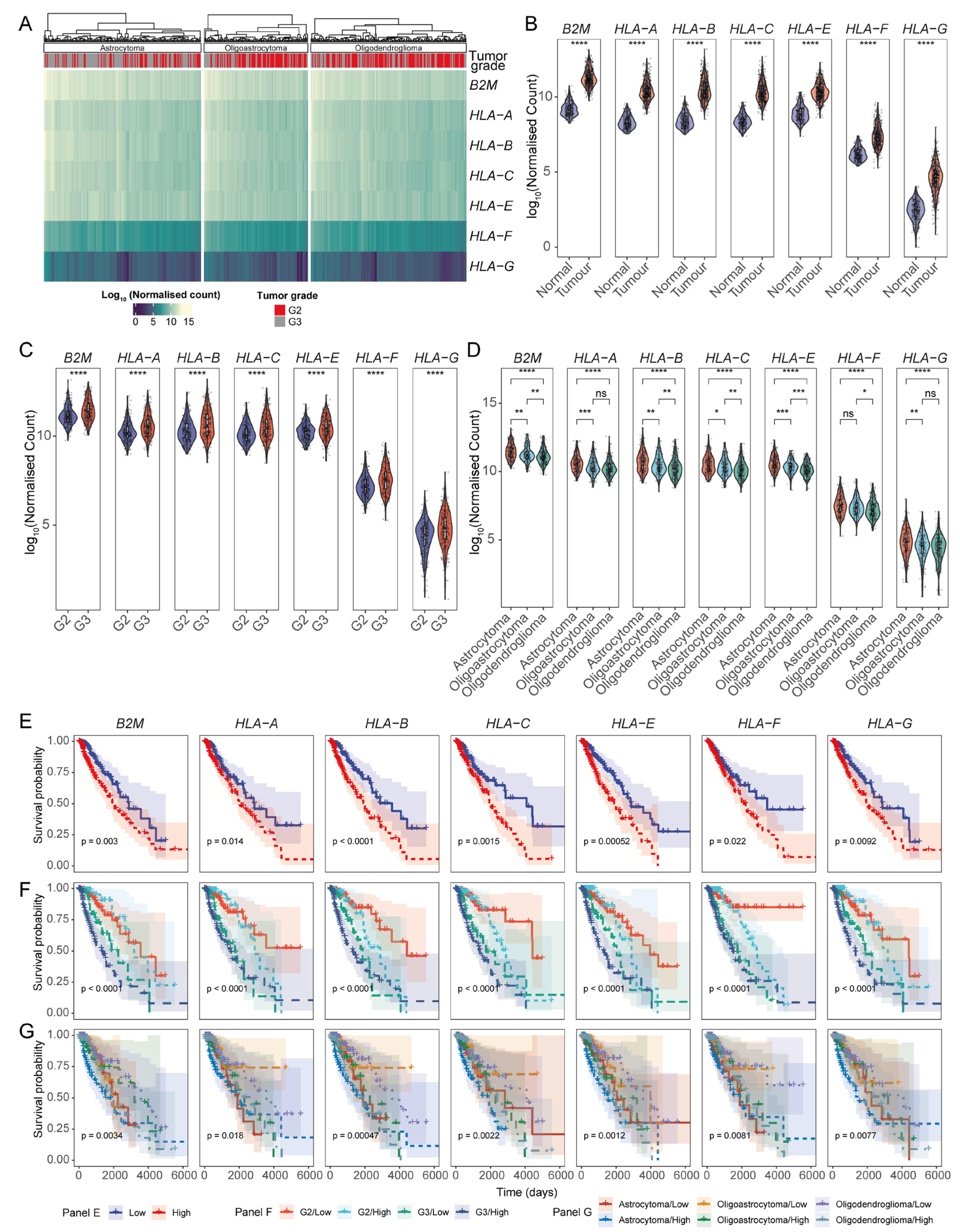
3.1. A Pan-Cancer Screen Reveals an Association Between the Differential Expression of HLA-I Transcripts and Cancer Patient Prognoses
3.2. Association of HLA-I Transcript Expression with Poor LGG Patient Prognosis Is Dependent on Tumour Grade
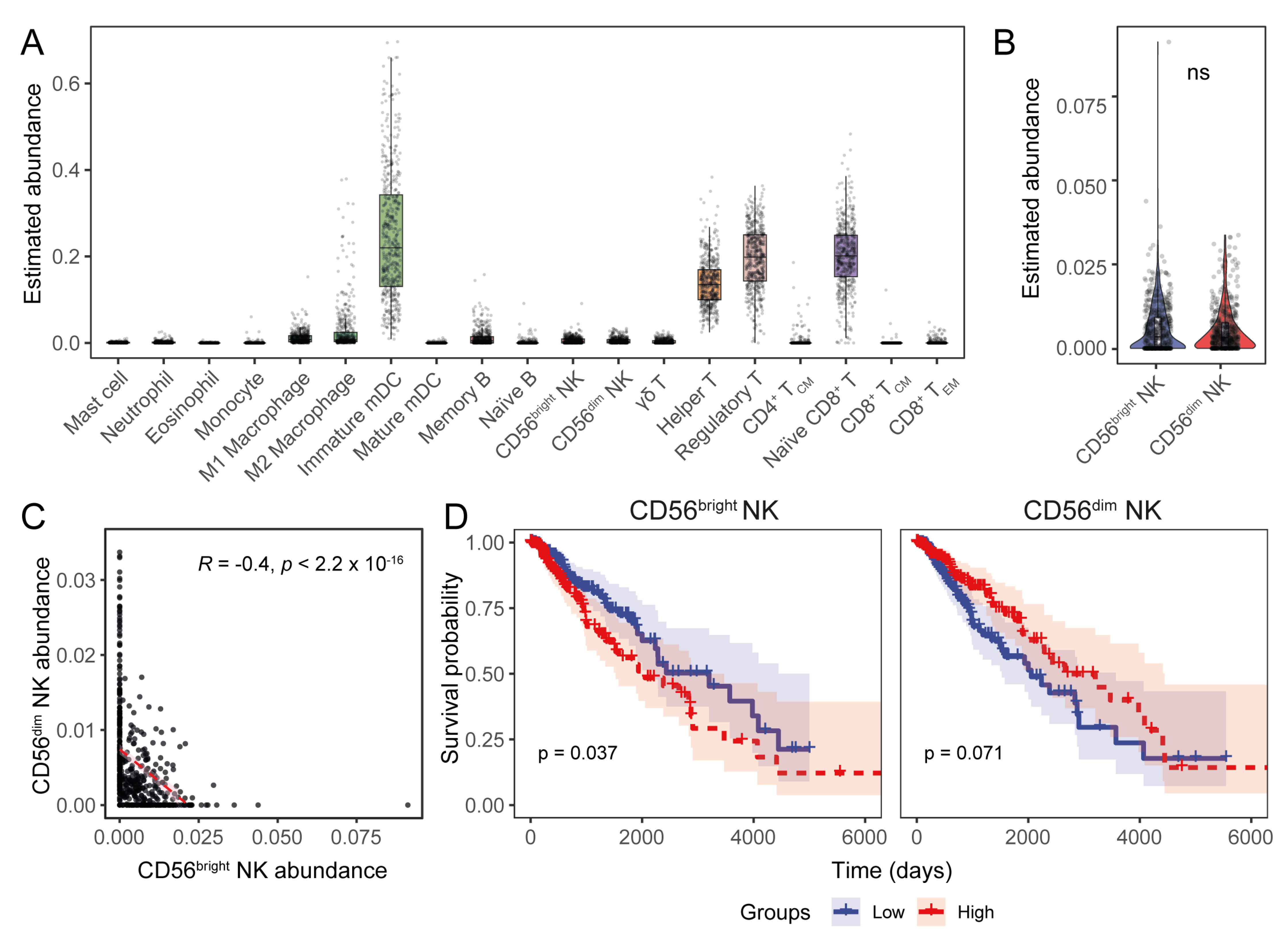
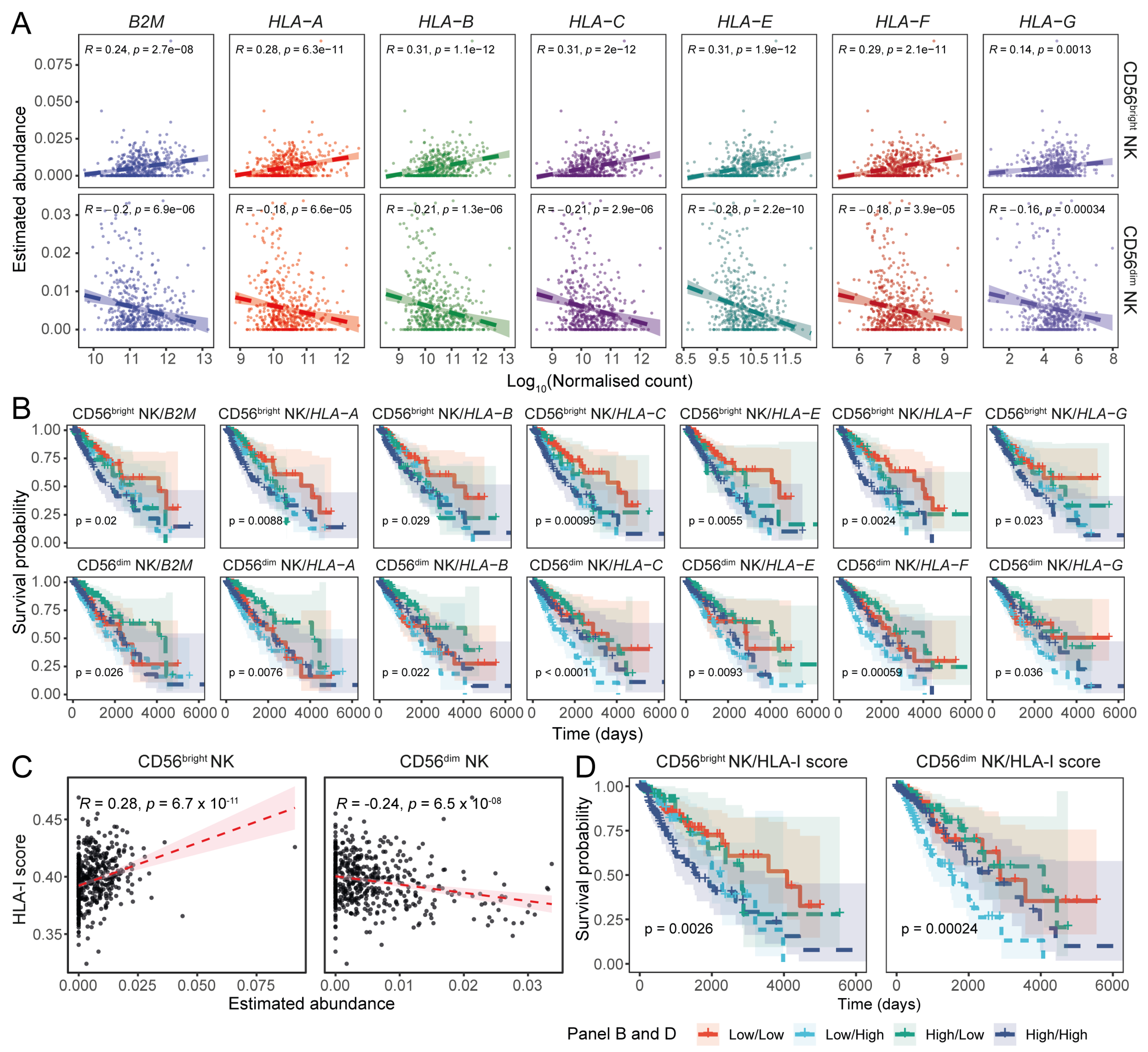
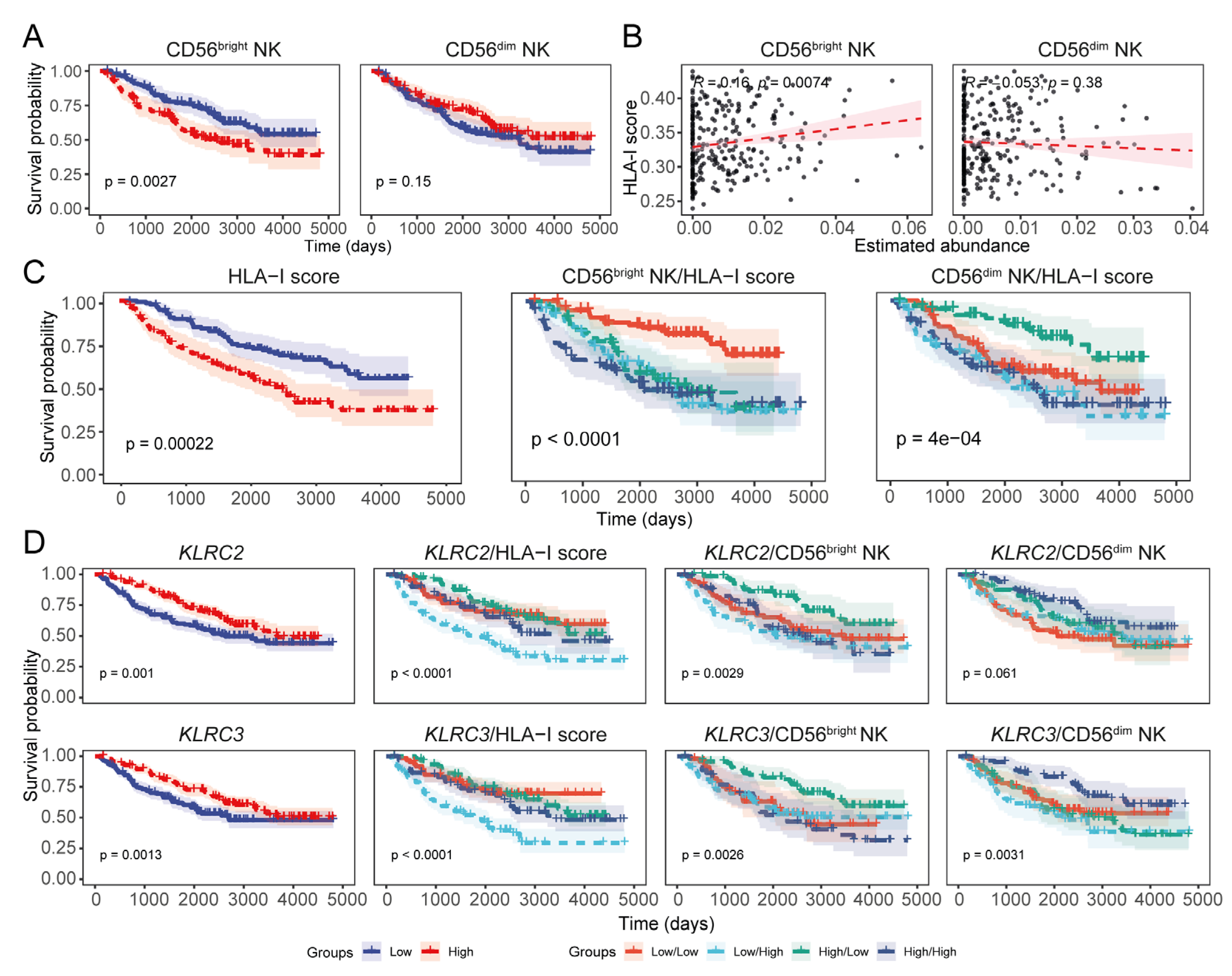
3.3. Expression of the Transcriptional Signature of CD56dim NK Cells Inversely Correlates with HLA-I Transcript Expression in LGGs
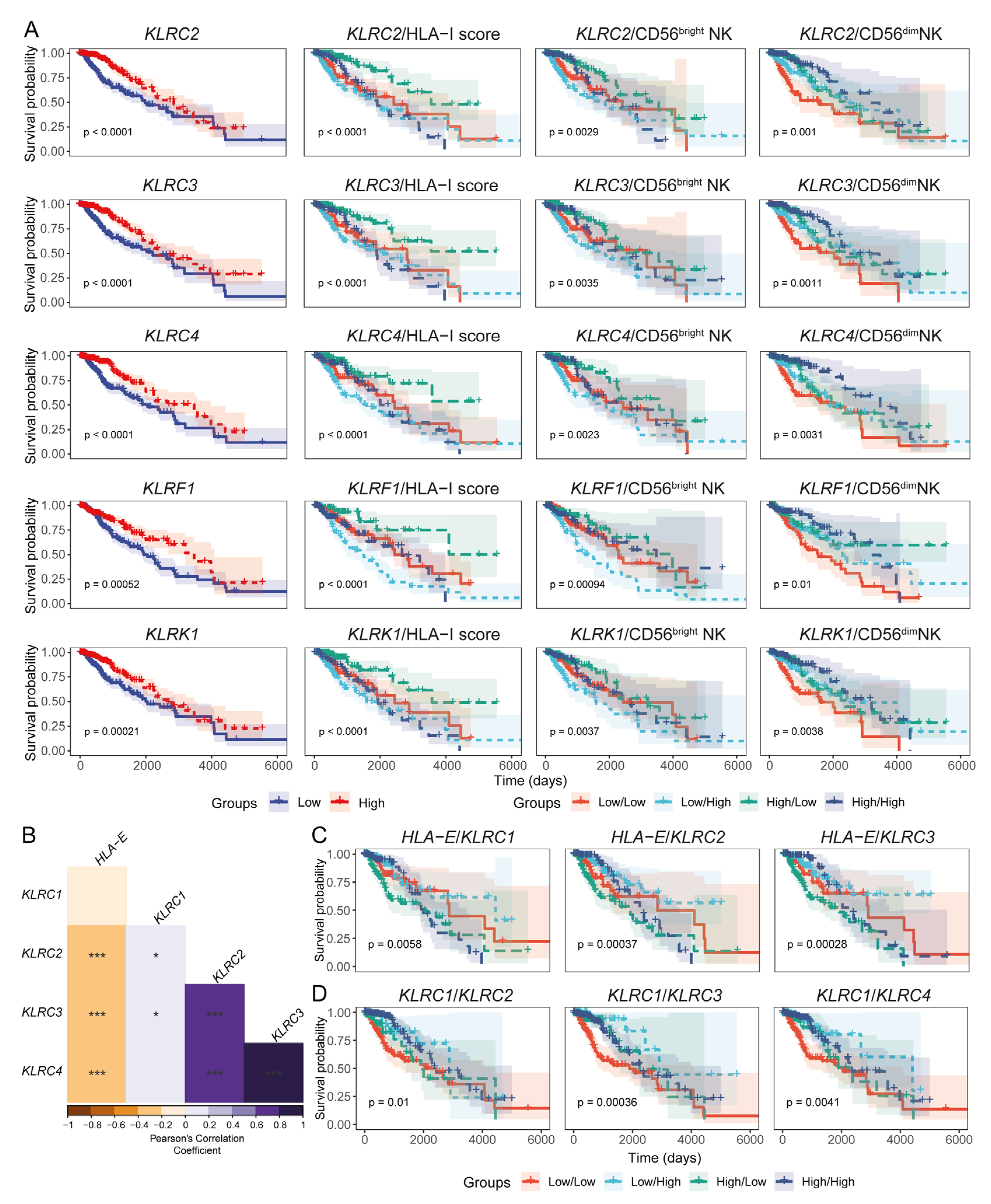
3.4. Expression Levels of HLA-I and NK Cell Receptor Transcripts Are Associated with LGG Patient Prognoses
3.5. Expression of HLA-I Transcripts Is Negatively Associated with the CD56dim NK Signature in the CGGA LGG Tissue Transcriptomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aiman, W.; Gasalberti, D.P.; Rayi, A. Low-Grade Gliomas. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2024. [Google Scholar]
- Rebecca, J.; Limb, C.G.; Leon, T.L. Natural History and Management Options of Low-Grade Glioma. In Neurosurgical Diseases; Thieme Medical Publishers, Inc.: New York, NY, USA, 2022. [Google Scholar] [CrossRef]
- Brown, T.J.; Bota, D.A.; van Den Bent, M.J.; Brown, P.D.; Maher, E.; Aregawi, D.; Liau, L.M.; Buckner, J.C.; Weller, M.; Berger, M.S.; et al. Management of low-grade glioma: A systematic review and meta-analysis. Neurooncol. Pract. 2019, 6, 249–258. [Google Scholar] [CrossRef]
- Haddad, A.F.; Young, J.S.; Oh, J.Y.; Okada, H.; Aghi, M.K. The immunology of low-grade gliomas. Neurosurg. Focus. 2022, 52, E2. [Google Scholar] [CrossRef]
- Song, L.R.; Weng, J.C.; Li, C.B.; Huo, X.L.; Li, H.; Hao, S.Y.; Wu, Z.; Wang, L.; Li, D.; Zhang, J.T. Prognostic and predictive value of an immune infiltration signature in diffuse lower-grade gliomas. JCI Insight 2020, 5, e133811. [Google Scholar] [CrossRef]
- Friebel, E.; Kapolou, K.; Unger, S.; Núñez, N.G.; Utz, S.; Rushing, E.J.; Regli, L.; Weller, M.; Greter, M.; Tugues, S. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell 2020, 181, 1626–1642.e20. [Google Scholar] [CrossRef]
- Heimberger, A.B.; Abou-Ghazal, M.; Reina-Ortiz, C.; Yang, D.S.; Sun, W.; Qiao, W.; Hiraoka, N.; Fuller, G.N. Incidence and prognostic impact of FoxP3+ regulatory T cells in human gliomas. Clin. Cancer Res. 2008, 14, 5166–5172. [Google Scholar] [CrossRef]
- Klemm, F.; Maas, R.R.; Bowman, R.L.; Kornete, M.; Soukup, K.; Nassiri, S.; Brouland, J.-P.; Iacobuzio-Donahue, C.A.; Brennan, C.; Tabar, V. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 2020, 181, 1643–1660.e17. [Google Scholar] [CrossRef]
- Müller, S.; Kohanbash, G.; Liu, S.J.; Alvarado, B.; Carrera, D.; Bhaduri, A.; Watchmaker, P.B.; Yagnik, G.; Di Lullo, E.; Malatesta, M. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 2017, 18, 234. [Google Scholar] [CrossRef]
- Venteicher, A.S.; Tirosh, I.; Hebert, C.; Yizhak, K.; Neftel, C.; Filbin, M.G.; Hovestadt, V.; Escalante, L.E.; Shaw, M.L.; Rodman, C. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 2017, 355, eaai8478. [Google Scholar] [CrossRef]
- Hicklin, D.J.; Marincola, F.M.; Ferrone, S. HLA class I antigen downregulation in human cancers: T-cell immunotherapy revives an old story. Mol. Med. Today 1999, 5, 178–186. [Google Scholar] [CrossRef]
- Yang, K.; Halima, A.; Chan, T.A. Antigen presentation in cancer—Mechanisms and clinical implications for immunotherapy. Nat. Rev. Clin. Oncol. 2023, 20, 604–623. [Google Scholar] [CrossRef]
- Wieczorek, M.; Abualrous, E.T.; Sticht, J.; Álvaro-Benito, M.; Stolzenberg, S.; Noé, F.; Freund, C. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front. Immunol. 2017, 8, 292. [Google Scholar] [CrossRef] [PubMed]
- Hazini, A.; Fisher, K.; Seymour, L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J. Immunother. Cancer 2021, 9, e002899. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, J.E.; Hsu, K.C. Natural killer cell education in human health and disease. Curr. Opin. Immunol. 2018, 50, 102–111. [Google Scholar] [CrossRef]
- Brodin, P.; Kärre, K.; Höglund, P. NK cell education: Not an on-off switch but a tunable rheostat. Trends Immunol. 2009, 30, 143–149. [Google Scholar] [CrossRef]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar] [CrossRef]
- Yokoyama, W.M.; Kim, S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol. Rev. 2006, 214, 143–154. [Google Scholar] [CrossRef]
- Raulet, D.H.; Vance, R.E. Self-tolerance of natural killer cells. Nat. Rev. Immunol. 2006, 6, 520–531. [Google Scholar] [CrossRef]
- Hofmeister, V.; Weiss, E.H. HLA-G modulates immune responses by diverse receptor interactions. Semin. Cancer Biol. 2003, 13, 317–323. [Google Scholar] [CrossRef]
- Kaiser, B.K.; Barahmand-pour, F.; Paulsene, W.; Medley, S.; Geraghty, D.E.; Strong, R.K. Interactions between NKG2x Immunoreceptors and HLA-E Ligands Display Overlapping Affinities and Thermodynamics1. J. Immunol. 2005, 174, 2878–2884. [Google Scholar] [CrossRef]
- Kaiser, B.K.; Pizarro, J.C.; Kerns, J.; Strong, R.K. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc. Natl. Acad. Sci. USA 2008, 105, 6696–6701. [Google Scholar] [CrossRef]
- Trowsdale, J.; Jones, D.C.; Barrow, A.D.; Traherne, J.A. Surveillance of cell and tissue perturbation by receptors in the LRC. Immunol. Rev. 2015, 267, 117–136. [Google Scholar] [CrossRef] [PubMed]
- Barrow, A.D.; Colonna, M. Exploiting NK Cell Surveillance Pathways for Cancer Therapy. Cancers 2019, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Freud, A.G.; Mundy-Bosse, B.L.; Yu, J.; Caligiuri, M.A. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity 2017, 47, 820–833. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.A.; Miller, J.S. Exploring the NK cell platform for cancer immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 85–100. [Google Scholar] [CrossRef]
- Bald, T.; Pedde, A.-M.; Corvino, D.; Böttcher, J.P. Chapter Two—The role of NK cell as central communicators in cancer immunity. In Advances in Immunology; Alt, F.W., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 61–88. [Google Scholar]
- Cózar, B.; Greppi, M.; Carpentier, S.; Narni-Mancinelli, E.; Chiossone, L.; Vivier, E. Tumor-Infiltrating Natural Killer Cells. Cancer Discov. 2021, 11, 34–44. [Google Scholar] [CrossRef]
- Guillerey, C. NK Cells in the Tumor Microenvironment. In Tumor Microenvironment: Hematopoietic Cells—Part B; Birbrair, A., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 69–90. [Google Scholar] [CrossRef]
- Larsen, S.K.; Gao, Y.; Basse, P.H. NK cells in the tumor microenvironment. Crit. Rev. Oncog. 2014, 19, 91–105. [Google Scholar] [CrossRef]
- Breznik, B.; Ko, M.-W.; Tse, C.; Chen, P.-C.; Senjor, E.; Majc, B.; Habič, A.; Angelillis, N.; Novak, M.; Župunski, V.; et al. Infiltrating natural killer cells bind, lyse and increase chemotherapy efficacy in glioblastoma stem-like tumorospheres. Commun. Biol. 2022, 5, 436. [Google Scholar] [CrossRef]
- Fares, J.; Davis, Z.B.; Rechberger, J.S.; Toll, S.A.; Schwartz, J.D.; Daniels, D.J.; Miller, J.S.; Khatua, S. Advances in NK cell therapy for brain tumors. NPJ Precis. Oncol. 2023, 7, 17. [Google Scholar] [CrossRef]
- Sedgwick, A.J.; Ghazanfari, N.; Constantinescu, P.; Mantamadiotis, T.; Barrow, A.D. The Role of NK Cells and Innate Lymphoid Cells in Brain Cancer. Front. Immunol. 2020, 11, 1549. [Google Scholar] [CrossRef]
- Sun, Y.; Sedgwick, A.J.; Palarasah, Y.; Mangiola, S.; Barrow, A.D. A Transcriptional Signature of PDGF-DD Activated Natural Killer Cells Predicts More Favorable Prognosis in Low-Grade Glioma. Front. Immunol. 2021, 12, 668391. [Google Scholar] [CrossRef]
- Barrow, A.D.; Edeling, M.A.; Trifonov, V.; Luo, J.; Goyal, P.; Bohl, B.; Bando, J.K.; Kim, A.H.; Walker, J.; Andahazy, M.; et al. Natural Killer Cells Control Tumor Growth by Sensing a Growth Factor. Cell 2018, 172, 534–548.e19. [Google Scholar] [CrossRef] [PubMed]
- Balatsoukas, A.; Rossignoli, F.; Shah, K. NK cells in the brain: Implications for brain tumor development and therapy. Trends Mol. Med. 2022, 28, 194–209. [Google Scholar] [CrossRef] [PubMed]
- Castriconi, R.; Daga, A.; Dondero, A.; Zona, G.; Poliani, P.L.; Melotti, A.; Griffero, F.; Marubbi, D.; Spaziante, R.; Bellora, F. NK cells recognize and kill human glioblastoma cells with stem cell-like properties. J. Immunol. 2009, 182, 3530–3539. [Google Scholar] [CrossRef] [PubMed]
- Poli, A.; Kmiecik, J.; Domingues, O.; Hentges, F.; Bléry, M.; Chekenya, M.; Boucraut, J.; Zimmer, J. NK Cells in Central Nervous System Disorders. J. Immunol. 2013, 190, 5355–5362. [Google Scholar] [CrossRef]
- Crane, C.A.; Han, S.J.; Barry, J.J.; Ahn, B.J.; Lanier, L.L.; Parsa, A.T. TGF-β downregulates the activating receptor NKG2D on NK cells and CD8+ T cells in glioma patients. Neuro-Oncol. 2009, 12, 7–13. [Google Scholar] [CrossRef]
- Chang, K.; Creighton, C.J.; Davis, C.; Donehower, L.; Drummond, J.; Wheeler, D.; Ally, A.; Balasundaram, M.; Birol, I.; Butterfield, Y.S.N.; et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef]
- Jo, J.; van den Bent, M.J.; Nabors, B.; Wen, P.Y.; Schiff, D. Surveillance imaging frequency in adult patients with lower-grade (WHO Grade 2 and 3) gliomas. Neuro-Oncol. 2022, 24, 1035–1047. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Al Kamran Khan, M.A.; Wu, J.; Sun, Y.; Barrow, A.D.; Papenfuss, A.T.; Mangiola, S. Cellsig plug-in enhances CIBERSORTx signature selection for multidataset transcriptomes with sparse multilevel modelling. Bioinformatics 2023, 39, btad685. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Mangiola, S.; Molania, R.; Dong, R.; Doyle, M.A.; Papenfuss, A.T. tidybulk: An R tidy framework for modular transcriptomic data analysis. Genome Biol. 2021, 22, 42. [Google Scholar] [CrossRef]
- Foroutan, M.; Bhuva, D.D.; Lyu, R.; Horan, K.; Cursons, J.; Davis, M.J. Single sample scoring of molecular phenotypes. BMC Bioinform. 2018, 19, 404. [Google Scholar] [CrossRef]
- Kassambara, A.; Kosinski, M.; Biecek, P. Survminer: Drawing Survival Curves Using ‘ggplot2’, R Package Version 0.5.0. Available online: https://github.com/kassambara/survminer (accessed on 23 September 2024).
- Mantel, N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother. Rep. 1966, 50, 163–170. [Google Scholar]
- Abdelfattah, N.; Kumar, P.; Wang, C.; Leu, J.-S.; Flynn, W.F.; Gao, R.; Baskin, D.S.; Pichumani, K.; Ijare, O.B.; Wood, S.L.; et al. Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target. Nat. Commun. 2022, 13, 767. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Khan, M.A.A.K.; Sedgwick, A.J.; Sun, Y.; Vivian, J.P.; Corbett, A.J.; Dolcetti, R.; Mantamadiotis, T.; Mangiola, S.; Barrow, A.D. Transcriptional signature of CD56bright NK cells predicts favourable prognosis in bladder cancer. Front. Immunol. 2025, 15, 1474652. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.-r.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Wilcoxon, F. Individual Comparisons by Ranking Methods. Biom. Bull. 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Kerdiles, Y.; Ugolini, S.; Vivier, E. T cell regulation of natural killer cells. J. Exp. Med. 2013, 210, 1065–1068. [Google Scholar] [CrossRef]
- Perez-Diez, A.; Joncker, N.T.; Choi, K.; Chan, W.F.N.; Anderson, C.C.; Lantz, O.; Matzinger, P. CD4 cells can be more efficient at tumor rejection than CD8 cells. Blood 2007, 109, 5346–5354. [Google Scholar] [CrossRef]
- Doorduijn, E.M.; Sluijter, M.; Salvatori, D.C.; Silvestri, S.; Maas, S.; Arens, R.; Ossendorp, F.; van der Burg, S.H.; van Hall, T. CD4+ T Cell and NK Cell Interplay Key to Regression of MHC Class Ilow Tumors upon TLR7/8 Agonist Therapy. Cancer Immunol. Res. 2017, 5, 642–653. [Google Scholar] [CrossRef]
- Prins, R.M.; Vo, D.D.; Khan-Farooqi, H.; Yang, M.-Y.; Soto, H.; Economou, J.S.; Liau, L.M.; Ribas, A. NK and CD4 Cells Collaborate to Protect against Melanoma Tumor Formation in the Brain1. J. Immunol. 2006, 177, 8448–8455. [Google Scholar] [CrossRef]
- Kyrysyuk, O.; Wucherpfennig, K.W. Designing Cancer Immunotherapies That Engage T Cells and NK Cells. Annu. Rev. Immunol. 2023, 41, 17–38. [Google Scholar] [CrossRef]
- Badrinath, S.; Dellacherie, M.O.; Li, A.; Zheng, S.; Zhang, X.; Sobral, M.; Pyrdol, J.W.; Smith, K.L.; Lu, Y.; Haag, S.; et al. A vaccine targeting resistant tumours by dual T cell plus NK cell attack. Nature 2022, 606, 992–998. [Google Scholar] [CrossRef]
- Rebuffet, L.; Melsen, J.E.; Escalière, B.; Basurto-Lozada, D.; Bhandoola, A.; Björkström, N.K.; Bryceson, Y.T.; Castriconi, R.; Cichocki, F.; Colonna, M.; et al. High-dimensional single-cell analysis of human natural killer cell heterogeneity. Nat. Immunol. 2024, 25, 1474–1488. [Google Scholar] [CrossRef]
- Wolf, N.K.; Kissiov, D.U.; Raulet, D.H. Roles of natural killer cells in immunity to cancer, and applications to immunotherapy. Nat. Rev. Immunol. 2023, 23, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Avril, T.; Vauleon, E.; Hamlat, A.; Saikali, S.; Etcheverry, A.; Delmas, C.; Diabira, S.; Mosser, J.; Quillien, V. Human glioblastoma stem-like cells are more sensitive to allogeneic NK and T cell-mediated killing compared with serum-cultured glioblastoma cells. Brain Pathol. 2012, 22, 159–174. [Google Scholar] [CrossRef]
- Poli, A.; Wang, J.; Domingues, O.; Planagumà, J.; Yan, T.; Rygh, C.B.; Skaftnesmo, K.O.; Thorsen, F.; McCormack, E.; Hentges, F. Targeting glioblastoma with NK cells and mAb against NG2/CSPG4 prolongs animal survival. Oncotarget 2013, 4, 1527. [Google Scholar] [CrossRef]
- Alizadeh, D.; Zhang, L.; Brown, C.E.; Farrukh, O.; Jensen, M.C.; Badie, B. Induction of anti-glioma natural killer cell response following multiple low-dose intracerebral CpG therapy. Clin. Cancer Res. 2010, 16, 3399–3408. [Google Scholar] [CrossRef]
- Friese, M.A.; Wischhusen, J.r.; Wick, W.; Weiler, M.; Eisele, G.n.; Steinle, A.; Weller, M. RNA interference targeting transforming growth factor-β enhances NKG2D-mediated antiglioma immune response, inhibits glioma cell migration and invasiveness, and abrogates tumorigenicity in vivo. Cancer Res. 2004, 64, 7596–7603. [Google Scholar] [CrossRef]
- Pellegatta, S.; Di Ianni, N.; Pessina, S.; Paterra, R.; Anghileri, E.; Eoli, M.; Finocchiaro, G. ABCC3 Expressed by CD56dim CD16+ NK Cells Predicts Response in Glioblastoma Patients Treated with Combined Chemotherapy and Dendritic Cell Immunotherapy. Int. J. Mol. Sci. 2019, 20, 5886. [Google Scholar] [CrossRef]
- Lu, J.; Li, H.; Chen, Z.; Fan, L.; Feng, S.; Cai, X.; Wang, H. Identification of 3 subpopulations of tumor-infiltrating immune cells for malignant transformation of low-grade glioma. Cancer Cell Int. 2019, 19, 265. [Google Scholar] [CrossRef]
- Weenink, B.; Draaisma, K.; Ooi, H.Z.; Kros, J.M.; Sillevis Smitt, P.A.E.; Debets, R.; French, P.J. Low-grade glioma harbors few CD8 T cells, which is accompanied by decreased expression of chemo-attractants, not immunogenic antigens. Sci. Rep. 2019, 9, 14643. [Google Scholar] [CrossRef]
- Chongsathidkiet, P.; Jackson, C.; Koyama, S.; Loebel, F.; Cui, X.; Farber, S.H.; Woroniecka, K.; Elsamadicy, A.A.; Dechant, C.A.; Kemeny, H.R.; et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat. Med. 2018, 24, 1459–1468. [Google Scholar] [CrossRef]
- Kohanbash, G.; Carrera, D.A.; Shrivastav, S.; Ahn, B.J.; Jahan, N.; Mazor, T.; Chheda, Z.S.; Downey, K.M.; Watchmaker, P.B.; Beppler, C.; et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J. Clin. Investig. 2017, 127, 1425–1437. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Sirota, M.; Butte, A.J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 2015, 6, 8971. [Google Scholar] [CrossRef] [PubMed]
- Touat, M.; Li, Y.Y.; Boynton, A.N.; Spurr, L.F.; Iorgulescu, J.B.; Bohrson, C.L.; Cortes-Ciriano, I.; Birzu, C.; Geduldig, J.E.; Pelton, K.; et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature 2020, 580, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Villanueva-Meyer, J.; Grimmer, M.R.; Hilz, S.; Solomon, D.A.; Choi, S.; Wahl, M.; Mazor, T.; Hong, C.; Shai, A.; et al. Temozolomide-induced hypermutation is associated with distant recurrence and reduced survival after high-grade transformation of low-grade IDH-mutant gliomas. Neuro-Oncol. 2021, 23, 1872–1884. [Google Scholar] [CrossRef]
- Mittelbronn, M.; Simon, P.; Löffler, C.; Capper, D.; Bunz, B.; Harter, P.; Schlaszus, H.; Schleich, A.; Tabatabai, G.; Goeppert, B.; et al. Elevated HLA-E levels in human glioblastomas but not in grade I to III astrocytomas correlate with infiltrating CD8+ cells. J. Neuroimmunol. 2007, 189, 50–58. [Google Scholar] [CrossRef]
- Kmiecik, J.; Poli, A.; Brons, N.H.C.; Waha, A.; Eide, G.E.; Enger, P.Ø.; Zimmer, J.; Chekenya, M. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. J. Neuroimmunol. 2013, 264, 71–83. [Google Scholar] [CrossRef]
- Bossard, C.; Bézieau, S.; Matysiak-Budnik, T.; Volteau, C.; Laboisse, C.L.; Jotereau, F.; Mosnier, J.F. HLA-E/β2 microglobulin overexpression in colorectal cancer is associated with recruitment of inhibitory immune cells and tumor progression. Int. J. Cancer 2012, 131, 855–863. [Google Scholar] [CrossRef]
- de Kruijf, E.M.; Sajet, A.; van Nes, J.G.; Natanov, R.; Putter, H.; Smit, V.T.; Liefers, G.J.; van den Elsen, P.J.; van de Velde, C.J.; Kuppen, P.J. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J. Immunol. 2010, 185, 7452–7459. [Google Scholar] [CrossRef]
- Gooden, M.; Lampen, M.; Jordanova, E.S.; Leffers, N.; Trimbos, J.B.; van der Burg, S.H.; Nijman, H.; van Hall, T. HLA-E expression by gynecological cancers restrains tumor-infiltrating CD8+ T lymphocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 10656–10661. [Google Scholar] [CrossRef]
- Guo, Z.-Y.; Lv, Y.-G.; Wang, L.; Shi, S.-J.; Yang, F.; Zheng, G.-X.; Wen, W.-H.; Yang, A.-G. Predictive value of HLA-G and HLA-E in the prognosis of colorectal cancer patients. Cell. Immunol. 2015, 293, 10–16. [Google Scholar] [CrossRef]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Söderström, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- van Montfoort, N.; Borst, L.; Korrer, M.J.; Sluijter, M.; Marijt, K.A.; Santegoets, S.J.; van Ham, V.J.; Ehsan, I.; Charoentong, P.; André, P. NKG2A blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell 2018, 175, 1744–1755.e15. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Song, J.; Zhang, H.; Liu, X.; Zuo, F.; Zhao, Y.; Zhao, Y.; Yin, X.; Guo, X.; Wu, X.; et al. Immune checkpoint HLA-E:CD94-NKG2A mediates evasion of circulating tumor cells from NK cell surveillance. Cancer Cell 2023, 41, 272–287.e9. [Google Scholar] [CrossRef] [PubMed]
- André, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Bléry, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743.e13. [Google Scholar] [CrossRef]
- Sætersmoen, M.; Kotchetkov, I.S.; Torralba-Raga, L.; Mansilla-Soto, J.; Sohlberg, E.; Krokeide, S.Z.; Hammer, Q.; Sadelain, M.; Malmberg, K.-J. Targeting HLA-E-overexpressing cancers with a NKG2A/C switch receptor. Med 2024, 6, 100521. [Google Scholar] [CrossRef]
- González-Tablas Pimenta, M.; Otero, Á.; Arandia Guzman, D.A.; Pascual-Argente, D.; Ruíz Martín, L.; Sousa-Casasnovas, P.; García-Martin, A.; Roa Montes de Oca, J.C.; Villaseñor-Ledezma, J.; Torres Carretero, L.; et al. Tumor cell and immune cell profiles in primary human glioblastoma: Impact on patient outcome. Brain Pathol. 2021, 31, 365–380. [Google Scholar] [CrossRef]
- Widodo, S.S.; Hutchinson, R.A.; Fang, Y.; Mangiola, S.; Neeson, P.J.; Darcy, P.K.; Barrow, A.D.; Hovens, C.M.; Dinevska, M.; Stylli, S.S.; et al. Toward precision immunotherapy using multiplex immunohistochemistry and in silico methods to define the tumor immune microenvironment. Cancer Immunol. Immunother. 2021, 70, 1811–1820. [Google Scholar] [CrossRef]
- Zhou, J.; Pei, X.; Yang, Y.; Wang, Z.; Gao, W.; Ye, R.; Zhang, X.; Liu, J.; Liu, Z.; Yang, X.; et al. Orphan nuclear receptor TLX promotes immunosuppression via its transcriptional activation of PD-L1 in glioma. J. Immunother. Cancer 2021, 9, e001937. [Google Scholar] [CrossRef]
- Sivonen, M.; Sirviö, K.A.; Wojciechowski, S.; Kailaanmäki, A.; Kaipainen, S.; Bailey, A.; Villalba, M.; Kekarainen, T. Cytokines impact natural killer cell phenotype and functionality against glioblastoma in vitro. Front. Immunol. 2023, 14, 1227064. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.A.A.K.; Peel, L.; Sedgwick, A.J.; Sun, Y.; Vivian, J.P.; Corbett, A.J.; Dolcetti, R.; Mantamadiotis, T.; Barrow, A.D. Reduced HLA-I Transcript Levels and Increased Abundance of a CD56dim NK Cell Signature Are Associated with Improved Survival in Lower-Grade Gliomas. Cancers 2025, 17, 1570. https://doi.org/10.3390/cancers17091570
Khan MAAK, Peel L, Sedgwick AJ, Sun Y, Vivian JP, Corbett AJ, Dolcetti R, Mantamadiotis T, Barrow AD. Reduced HLA-I Transcript Levels and Increased Abundance of a CD56dim NK Cell Signature Are Associated with Improved Survival in Lower-Grade Gliomas. Cancers. 2025; 17(9):1570. https://doi.org/10.3390/cancers17091570
Chicago/Turabian StyleKhan, Md Abdullah Al Kamran, Lorenza Peel, Alexander J. Sedgwick, Yuhan Sun, Julian P. Vivian, Alexandra J. Corbett, Riccardo Dolcetti, Theo Mantamadiotis, and Alexander D. Barrow. 2025. "Reduced HLA-I Transcript Levels and Increased Abundance of a CD56dim NK Cell Signature Are Associated with Improved Survival in Lower-Grade Gliomas" Cancers 17, no. 9: 1570. https://doi.org/10.3390/cancers17091570
APA StyleKhan, M. A. A. K., Peel, L., Sedgwick, A. J., Sun, Y., Vivian, J. P., Corbett, A. J., Dolcetti, R., Mantamadiotis, T., & Barrow, A. D. (2025). Reduced HLA-I Transcript Levels and Increased Abundance of a CD56dim NK Cell Signature Are Associated with Improved Survival in Lower-Grade Gliomas. Cancers, 17(9), 1570. https://doi.org/10.3390/cancers17091570







