Exploring microRNAs in Bile Duct Stents as Diagnostic Biomarkers for Biliary Pathologies
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Samples Collection
2.3. RNA Isolation and Quantitative Real-Time PCR
2.4. Statistical Analysis
3. Results
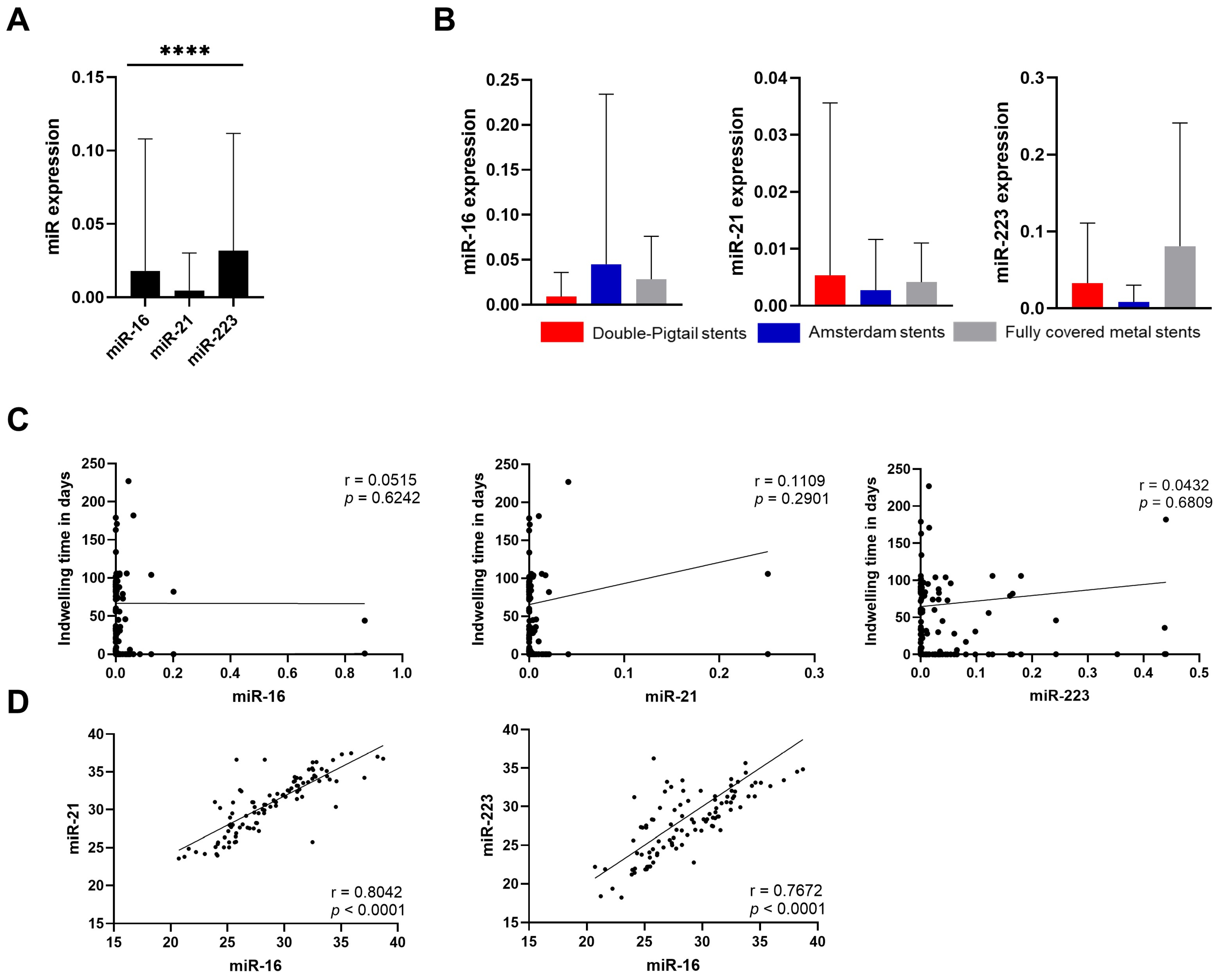
3.1. Preanalytical Evaluation
3.2. miRNA in Malignant Conditions
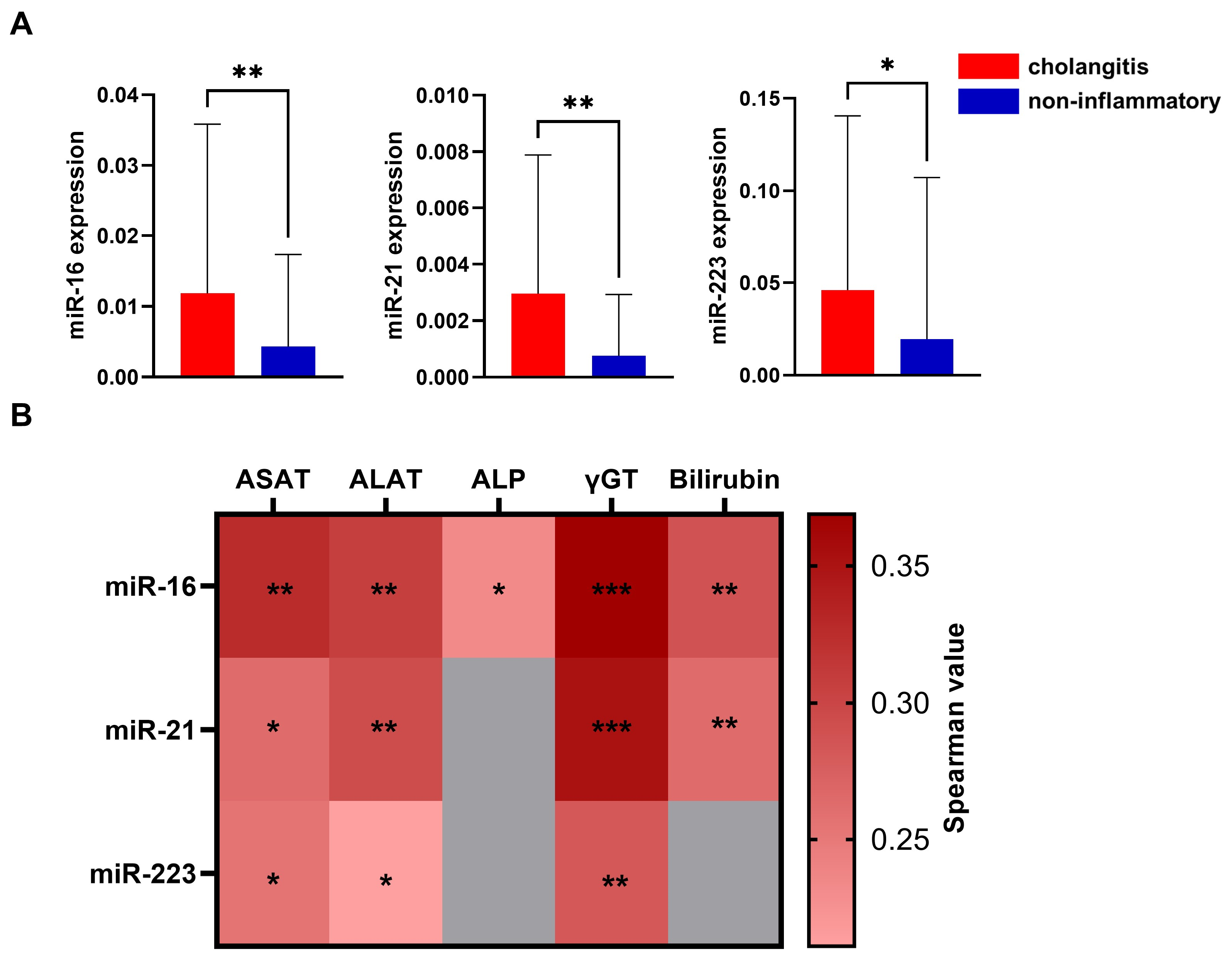
3.3. miRNA in Inflammation
3.4. Association of miRNA Levels with Laboratory Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALAT | Alanine aminotransferase |
| ALP | Alkaline phosphatase |
| ASAT | Aspartate aminotranferase |
| CA19-9 | Carbohydrate-Antigen 19-9 |
| CCA | Cholangiocarcinoma |
| CRP | C-reactive Protein |
| DNA | Deoxyribonucleic acid |
| ERC | Endoscopic retrograde cholangiography |
| FGFR | fibroblast growth factor receptors |
| γGT | Gamma-glutamyl transferase |
| IDH | Isocitrate dehydrogenase |
| KRAS | Kirsten rat sarcoma virus |
| miRNA | microRNA |
| PCR | Polymerase chain reaction |
| PMN | Polymorphonuclear leukocytes |
| PSC | Primary sclerosing cholangitis |
| RNA | Ribonucleic acid |
References
- An, Z.; Braseth, A.L.; Sahar, N. Acute Cholangitis: Causes, Diagnosis, and Management. Gastroenterol. Clin. N. Am. 2021, 50, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Zweigle, J.; Patel, P.; Tedder, B.; Khan, R.; Agrawal, S. Survival Analysis of Extrahepatic Cholangiocarcinoma Based on Surveillance, Epidemiology, and End Results Database. Ann. Hepatobiliary Pancreat. Surg. 2023, 27, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.; Gores, G.J. Pathogenesis, Diagnosis, and Management of Cholangiocarcinoma. Gastroenterology 2013, 145, 1215–1229. [Google Scholar] [CrossRef]
- Liau, J.Y.; Tsai, J.H.; Yuan, R.H.; Chang, C.N.; Lee, H.J.; Jeng, Y.M. Morphological Subclassification of Intrahepatic Cholangiocarcinoma: Etiological, Clinicopathological, and Molecular Features. Mod. Pathol. 2014, 27, 1163–1173. [Google Scholar] [CrossRef]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next Horizon in Mechanisms and Management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [PubMed]
- Liang, B.; Zhong, L.; He, Q.; Wang, S.; Pan, Z.; Wang, T.; Zhao, Y. Diagnostic Accuracy of Serum CA19-9 in Patients with Cholangiocarcinoma: A Systematic Review and Meta-Analysis. Med. Sci. Monit. 2015, 21, 3555–3563. [Google Scholar] [CrossRef]
- Lertkhachonsuk, A.A.; Buranawongtrakoon, S.; Lekskul, N.; Rermluk, N.; Wee-Stekly, W.W.; Charakorn, C. Serum CA19-9, CA-125 and CEA as Tumor Markers for Mucinous Ovarian Tumors. J. Obstet. Gynaecol. Res. 2020, 46, 2287–2291. [Google Scholar] [CrossRef]
- Furuya, N.; Kawa, S.; Tokoo, M.; Mukawa, K.; Maejima, S.; Oguchi, H. Comparative Study of CA242 and CA19-9 in Chronic Pancreatitis. Br. J. Cancer 1996, 73, 372–376. [Google Scholar] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a Big Role in Gene Regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in Cancer: Biomarkers, Functions and Therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. MiRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Link, J.; Thon, C.; Petkevicius, V.; Steponaitiene, R.; Malfertheiner, P.; Kupcinskas, J.; Link, A. The Translational Impact of Plant-Derived Xeno-MiRNA MiR-168 in Gastrointestinal Cancers and Preneoplastic Conditions. Diagnostics 2023, 13, 2701. [Google Scholar] [CrossRef] [PubMed]
- Link, J.; Thon, C.; Schanze, D.; Steponaitiene, R.; Kupcinskas, J.; Zenker, M.; Canbay, A.; Malfertheiner, P.; Link, A. Food-Derived Xeno-MicroRNAs: Influence of Diet and Detectability in Gastrointestinal Tract-Proof-of-Principle Study. Mol. Nutr. Food Res. 2019, 63, e1800076. [Google Scholar] [CrossRef] [PubMed]
- Cote, G.A.; Gore, A.J.; McElyea, S.D.; Heathers, L.E.; Xu, H.; Sherman, S.; Korc, M. A Pilot Study to Develop a Diagnostic Test for Pancreatic Ductal Adenocarcinoma Based on Differential Expression of Select MiRNA in Plasma and Bile. Am. J. Gastroenterol. 2014, 109, 1942–1952. [Google Scholar] [CrossRef]
- Selaru, F.M.; Olaru, A.V.; Kan, T.; David, S.; Cheng, Y.; Mori, Y.; Yang, J.; Paun, B.; Jin, Z.; Agarwal, R.; et al. MicroRNA-21 Is Overexpressed in Human Cholangiocarcinoma and Regulates Programmed Cell Death 4 and Tissue Inhibitor of Metalloproteinase 3. Hepatology 2009, 49, 1595–1601. [Google Scholar] [CrossRef]
- Jiao, P.; Wang, X.-P.; Luoreng, Z.-M.; Yang, J.; Jia, L.; Ma, Y.; Wei, D.-W. MiR-223: An Effective Regulator of Immune Cell Differentiation and Inflammation. Int. J. Biol. Sci. 2021, 17, 2308–2322. [Google Scholar] [CrossRef]
- Haneklaus, M.; Gerlic, M.; O’Neill, L.A.J.; Masters, S.L. MiR-223: Infection, Inflammation and Cancer. J. Intern. Med. 2013, 274, 215–226. [Google Scholar] [CrossRef]
- Schönauen, K.; Le, N.; von Arnim, U.; Schulz, C.; Malfertheiner, P.; Link, A. Circulating and Fecal MicroRNAs as Biomarkers for Inflammatory Bowel Diseases. Inflamm. Bowel. Dis. 2018, 24, 1547–1557. [Google Scholar] [CrossRef]
- Li, L.; Masica, D.; Ishida, M.; Tomuleasa, C.; Umegaki, S.; Kalloo, A.N.; Georgiades, C.; Singh, V.K.; Khashab, M.; Amateau, S.; et al. Human Bile Contains MicroRNA-Laden Extracellular Vesicles That Can Be Used for Cholangiocarcinoma Diagnosis. Hepatology 2014, 60, 896–907. [Google Scholar] [CrossRef]
- Li, L.; Piontek, K.B.; Kumbhari, V.; Ishida, M.; Selaru, F.M. Isolation and Profiling of MicroRNA-Containing Exosomes from Human Bile. J. Vis. Exp. 2016, 13, 54036. [Google Scholar] [CrossRef]
- Nakashiki, S.; Miuma, S.; Mishima, H.; Masumoto, H.; Hidaka, M.; Soyama, A.; Kanda, Y.; Fukushima, M.; Haraguchi, M.; Sasaki, R.; et al. Bile Extracellular Vesicles from End-Stage Liver Disease Patients Show Altered MicroRNA Content. Hepatol. Int. 2021, 15, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Kiriyama, S.; Kozaka, K.; Takada, T.; Strasberg, S.M.; Pitt, H.A.; Gabata, T.; Hata, J.; Liau, K.-H.; Miura, F.; Horiguchi, A.; et al. Tokyo Guidelines 2018: Diagnostic Criteria and Severity Grading of Acute Cholangitis (with Videos). J. Hepato-Biliary-Pancreat. Sci. 2018, 25, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Lederer, T.; Hipler, N.M.; Thon, C.; Kupcinskas, J.; Link, A. Comparison of Fecal MicroRNA Isolation Using Various Total RNA Isolation Kits. Genes 2024, 15, 498. [Google Scholar] [CrossRef]
- Schindler, P.; Kupcinskas, J.; Juzenas, S.; Skieceviciene, J.; Salteniene, V.; Schulz, C.; Weigt, J.; Malfertheiner, P.; Link, A. Expression of MicroRNAs in the Ascites of Patients with Peritoneal Carcinomatosis and Peritonitis. Cancer Cytopathol. 2018, 126, 353–363. [Google Scholar] [CrossRef]
- Link, A.; Becker, V.; Goel, A.; Wex, T.; Malfertheiner, P. Feasibility of Fecal MicroRNAs as Novel Biomarkers for Pancreatic Cancer. PLoS ONE 2012, 7, e42933. [Google Scholar] [CrossRef]
- Lange, T.; Stracke, S.; Rettig, R.; Lendeckel, U.; Kuhn, J.; Schlüter, R.; Rippe, V.; Endlich, K.; Endlich, N. Identification of MiR-16 as an Endogenous Reference Gene for the Normalization of Urinary Exosomal MiRNA Expression Data from CKD Patients. PLoS ONE 2017, 12, e0183435. [Google Scholar] [CrossRef]
- Link, A.; Balaguer, F.; Shen, Y.; Nagasaka, T.; Lozano, J.J.; Boland, C.R.; Goel, A. Fecal MicroRNAs as Novel Biomarkers for Colon Cancer Screening. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1766–1774. [Google Scholar] [CrossRef]
- Liu, C.-H.; Huang, Q.; Jin, Z.-Y.; Xie, F.; Zhu, C.-L.; Liu, Z.; Wang, C. Circulating MicroRNA-21 as a Prognostic, Biological Marker in Cholangiocarcinoma. J. Cancer Res. Ther. 2018, 14, 220–225. [Google Scholar] [CrossRef]
- Link, A.; Kupcinskas, J. MicroRNAs as Non-Invasive Diagnostic Biomarkers for Gastric Cancer: Current Insights and Future Perspectives. World J. Gastroenterol. 2018, 24, 3313–3329. [Google Scholar] [CrossRef]
- Ge, X.; Tang, L.; Wang, Y.; Wang, N.; Zhou, J.; Deng, X.; Zhong, Y.; Li, Q.; Wang, F.; Jiang, G.; et al. The Diagnostic Value of Exosomal MiRNAs in Human Bile of Malignant Biliary Obstructions. Dig. Liver Dis. 2021, 53, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Shigehara, K.; Yokomuro, S.; Ishibashi, O.; Mizuguchi, Y.; Arima, Y.; Kawahigashi, Y.; Kanda, T.; Akagi, I.; Tajiri, T.; Yoshida, H.; et al. Real-Time PCR-Based Analysis of the Human Bile MicroRNAome Identifies MiR-9 as a Potential Diagnostic Biomarker for Biliary Tract Cancer. PLoS ONE 2011, 6, e23584. [Google Scholar] [CrossRef]
- Vaishnavi, C.; Samanta, J.; Kochhar, R. Characterization of Biofilms in Biliary Stents and Potential Factors Involved in Occlusion. World J. Gastroenterol. 2018, 24, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Uchihata, Y.; Arihiro, K.; Kaneko, Y.; Shimizu, T.; Marubashi, Y.; Aoki, C.; Murakami, T.; Ochi, M.; Niihara, N.; Ohtsuka, K.; et al. Analysis of MicroRNA in Bile Cytologic Samples Is Useful for Detection and Diagnosis of Extrahepatic Cholangiocarcinoma. Am. J. Clin. Pathol. 2022, 158, 122–131. [Google Scholar] [CrossRef]
- Bose, K.; Scurt, F.G.; Thon, C.; Franke, S.; Schulz, C.; Malfertheiner, P.; Link, A. Factors Affecting Performance of DNA Methylation as a Potential Biomarker in Ascites for Peritonitis and Peritoneal Carcinomatosis. J. Gastrointest. Liver Dis. 2023, 32, 206–215. [Google Scholar] [CrossRef]
- Han, J.-Y.; Ahn, K.S.; Kim, T.-S.; Kim, Y.H.; Cho, K.B.; Shin, D.W.; Baek, W.-K.; Suh, S.-I.; Jang, B.-C.; Kang, K.J. Liquid Biopsy from Bile-Circulating Tumor DNA in Patients with Biliary Tract Cancer. Cancers 2021, 13, 4581. [Google Scholar] [CrossRef]
- Casadio, M.; Biancaniello, F.; Overi, D.; Venere, R.; Carpino, G.; Gaudio, E.; Alvaro, D.; Cardinale, V. Molecular Landscape and Therapeutic Strategies in Cholangiocarcinoma: An Integrated Translational Approach towards Precision Medicine. Int. J. Mol. Sci. 2021, 22, 5613. [Google Scholar] [CrossRef]

| Patients | 100 | |
|---|---|---|
| Age in years | 65.50 ± 12.57 | |
| Sex | Male | 71 (71.00%) |
| Female | 29 (29.00%) | |
| Stent types | Double pigtail | 69 (69.00%) |
| Amsterdam | 21 (21.00%) | |
| Metal stents | 7 (7.00%) | |
| No information | 3 (3.00%) | |
| Indwelling time in days | 71.47 ± 63.55 | |
| Reason for stent therapy | CCA | 22 |
| PCA | 5 | |
| CRC | 5 | |
| Other malignancies | 8 | |
| Non-malignant stenosis | 60 | |
| Patients with cholangitis | 42 | |
| Control group | 25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hipler, N.M.; Thon, C.; Lehr, K.; Furnari, M.; Obst, W.; Keitel, V.; Weigt, J.; Link, A. Exploring microRNAs in Bile Duct Stents as Diagnostic Biomarkers for Biliary Pathologies. Cancers 2025, 17, 1171. https://doi.org/10.3390/cancers17071171
Hipler NM, Thon C, Lehr K, Furnari M, Obst W, Keitel V, Weigt J, Link A. Exploring microRNAs in Bile Duct Stents as Diagnostic Biomarkers for Biliary Pathologies. Cancers. 2025; 17(7):1171. https://doi.org/10.3390/cancers17071171
Chicago/Turabian StyleHipler, Noam Mathias, Cosima Thon, Konrad Lehr, Manuele Furnari, Wilfried Obst, Verena Keitel, Jochen Weigt, and Alexander Link. 2025. "Exploring microRNAs in Bile Duct Stents as Diagnostic Biomarkers for Biliary Pathologies" Cancers 17, no. 7: 1171. https://doi.org/10.3390/cancers17071171
APA StyleHipler, N. M., Thon, C., Lehr, K., Furnari, M., Obst, W., Keitel, V., Weigt, J., & Link, A. (2025). Exploring microRNAs in Bile Duct Stents as Diagnostic Biomarkers for Biliary Pathologies. Cancers, 17(7), 1171. https://doi.org/10.3390/cancers17071171







