An Overview of Artificial Intelligence in Gynaecological Pathology Diagnostics
Simple Summary
Abstract
1. Introduction
2. Methods
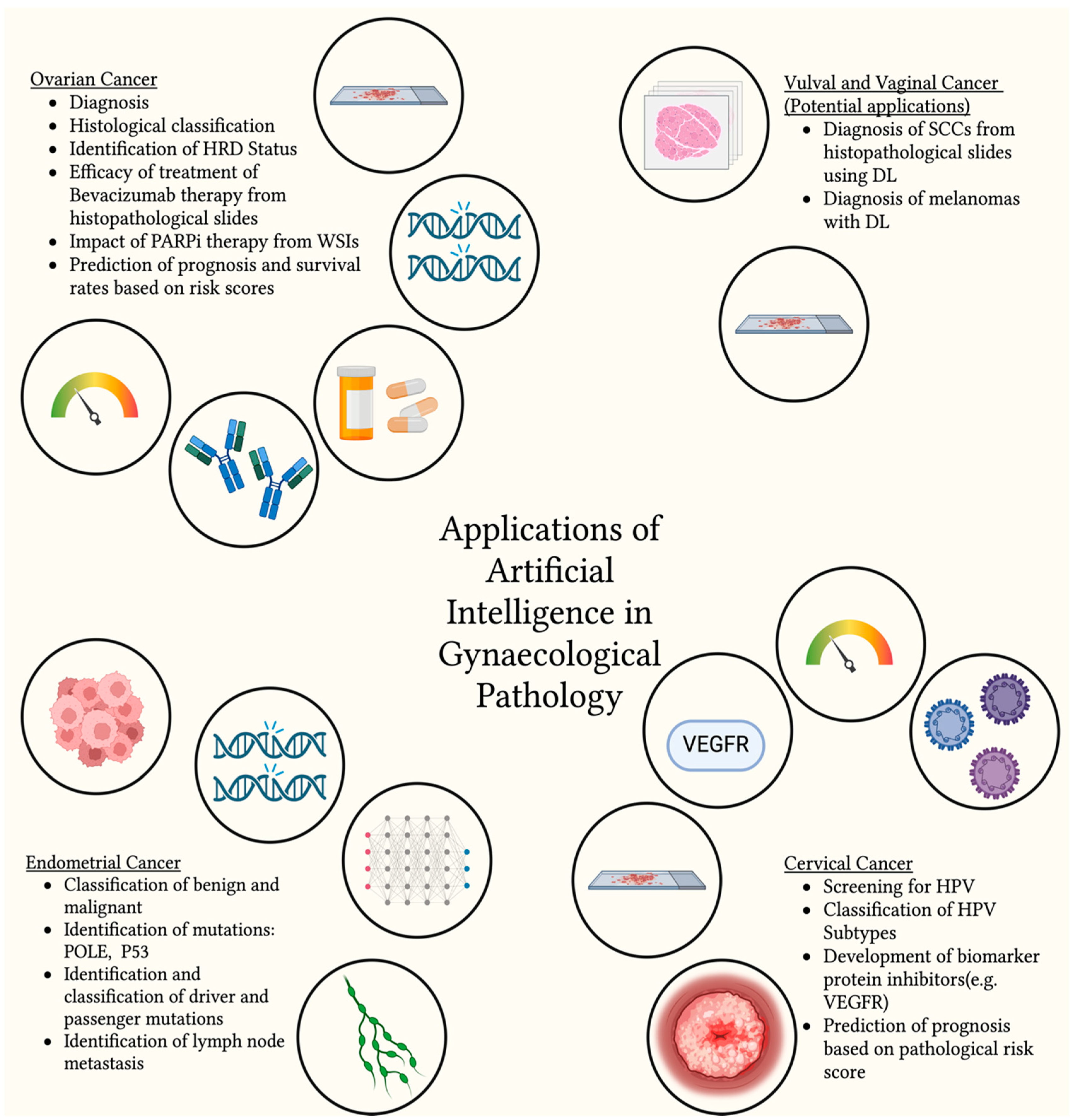
3. Ovarian Cancer
4. Endometrial Cancer
5. Cervical Cancer
6. Vulval and Vaginal Cancers
7. Discussion
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| WSI | Whole Slide Imaging |
| FDA | Food and Drug Administration |
| DL | Deep Learning |
| HRD | Homologous Recombinant Deficiency |
| PARPi | Poly-ADP-ribose polymerase inhibitor |
| CNN | Convolutional Neural Networks |
| EMA | European Medicines Agency |
| OCDPI | Ovarian Cancer Digital Pathology Index |
| POLE | Polymerase ε |
| MMRd | Mismatch Repair Deficient |
| NSMP | No Specific Molecular Type |
| WHO | World Health Organisation |
| CLAM | Clustering-constrained Attention-based Multiple instance learning |
| AUROC | Area Under the Receiver Operating Curve |
| PORTEC | Post Operative Radiation Therapy in Endometrial Cancer |
| TLS | Tertiary Lymphoid Structures |
| HECTOR | Histopathology based Endometrial Cancer Tailored Outcome Risk |
| MIL | Multiple Instance Learning |
| HPV | Human Papilloma Virus |
| CIN | Cervical Intraepithelial Neoplasia |
| CGIN | Cervical Glandular Intraepithelial Neoplasia |
| PAT | Pap Smear Analysis Tool |
| RNN | Recurrent Neural Network |
| PLCO | Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial |
| SCC | Squamous Cell Carcinoma |
References
- Go, H. Digital Pathology and Artificial Intelligence Applications in Pathology. Brain Tumor Res. Treat. 2022, 10, 76. [Google Scholar] [CrossRef]
- Cornish, T.C.; Swapp, R.E.; Kaplan, K.J. Whole-slide imaging: Routine pathologic diagnosis. Adv. Anat. Pathol. 2012, 19, 152–159. [Google Scholar] [CrossRef]
- Kumar, N.; Gupta, R.; Gupta, S. Whole Slide Imaging (WSI) in Pathology: Current Perspectives and Future Directions. J. Digit. Imaging 2020, 33, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Philip, A.K.; Samuel, B.A.; Bhatia, S.; Khalifa, S.A.M.; El-Seedi, H.R. Artificial Intelligence and Precision Medicine: A New Frontier for the Treatment of Brain Tumors. Life 2022, 13, 24. [Google Scholar] [CrossRef]
- Walsh, E.; Orsi, N.M. The current troubled state of the global pathology workforce: A concise review. Diagn. Pathol. 2024, 19, 163. [Google Scholar] [CrossRef] [PubMed]
- Sayed, S.; Lukande, R.; Fleming, K.A. Providing Pathology Support in Low-Income Countries. J. Glob. Oncol. 2015, 1, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.J.; Bottoms, D.; Treanor, D. Future-proofing pathology: The case for clinical adoption of digital pathology. J. Clin. Pathol. 2017, 70, 1010–1018. [Google Scholar] [CrossRef]
- Shanes, J.G.; Ghali, J.; Billingham, M.E.; Ferrans, V.J.; Fenoglio, J.J.; Edwards, W.D.; Tsai, C.C.; Saffitz, J.E.; Isner, J.; Furner, S. Interobserver variability in the pathologic interpretation of endomyocardial biopsy results. Circulation 1987, 75, 401–405. [Google Scholar] [CrossRef]
- Serag, A.; Ion-Margineanu, A.; Qureshi, H.; McMillan, R.; Saint Martin, M.J.; Diamond, J.; O’Reilly, P.; Hamilton, P. Translational AI and Deep Learning in Diagnostic Pathology. Front. Med. 2019, 6, 185. [Google Scholar] [CrossRef]
- Griffin, J.; Treanor, D. Digital pathology in clinical use: Where are we now and what is holding us back? Histopathology 2017, 70, 134–145. [Google Scholar] [CrossRef]
- Sarwar, S.; Dent, A.; Faust, K.; Richer, M.; Djuric, U.; Van Ommeren, R.; Diamandis, P. Physician perspectives on integration of artificial intelligence into diagnostic pathology. NPJ Digit. Med. 2019, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Nakayama, K.I. Artificial intelligence in oncology. Cancer Sci. 2020, 111, 1452–1460. [Google Scholar] [CrossRef] [PubMed]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef]
- Nam, J.G.; Hwang, E.J.; Kim, J.; Park, N.; Lee, E.H.; Kim, H.J.; Nam, M.; Lee, J.H.; Park, C.M.; Goo, J.M. AI Improves Nodule Detection on Chest Radiographs in a Health Screening Population: A Randomized Controlled Trial. Radiology 2023, 307, e221894. [Google Scholar] [CrossRef]
- Chen, D.; Fu, M.; Chi, L.; Lin, L.; Cheng, J.; Xue, W.; Long, C.; Jiang, W.; Dong, X.; Sui, J.; et al. Prognostic and predictive value of a pathomics signature in gastric cancer. Nat. Commun. 2022, 13, 6903. [Google Scholar] [CrossRef]
- da Silva, L.M.; Pereira, E.M.; Salles, P.G.; Godrich, R.; Ceballos, R.; Kunz, J.D.; Casson, A.; Viret, J.; Chandarlapaty, S.; Gil Ferreira, C.; et al. Independent real-world application of a clinical-grade automated prostate cancer detection system. J. Pathol. 2021, 254, 147. [Google Scholar] [CrossRef] [PubMed]
- Akazawa, M.; Hashimoto, K. Artificial Intelligence in Ovarian Cancer Diagnosis. Anticancer Res. 2020, 40, 4795–4800. [Google Scholar] [CrossRef]
- Wang, Y.L.; Gao, S.; Xiao, Q.; Li, C.; Grzegorzek, M.; Zhang, Y.Y.; Li, X.H.; Kang, Y.; Liu, F.H.; Huang, D.H.; et al. Role of artificial intelligence in digital pathology for gynecological cancers. Comput. Struct. Biotechnol. J. 2024, 24, 205–212. [Google Scholar] [CrossRef]
- Rewcastle, E.; Gudlaugsson, E.; Lillesand, M.; Skaland, I.; Baak, J.P.; Janssen, E.A. Automated Prognostic Assessment of Endometrial Hyperplasia for Progression Risk Evaluation Using Artificial Intelligence. Mod. Pathol. 2023, 36, 100116. [Google Scholar] [CrossRef]
- Zhang, X.; Ba, W.; Zhao, X.; Wang, C.; Li, Q.; Zhang, Y.; Lu, S.; Wang, L.; Wang, S.; Song, Z.; et al. Clinical-grade endometrial cancer detection system via whole-slide images using deep learning. Front. Oncol. 2022, 12, 1040238. [Google Scholar] [CrossRef]
- Cancer Today. Available online: https://gco.iarc.fr/today/en (accessed on 7 September 2024).
- Havasi, A.; Cainap, S.S.; Havasi, A.T.; Cainap, C. Ovarian Cancer—Insights into Platinum Resistance and Overcoming It. Medicina 2023, 59, 544. [Google Scholar] [CrossRef]
- McGregor, S.M. Pathologic Classification of Ovarian Cancer. Methods Mol. Biol. 2022, 2424, 11–40. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Primers 2016, 2, 16061. [Google Scholar] [CrossRef]
- Wu, M.; Yan, C.; Liu, H.; Liu, Q. Automatic classification of ovarian cancer types from cytological images using deep convolutional neural networks. Biosci. Rep. 2018, 38, BSR20180289. [Google Scholar] [CrossRef] [PubMed]
- Breen, J.; Allen, K.; Zucker, K.; Adusumilli, P.; Scarsbrook, A.; Hall, G.; Orsi, N.M.; Ravikumar, N. Artificial intelligence in ovarian cancer histopathology: A systematic review. NPJ Precis. Oncol. 2023, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- BenTaieb, A.; Li-Chang, H.; Huntsman, D.; Hamarneh, G. A structured latent model for ovarian carcinoma subtyping from histopathology slides. Med. Image Anal. 2017, 39, 194–205. [Google Scholar] [CrossRef]
- Farahani, H.; Boschman, J.; Farnell, D.; Darbandsari, A.; Zhang, A.; Ahmadvand, P.; Jones, S.J.M.; Huntsman, D.; Köbel, M.; Gilks, C.B.; et al. Deep learning-based histotype diagnosis of ovarian carcinoma whole-slide pathology images. Mod. Pathol. 2022, 35, 1983–1990. [Google Scholar] [CrossRef]
- Huttunen, M.J.; Hassan, A.; McCloskey, C.W.; Fasih, S.; Upham, J.; Vanderhyden, B.C.; Boyd, R.W.; Murugkar, S. Automated classification of multiphoton microscopy images of ovarian tissue using deep learning. J. Biomed. Opt. 2018, 23, 066002. [Google Scholar] [CrossRef]
- Manchana, T.; Phoolcharoen, N.; Tantbirojn, P. BRCA mutation in high grade epithelial ovarian cancers. Gynecol. Oncol. Rep. 2019, 29, 102. [Google Scholar] [CrossRef]
- Miller, R.E.; Elyashiv, O.; El-Shakankery, K.H.; Ledermann, J.A. Ovarian Cancer Therapy: Homologous Recombination Deficiency as a Predictive Biomarker of Response to PARP Inhibitors. OncoTargets Ther. 2022, 15, 1105. [Google Scholar] [CrossRef]
- Bourgade, R.; Rabilloud, N.; Perennec, T.; Pécot, T.; Garrec, C.; Guédon, A.F.; Delnatte, C.; Bézieau, S.; Lespagnol, A.; de Tayrac, M.; et al. Deep Learning for Detecting BRCA Mutations in High-Grade Ovarian Cancer Based on an Innovative Tumor Segmentation Method From Whole Slide Images. Mod. Pathol. 2023, 36, 100304. [Google Scholar] [CrossRef] [PubMed]
- Abkevich, V.; Timms, K.M.; Hennessy, B.T.; Potter, J.; Carey, M.S.; Meyer, L.A.; Smith-McCune, K.; Broaddus, R.; Lu, K.H.; Chen, J.; et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer 2012, 107, 1776–1782. [Google Scholar] [CrossRef]
- Westphalen, C.B.; Fine, A.D.; André, F.; Ganesan, S.; Heinemann, V.; Rouleau, E.; Turnbull, C.; Palacios, L.G.; Lopez, J.-A.; Sokol, E.S.; et al. Pan-cancer Analysis of Homologous Recombination Repair-associated Gene Alterations and Genome-wide Loss-of-Heterozygosity Score. Clin. Cancer Res. 2022, 28, 1412–1421. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.; Leary, A.; Scott, C.; Serra, V.; Lord, C.; Bowtell, D.; Chang, D.; Garsed, D.; Jonkers, J.; Ledermann, J.; et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 2020, 31, 1606–1622. [Google Scholar] [CrossRef] [PubMed]
- Shafi, G.; PM, S.; Ulle, A.; Srinivasan, K.; Vasudevan, A.; Jadhav, V.; Joshi, S.; Raut, N.V.; Khandare, J.; Uttarwar, M.; et al. AI-enabled identification prediction of homologous recombination deficiency (HRD) from histopathology images. J. Clin. Oncol. 2022, 40, 3019. [Google Scholar] [CrossRef]
- Wang, C.W.; Chang, C.C.; Lee, Y.C.; Lin, Y.J.; Lo, S.C.; Hsu, P.C.; Liou, Y.A.; Wang, C.H.; Chao, T.K. Weakly supervised deep learning for prediction of treatment effectiveness on ovarian cancer from histopathology images. Comput. Med. Imaging Graph. 2022, 99, 102093. [Google Scholar] [CrossRef]
- Laury, A.R.; Blom, S.; Ropponen, T.; Virtanen, A.; Carpén, O.M. Artificial intelligence-based image analysis can predict outcome in high-grade serous carcinoma via histology alone. Sci. Rep. 2021, 11, 19165. [Google Scholar] [CrossRef]
- Parpinel, G.; Laudani, M.E.; Piovano, E.; Zola, P.; Lecuru, F. The Use of Artificial Intelligence for Complete Cytoreduction Prediction in Epithelial Ovarian Cancer: A Narrative Review. Cancer Control 2023, 30, 10732748231159553. [Google Scholar] [CrossRef]
- Laury, A.R.; Zheng, S.; Aho, N.; Fallegger, R.; Hänninen, S.; Saez-Rodriguez, J.; Tanevski, J.; Youssef, O.; Tang, J.; Carpén, O.M. Opening the Black Box: Spatial Transcriptomics and the Relevance of Artificial Intelligence-Detected Prognostic Regions in High-Grade Serous Carcinoma. Mod. Pathol. 2024, 37, 100508. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Zhuo, L.; Sun, K.; Meng, F.; Zhou, M.; Sun, J. Prediction of prognosis and treatment response in ovarian cancer patients from histopathology images using graph deep learning: A multicenter retrospective study. Eur. J. Cancer 2024, 199, 113532. [Google Scholar] [CrossRef]
- Wu, M.; Zhu, C.; Yang, J.; Cheng, S.; Yang, X.; Gu, S.; Xu, S.; Wu, Y.; Shen, W.; Huang, S.; et al. Exploring prognostic indicators in the pathological images of ovarian cancer based on a deep survival network. Front. Genet. 2023, 13, 1069673. [Google Scholar] [CrossRef] [PubMed]
- Cancer Today. Available online: https://gco.iarc.fr/today/en/dataviz/pie?mode=cancer&group_populations=1&sexes=2 (accessed on 1 June 2024).
- Sankaranarayanan, R.; Ferlay, J. Worldwide Burden of Gynecological Cancer. In Handbook of Disease Burdens and Quality of Life Measures; Springer: New York, NY, USA, 2010; pp. 803–823. [Google Scholar] [CrossRef]
- Bell, D.W.; Ara, O.; O’Hara, A.J. The genomics and genetics of endometrial cancer. Adv. Genom. Genet. 2012, 2, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.; Le Gallo, M.; Bell, D.W. The Mutational Landscape of Endometrial Cancer. Curr. Opin. Genet. Dev. 2015, 30, 25. [Google Scholar] [CrossRef] [PubMed]
- Emons, G.; Beckmann, M.W.; Schmidt, D.; Mallmann, P. New WHO Classification of Endometrial Hyperplasias. Geburtshilfe Frauenheilkd 2015, 75, 135. [Google Scholar] [CrossRef]
- Fell, C.; Mohammadi, M.; Morrison, D.; Arandjelović, O.; Syed, S.; Konanahalli, P.; Bell, S.; Bryson, G.; Harrison, D.J.; Harris-Birtill, D. Detection of malignancy in whole slide images of endometrial cancer biopsies using artificial intelligence. PLoS ONE 2023, 18, e0282577. [Google Scholar] [CrossRef]
- Zhao, F.; Dong, D.; Du, H.; Guo, Y.; Su, X.; Wang, Z.; Xie, X.; Wang, M.; Zhang, H.; Cao, X.; et al. Diagnosis of endometrium hyperplasia and screening of endometrial intraepithelial neoplasia in histopathological images using a global-to-local multi-scale convolutional neural network. Comput. Methods Programs Biomed. 2022, 221, 106906. [Google Scholar] [CrossRef]
- Sun, H.; Zeng, X.; Xu, T.; Peng, G.; Ma, Y. Computer-Aided Diagnosis in Histopathological Images of the Endometrium Using a Convolutional Neural Network and Attention Mechanisms. IEEE J. Biomed. Health Inform. 2020, 24, 1664–1676. [Google Scholar] [CrossRef]
- Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 2021, 5, 555. [Google Scholar] [CrossRef]
- Mohammadi, M.; Cooper, J.; Arandelović, O.; Fell, C.; Morrison, D.; Syed, S.; Konanahalli, P.; Bell, S.; Bryson, G.; Harrison, D.J.; et al. Weakly supervised learning and interpretability for endometrial whole slide image diagnosis. Exp. Biol. Med. 2022, 247, 2025. [Google Scholar] [CrossRef]
- Goyal, M.; Tafe, L.J.; Feng, J.X.; Muller, K.E.; Hondelink, L.; Bentz, J.L.; Hassanpour, S. Deep Learning for Grading Endometrial Cancer. Am. J. Pathol. 2024, 194, 1701–1711. [Google Scholar] [CrossRef]
- Fremond, S.; Andani, S.; Wolf, J.B.; Dijkstra, J.; Melsbach, S.; Jobsen, J.J.; Brinkhuis, M.; Roothaan, S.; Jurgenliemk-Schulz, I.; Lutgens, L.C.H.W.; et al. Interpretable deep learning model to predict the molecular classification of endometrial cancer from haematoxylin and eosin-stained whole-slide images: A combined analysis of the PORTEC randomised trials and clinical cohorts. Lancet Digit. Health 2023, 5, e71–e82. [Google Scholar] [CrossRef] [PubMed]
- Stan, A.; Bosart, K.; Kaur, M.; Vo, M.; Escorcia, W.; Yoder, R.J.; Bouley, R.A.; Petreaca, R.C. Detection of driver mutations and genomic signatures in endometrial cancers using artificial intelligence algorithms. PLoS ONE 2024, 19, e0299114. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, S.F.; Bao, W. Molecular subtypes of endometrial cancer: Implications for adjuvant treatment strategies. Int. J. Gynecol. Obstet. 2024, 164, 436–459. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Liu, W.; DeLair, D.; Razavian, N.; Fenyö, D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2021, 2, 100400. [Google Scholar] [CrossRef]
- Fridman, W.H.; Meylan, M.; Pupier, G.; Calvez, A.; Hernandez, I.; Sautès-Fridman, C. Tertiary lymphoid structures and B cells: An intratumoral immunity cycle. Immunity 2023, 56, 2254–2269. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.; Hamanishi, J.; Ukita, M.; Yamanoi, K.; Takamatsu, S.; Abiko, K.; Murakami, R.; Miyamoto, T.; Suzuki, H.; Ueda, A.; et al. Tertiary lymphoid structures are associated with favorable survival outcomes in patients with endometrial cancer. Cancer Immunol. Immunother. 2022, 71, 1431–1442. [Google Scholar] [CrossRef]
- Suzuki, H.; Hamada, K.; Hamanishi, J.; Ueda, A.; Murakami, R.; Taki, M.; Mizuno, R.; Watanabe, K.; Sato, H.; Hosoe, Y.; et al. Artificial intelligence-based spatial analysis of tertiary lymphoid structures and clinical significance for endometrial cancer. Cancer Immunol. Immunother. 2025, 74, 84. [Google Scholar] [CrossRef]
- Van Den Heerik, A.S.V.M.; Horeweg, N.; De Boer, S.M.; Bosse, T.; Creutzberg, C.L. Adjuvant therapy for endometrial cancer in the era of molecular classification: Radiotherapy, chemoradiation and novel targets for therapy. Int. J. Gynecol. Cancer 2021, 31, 594. [Google Scholar] [CrossRef]
- Bogani, G.; Monk, B.; Powell, M.; Westin, S.; Slomovitz, B.; Moore, K.; Eskander, R.N.; Raspagliesi, F.; Barretina-Ginesta, M.-P.; Colombo, N.; et al. Adding immunotherapy to first-line treatment of advanced and metastatic endometrial cancer. Ann. Oncol. 2024, 35, 414–428. [Google Scholar] [CrossRef]
- Karpel, H.C.; Slomovitz, B.; Coleman, R.L.; Pothuri, B. Treatment options for molecular subtypes of endometrial cancer in 2023. Curr. Opin. Obstet. Gynecol. 2023, 35, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Zhao, Y.; Chen, J.; Zhao, T.; Mei, J.; Fan, Y.; Lin, Z.; Yao, J.; Bu, H. A deep learning model for lymph node metastasis prediction based on digital histopathological images of primary endometrial cancer. Quant. Imaging Med. Surg. 2023, 13, 1899–1913. [Google Scholar] [CrossRef]
- Volinsky-Fremond, S.; Horeweg, N.; Andani, S.; Wolf, J.B.; Lafarge, M.W.; de Kroon, C.D.; Ørtoft, G.; Høgdall, E.; Dijkstra, J.; Jobsen, J.J.; et al. Prediction of recurrence risk in endometrial cancer with multimodal deep learning. Nat. Med. 2024, 30, 1962–1973. [Google Scholar] [CrossRef] [PubMed]
- Cervical Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cervical-cancer (accessed on 5 July 2024).
- Fontham, E.T.H.; Wolf, A.M.D.; Church, T.R.; Etzioni, R.; Flowers, C.R.; Herzig, A.; Guerra, C.E.; Oeffinger, K.C.; Shih, Y.T.; Walter, L.C.; et al. Cervical cancer screening for individuals at average risk: 2020 guideline update from the American Cancer Society. CA Cancer J. Clin. 2020, 70, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Cancer Research UK. Cervical Cancer Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/cervical-cancer#heading-Zero (accessed on 5 July 2024).
- Sawaya, G.F.; Smith-Mccune, K.; Kuppermann, M. Cervical Cancer Screening: More Choices in 2019. JAMA 2019, 321, 2018–2019. [Google Scholar] [CrossRef]
- Casas, C.P.R.; Albuquerque, R.d.C.R.d.; Loureiro, R.B.; Gollner, A.M.; de Freitas, M.G.; Duque, G.P.D.N.; Viscondi, J.Y.K. Cervical cancer screening in low- and middle-income countries: A systematic review of economic evaluation studies. Clinics 2022, 77, 100080. [Google Scholar] [CrossRef]
- Arbyn, M.; Ronco, G.; Anttila, A.; Meijer, C.J.; Poljak, M.; Ogilvie, G.; Koliopoulos, G.; Naucler, P.; Sankaranarayanan, R.; Peto, J. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine 2012, 30 (Suppl. S5), F88–F99. [Google Scholar] [CrossRef]
- Nahvijou, A.; Daroudi, R.; Tahmasebi, M.; Hashemi, F.A.; Hemami, M.R.; Sari, A.A.; Marenani, A.B.; Zendehdel, K. Cost-Effectiveness of Different Cervical Screening Strategies in Islamic Republic of Iran: A Middle-Income Country with a Low Incidence Rate of Cervical Cancer. PLoS ONE 2016, 11, 156705. [Google Scholar] [CrossRef]
- Li, Y.; Ma, L.; Yang, C.; Chen, Z.; Zhao, Y.; Dang, L.; Lang, J.; Qiao, Y. A study on service capacity of primary medical and health institutions for cervical cancer screening in urban and rural areas in China. Chin. J. Cancer Res. 2019, 31, 838. [Google Scholar] [CrossRef]
- Pantanowitz, L. Improving the Pap test with artificial intelligence. Cancer Cytopathol. 2022, 130, 402–404. [Google Scholar] [CrossRef]
- Holmström, O.; Linder, N.; Kaingu, H.; Mbuuko, N.; Mbete, J.; Kinyua, F.; Törnquist, S.; Muinde, M.; Krogerus, L.; Lundin, M.; et al. Point-of-Care Digital Cytology With Artificial Intelligence for Cervical Cancer Screening in a Resource-Limited Setting. JAMA Netw. Open 2021, 4, e211740. [Google Scholar] [CrossRef]
- Ramirez, C.A.M.; Greenop, M.; Almoshawah, Y.A.; Hirsch, P.L.M.; Rehman, I.U. Advancing cervical cancer diagnosis and screening with spectroscopy and machine learning. Expert Rev. Mol. Diagn. 2023, 23, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Wong, O.G.W.; Ng, I.F.Y.; Tsun, O.K.L.; Pang, H.H.; Ip, P.P.C.; Cheung, A.N.Y. Machine Learning Interpretation of Extended Human Papillomavirus Genotyping by Onclarity in an Asian Cervical Cancer Screening Population. J. Clin. Microbiol. 2019, 57, 997–1016. [Google Scholar] [CrossRef]
- Zhu, X.; Yao, Q.; Dai, W.; Ji, L.; Yao, Y.; Pang, B.; Turic, B.; Yao, L.; Liu, Z. Cervical cancer screening aided by artificial intelligence, China. Bull. World Health Organ. 2023, 101, 381. [Google Scholar] [CrossRef]
- Bao, H.; Sun, X.; Zhang, Y.; Pang, B.; Li, H.; Zhou, L.; Wu, F.; Cao, D.; Wang, J.; Turic, B.; et al. The artificial intelligence-assisted cytology diagnostic system in large-scale cervical cancer screening: A population-based cohort study of 0.7 million women. Cancer Med. 2020, 9, 6896–6906. [Google Scholar] [CrossRef] [PubMed]
- Xue, P.; Ng, M.T.A.; Qiao, Y. The challenges of colposcopy for cervical cancer screening in LMICs and solutions by artificial intelligence. BMC Med. 2020, 18, 169. [Google Scholar] [CrossRef]
- Nakisige, C.; de Fouw, M.; Kabukye, J.; Sultanov, M.; Nazrui, N.; Rahman, A.; de Zeeuw, J.; Koot, J.; Rao, A.P.; Prasad, K.; et al. Artificial intelligence and visual inspection in cervical cancer screening. Int. J. Gynecol. Cancer 2023, 33, 1515. [Google Scholar] [CrossRef]
- Cardoza-Favarato, L.L.G.; Papillomavirus, H. Encyclopedia of Child and Adolescent Health, 1st ed.; Academic Press: Cambridge, MA, USA, 2023; Volume 1, pp. 953–964. [Google Scholar] [CrossRef]
- Pathania, D.; Landeros, C.; Rohrer, L.; D’Agostino, V.; Hong, S.; Degani, I.; Avila-Wallace, M.; Pivovarov, M.; Randall, T.; Weissleder, R.; et al. Point-of-care cervical cancer screening using deep learning-based microholography. Theranostics 2019, 9, 8438. [Google Scholar] [CrossRef]
- Tian, R.; Cui, Z.; He, D.; Tian, X.; Gao, Q.; Ma, X.; Yang, J.-R.; Wu, J.; Das, B.C.; Severinov, K.; et al. Risk stratification of cervical lesions using capture sequencing and machine learning method based on HPV and human integrated genomic profiles. Carcinogenesis 2019, 40, 1220–1228. [Google Scholar] [CrossRef]
- Thrall, M.J. Automated screening of Papanicolaou tests: A review of the literature. Diagn. Cytopathol. 2019, 47, 20–27. [Google Scholar] [CrossRef]
- Wang, C.W.; Liou, Y.A.; Lin, Y.J.; Chang, C.C.; Chu, P.H.; Lee, Y.C.; Wang, C.H.; Chao, T.K. Artificial intelligence-assisted fast screening cervical high grade squamous intraepithelial lesion and squamous cell carcinoma diagnosis and treatment planning. Sci. Rep. 2021, 11, 16244. [Google Scholar] [CrossRef]
- William, W.; Ware, A.; Basaza-Ejiri, A.H.; Obungoloch, J. A pap-smear analysis tool (PAT) for detection of cervical cancer from pap-smear images. Biomed. Eng. Online 2019, 18, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Chen, F.; Shi, J.J.; Huang, Y.L.; Wang, M. Convolutional Neural Networks for Classifying Cervical Cancer Types Using Histological Images. J. Digit. Imaging 2023, 36, 441. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Liu, S.; Yu, J.; Rao, G.; Xiao, Y.; Han, W.; Zhu, W.; Lv, X.; Li, N.; Cai, J.; et al. Robust whole slide image analysis for cervical cancer screening using deep learning. Nat. Commun. 2021, 12, 5639. [Google Scholar] [CrossRef]
- Chu, R.; Zhang, Y.; Qiao, X.; Xie, L.; Chen, W.; Zhao, Y.; Xu, Y.; Yuan, Z.; Liu, X.; Yin, A.; et al. Risk Stratification of Early-Stage Cervical Cancer with Intermediate-Risk Factors: Model Development and Validation Based on Machine Learning Algorithm. Oncologist 2021, 26, e2217. [Google Scholar] [CrossRef] [PubMed]
- Obrzut, B.; Kusy, M.; Semczuk, A.; Obrzut, M.; Kluska, J. Prediction of 5–year overall survival in cervical cancer patients treated with radical hysterectomy using computational intelligence methods. BMC Cancer 2017, 17, 840. [Google Scholar] [CrossRef]
- Chen, C.; Cao, Y.; Li, W.; Liu, Z.; Liu, P.; Tian, X.; Sun, C.; Wang, W.; Gao, H.; Kang, S.; et al. The pathological risk score: A new deep learning-based signature for predicting survival in cervical cancer. Cancer Med. 2023, 12, 1051. [Google Scholar] [CrossRef]
- Cancer Today. Available online: https://gco.iarc.who.int/today/en/fact-sheets-cancers (accessed on 1 November 2024).
- Mascarenhas, M.; Alencoão, I.; Carinhas, M.J.; Martins, M.; Ribeiro, T.; Mendes, F.; Cardoso, P.; Almeida, M.J.; Mota, J.; Fernandes, J.; et al. Artificial Intelligence and Colposcopy: Automatic Identification of Vaginal Squamous Cell Carcinoma Precursors. Cancers 2024, 16, 3540. [Google Scholar] [CrossRef]
- Yang, S.; Li, S.; Liu, J.; Sun, X.; Cen, Y.; Ren, R.; Ying, S.; Chen, Y.; Zhao, Z.; Liao, W. Histopathology-Based Diagnosis of Oral Squamous Cell Carcinoma Using Deep Learning. J. Dent. Res. 2022, 101, 1321–1327. [Google Scholar] [CrossRef]
- Wang, L.; Ding, L.; Liu, Z.; Sun, L.; Chen, L.; Jia, R.; Dai, X.; Cao, J.; Ye, J. Automated identification of malignancy in whole-slide pathological images: Identification of eyelid malignant melanoma in gigapixel pathological slides using deep learning. Br. J. Ophthalmol. 2020, 104, 318–323. [Google Scholar] [CrossRef]
- Duenweg, S.R.; Bobholz, S.A.; Lowman, A.K.; Stebbins, M.A.; Winiarz, A.; Nath, B.; Kyereme, F.; Iczkowski, K.A.; LaViolette, P.S. Whole slide imaging (WSI) scanner differences influence optical and computed properties of digitized prostate cancer histology. J. Pathol. Inform. 2023, 14, 100321. [Google Scholar] [CrossRef]
- Prezja, F.; Pölönen, I.; Äyrämö, S.; Ruusuvuori, P.; Kuopio, T. H&E Multi-Laboratory Staining Variance Exploration with Machine Learning. Appl. Sci. 2022, 12, 7511. [Google Scholar] [CrossRef]
- Sah, S.; Chen, L.; Houghton, J.; Kemppainen, J.; Marko, A.C.; Zeigler, R.; Latham, G.J. Functional DNA quantification guides accurate next-generation sequencing mutation detection in formalin-fixed, paraffin-embedded tumor biopsies. Genome. Med. 2013, 5, 77. [Google Scholar] [CrossRef]
- Sharmanska, V.; Quadrianto, N.; Lampert, C.H. Learning to rank using privileged information. In Proceedings of the IEEE International Conference on Computer Vision, Sydney, Australia, 1–8 December 2013; pp. 825–832. [Google Scholar] [CrossRef]
- Celi, L.A.; Cellini, J.; Charpignon, M.-L.; Dee, E.C.; Dernoncourt, F.; Eber, R.; Mitchell, W.G.; Moukheiber, L.; Schirmer, J.; Situ, J.; et al. Sources of bias in artificial intelligence that perpetuate healthcare disparities—A global review. PLoS Digit. Health 2022, 1, e0000022. [Google Scholar] [CrossRef] [PubMed]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Ye, J.; Zhou, Q.; Long, L.R.; Antani, S.; Xue, Z.; Cornwell, C.; Zaino, R.; Cheng, K.C.; Huang, X. Selective Synthetic Augmentation with HistoGAN for Improved Histopathology Image Classification. Med. Image. Anal. 2021, 67, 101816. [Google Scholar] [CrossRef]
- Choi, Y.; Yu, W.; Nagarajan, M.B.; Teng, P.; Goldin, J.G.; Raman, S.S.; Enzmann, D.R.; Kim, G.H.J.; Brown, M.S. Translating AI to Clinical Practice: Overcoming Data Shift with Explainability. Radiographics 2023, 43, e220105. [Google Scholar] [CrossRef]
- Border, S.P.; Sarder, P. From What to Why, the Growing Need for a Focus Shift Toward Explainability of AI in Digital Pathology. Front. Physiol. 2021, 12, 821217. [Google Scholar] [CrossRef]
- King, H.; Williams, B.; Treanor, D.; Randell, R. How for whom and in what contexts will artificial intelligence be adopted in pathology? A realist interview study. J. Am. Med. Inform. Assoc. 2023, 30, 529–538. [Google Scholar] [CrossRef]
- Hanna, M.G.; Ardon, O.; Reuter, V.E.; Sirintrapun, S.J.; England, C.; Klimstra, D.S.; Hameed, M.R. Integrating digital pathology into clinical practice. Mod. Pathol. 2022, 35, 152–164. [Google Scholar] [CrossRef]
| Paper Authors | Morphological Subtyping | Molecular Subtyping | Prognostication | Data Source | Training/Validation Test Set Size | External Validation |
|---|---|---|---|---|---|---|
| Ovarian Cancer | ||||||
| Wu et al. [25] | No | No | No | First Affiliated Hospital of Xinjiang Medical University | Original-Training: 5914; Validation: 1478 Augmented-Training: 65,050; Validation: 16,262 | No |
| BenTaieb et al. [27] | Yes | No | No | Unclear | Training set size: 73 | No |
| Farahani et al. [28] | Yes | No | No | OVCARE Archives, University of Calgary | Training set size: 948 Validation test set size: 60 | No |
| Bourgade et al. [32] | No | Yes—BRCA | No | University Hospitals of Nantes and Rennes, TCGA | Training set size: 1,040,149 tumour tiles Validation test set size: 111,727 tumour tiles | Yes |
| Shafi et al. [36] | No | Yes—HRD | No | Unclear | Training set size: 150 | No |
| Wang et al. [37] | No | No | Yes | Tri-Service General Hospital and the National Defense Medical Center, Taipei, Taiwan | Training data set size: 187; Testing data set size: 101 | No |
| Laury et al. [38] | Yes | No | No | HUS Helsinki University Hospital | Training set size: 205 Test set size: 22 | No |
| Laury et al. [40] | Yes | Yes—JUN | Yes | Helsinki Biobank | Training set: 205; Validation set: 22 | No |
| Yang et al. [41] | Yes | No | Yes | TCGA-OV, Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) and Harbin Medical University Cancer Hospital | 2449 slides | Yes |
| Wu et al. [42] | No | Yes—HRD, BRCA | Yes | TCGA-OV | Training data set: 72 Test data set: 18 | No |
| Endometrial Cancer | ||||||
| Fell et al. [48] | No | No | No | NHS Greater Glasgow and Clyde Biorepository and Pathology Tissue Resource | Training data set: 1248 Validation data set: 616 Test data set: 863 | No |
| Zhao et al. [49] | Yes | No | No | Unclear | Training data set: 6248; Validation data set: 1564; External validation data set: 1631 | Yes |
| Sun et al. [50] | Yes | No | No | Third Affiliated Hospital of Zhengzhou University | Data set size: 3302; External validation data set: 200 | Yes |
| Mohammadi et al. [52] | Yes | No | No | iCAIRD | Training data set: 998 Validation data set: 466 Test data set: 864 | No |
| Goyal et al. [53] | Yes | No | No | Dartmouth Health, TCGA | Training data set: 929; Validation data set: 100 | Yes |
| Fremond et al. [54] | No | Yes—POLE, p53abn, MMRd, NSMP | Yes | PORTEC-1, PORTEC-2, PORTEC-3, TCGA, TransPORTEC pilot study, Medisch Spectrum Twente cohort | Training set data size: 1240; Test set data size: 393 | No |
| Hong et al. [58] | Yes | Yes (multiple) | Yes | TCGA, Clinical Proteomic Tumor Analysis Consortium, NYU Hospitals | Data set size: 496 | Yes |
| Suzuki et al. [61] | No | No | Yes | Kyoto Cohort, ICI Cohort, TCGA | Data set size: 966 | No |
| Feng et al. [65] | Yes | No | Yes | West China Second University Hospital, Qingdao University, Affiliated Yantai Yu Huang Ding Hospital, Beijing Maternal and Child Health Care Hospital | Internal data set size: 2104 External data set size: 533 | Yes |
| Volinsky-Fremond et al. [66] | Yes | Yes | Yes | PORTEC 1,2,3, University Medical Center Groningen, Leiden University Medical Center | Test data set: 353; Training data set: 1408; External validation data set: 310 | Yes |
| Cervical Cancer | ||||||
| Holmström et al. [76] | Yes | No | No | Kinondo Kwetu Health Services Clinic, Kinondo, Kwale County | Training data set: 360; Validation data set: 361 | No |
| Wong et al. [78] | Yes | No | No | Cervical Cytology Laboratory, Department of Pathology, The University of Hong Kong | Training data set: 485; Validation data set: 120 | No |
| Bao et al. [80] | Yes | No | No | Hubei, China | Training data set: 103,793 | No |
| Nakisige et al. [82] | Yes | No | No | Uganda Cancer Institute, International Agency for Research on Cancer, Leiden University Medical Center | Training data set: 70; Test data set: 20; Validation data set: 10 | No |
| Pathania et al. [84] | Yes | No | No | Unclear | Training data set: 13,000 | Yes |
| Tian et al. [85] | Yes | Yes | No | The First Affiliated Hospital of Sun Yat-sen University | 30 Samples | No |
| Wang et al. [87] | Yes | No | No | Department of Pathology, Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan | Training data set: 97; Test data set: 46 | No |
| William et al. [88] | Yes | No | No | Mbarara Regional Referral Hospital | Dataset 1: 917; Dataset 2: 497; Dataset 3: 60 | No |
| Cheng et al. [90] | Yes | No | No | Multiple hospitals | Training set size: 46,810; Test set size: 6617; Validation set size: 10,229 | No |
| Chu et al. [91] | Yes | No | Yes | Qilu Hospital of Shandong University | Training data set: 385; Validation data set: 96 | No |
| Obrzut et al. [92] | No | No | Yes | Department of Obstetrics and Gynaecology of the Rzeszow State Hospital in Poland | Unclear | No |
| Chen et al. [93] | No | No | Yes | Multiple hospitals | Training data set: 836; Validation data set: 354 | No |
| Mascarenhas et al. [95] | Yes | No | No | Tertiary Care Centre (Centro Materno Infantil do Norte) | Training/Validation data sets: 51,525; Test data set size: 5725 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joshua, A.; Allen, K.E.; Orsi, N.M. An Overview of Artificial Intelligence in Gynaecological Pathology Diagnostics. Cancers 2025, 17, 1343. https://doi.org/10.3390/cancers17081343
Joshua A, Allen KE, Orsi NM. An Overview of Artificial Intelligence in Gynaecological Pathology Diagnostics. Cancers. 2025; 17(8):1343. https://doi.org/10.3390/cancers17081343
Chicago/Turabian StyleJoshua, Anna, Katie E. Allen, and Nicolas M. Orsi. 2025. "An Overview of Artificial Intelligence in Gynaecological Pathology Diagnostics" Cancers 17, no. 8: 1343. https://doi.org/10.3390/cancers17081343
APA StyleJoshua, A., Allen, K. E., & Orsi, N. M. (2025). An Overview of Artificial Intelligence in Gynaecological Pathology Diagnostics. Cancers, 17(8), 1343. https://doi.org/10.3390/cancers17081343







