Developing a Predictive Model for Significant Prostate Cancer Detection in Prostatic Biopsies from Seven Clinical Variables: Is Machine Learning Superior to Logistic Regression?
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
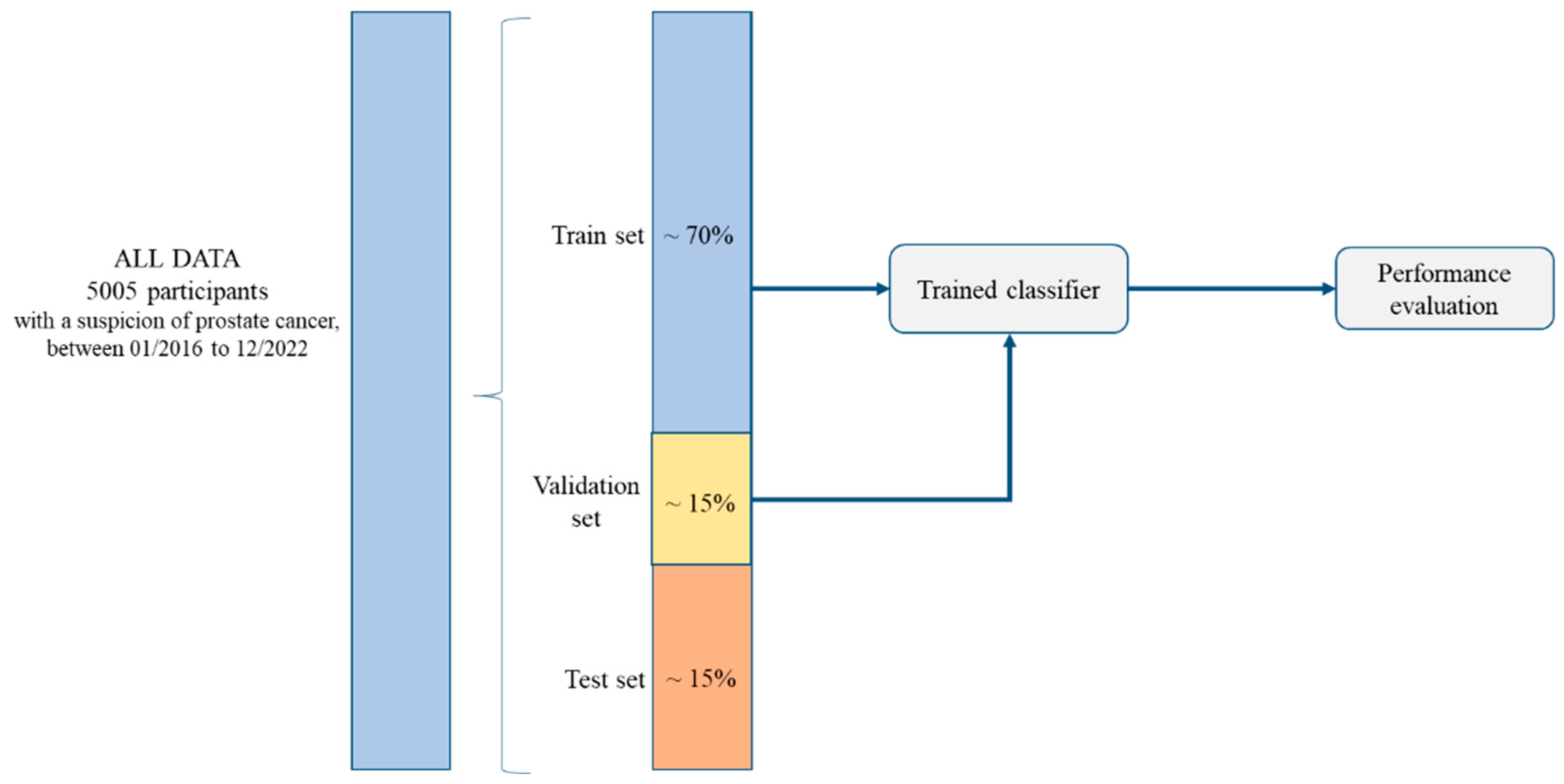
2.1. Study Design and Participants
2.2. Diagnostic Approach for Significant Prostate Cancer
2.3. Predictive Variables Included in the Models and Outcome Variable
2.4. Algorithms Used for Model Development
2.5. Statistical Analyses, Algorithm Performance, and Interpretation
3. Results
3.1. Participant Characteristics
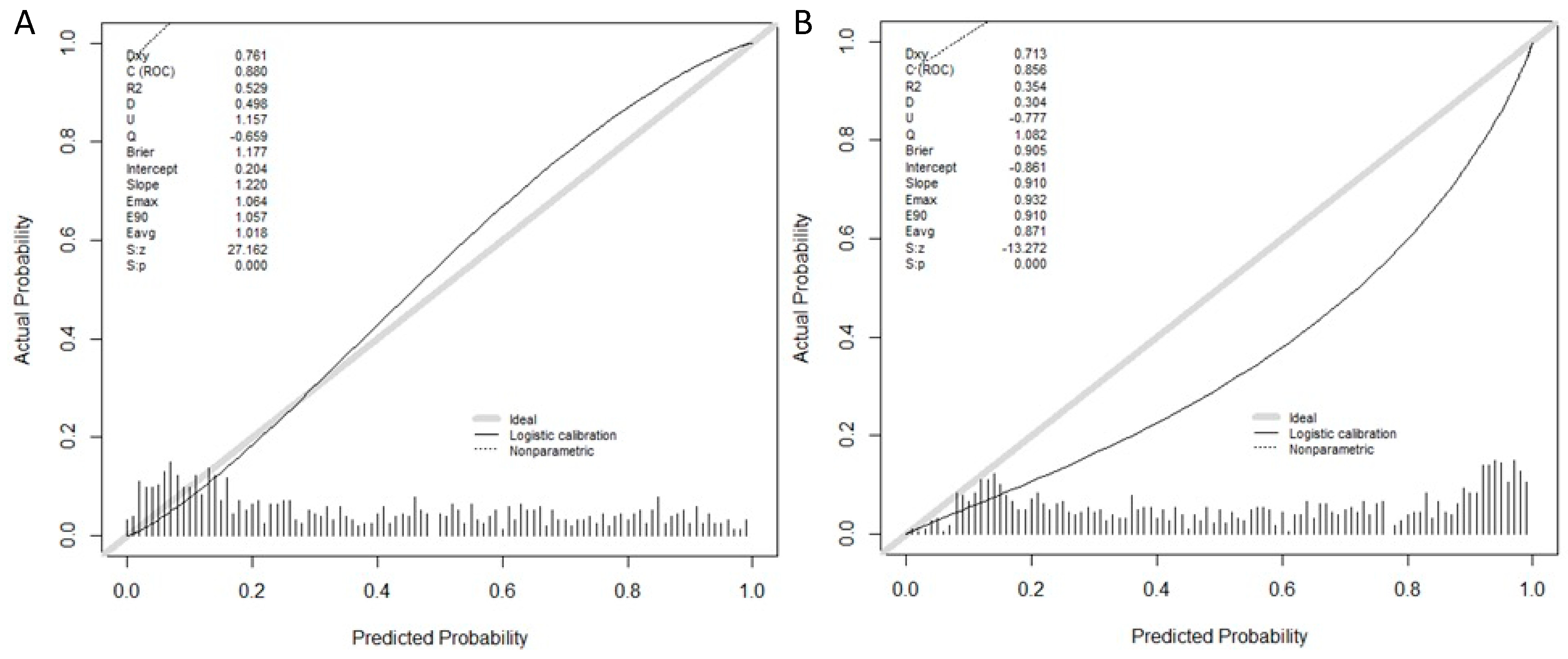
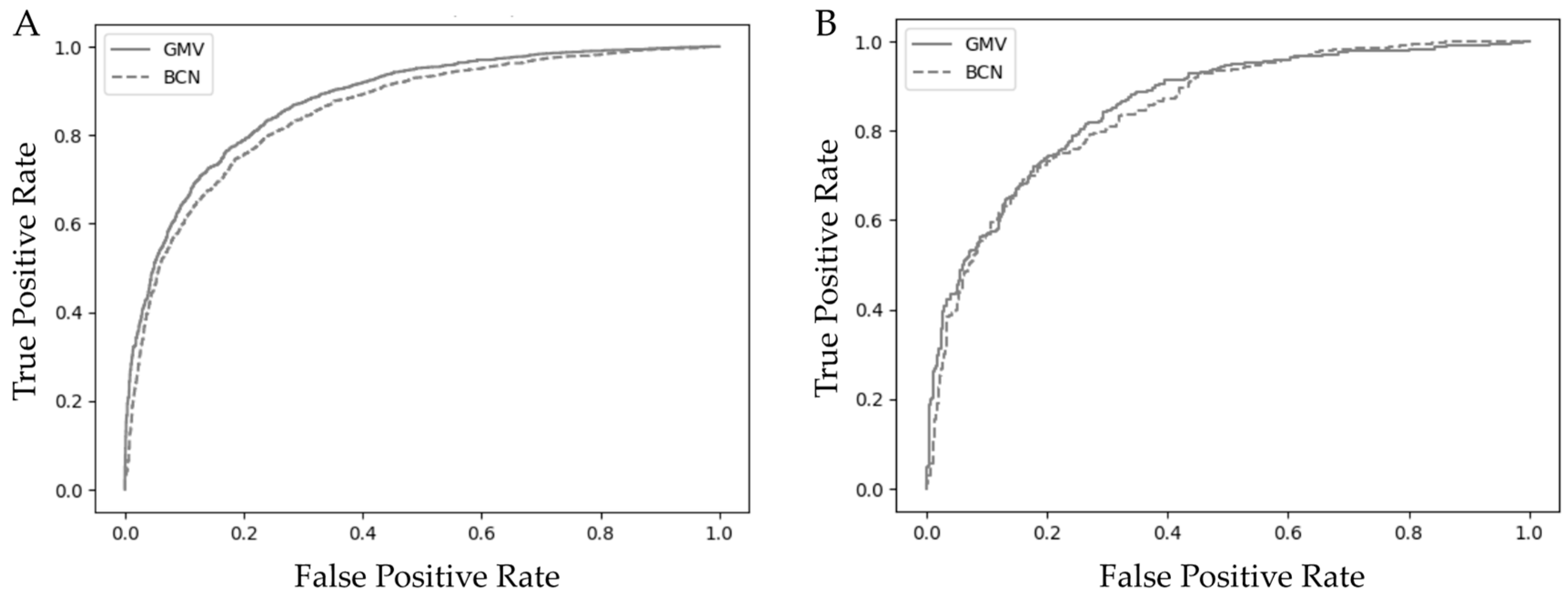
3.2. Model Performance, Calibration, and Validation of the GMV and BCN Predictive Models
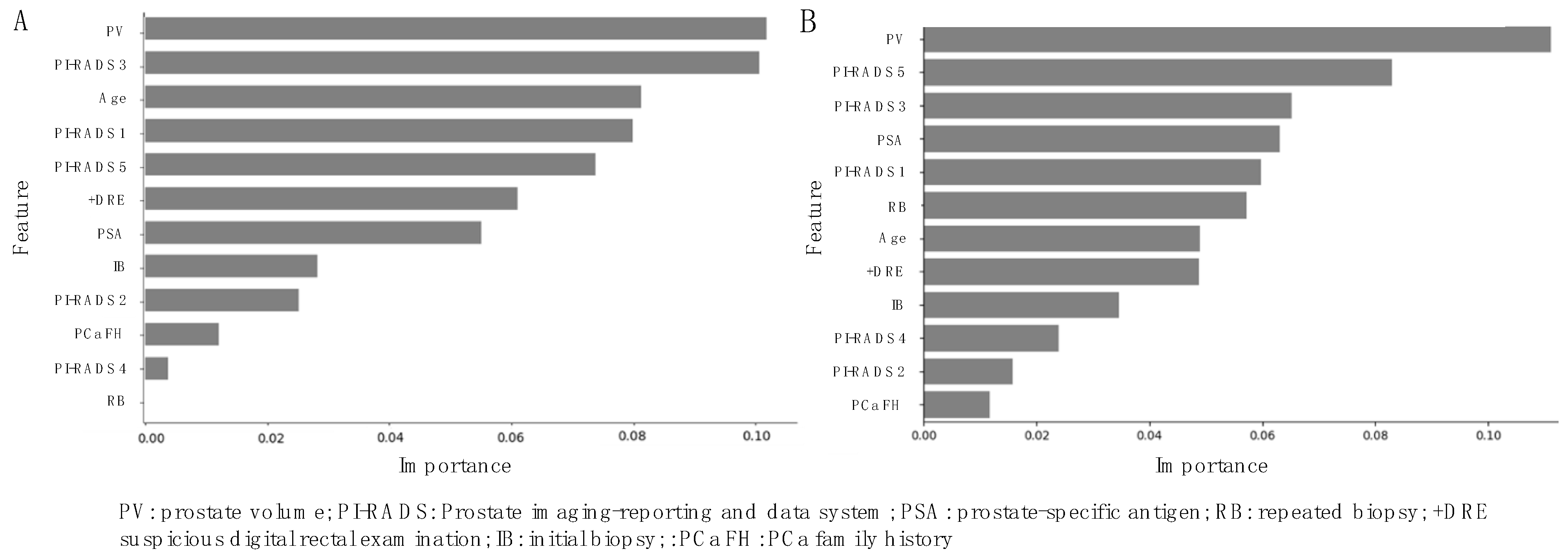
3.3. Variable Importance Interpretation with SHapley Additive exPlanations (SHAP)
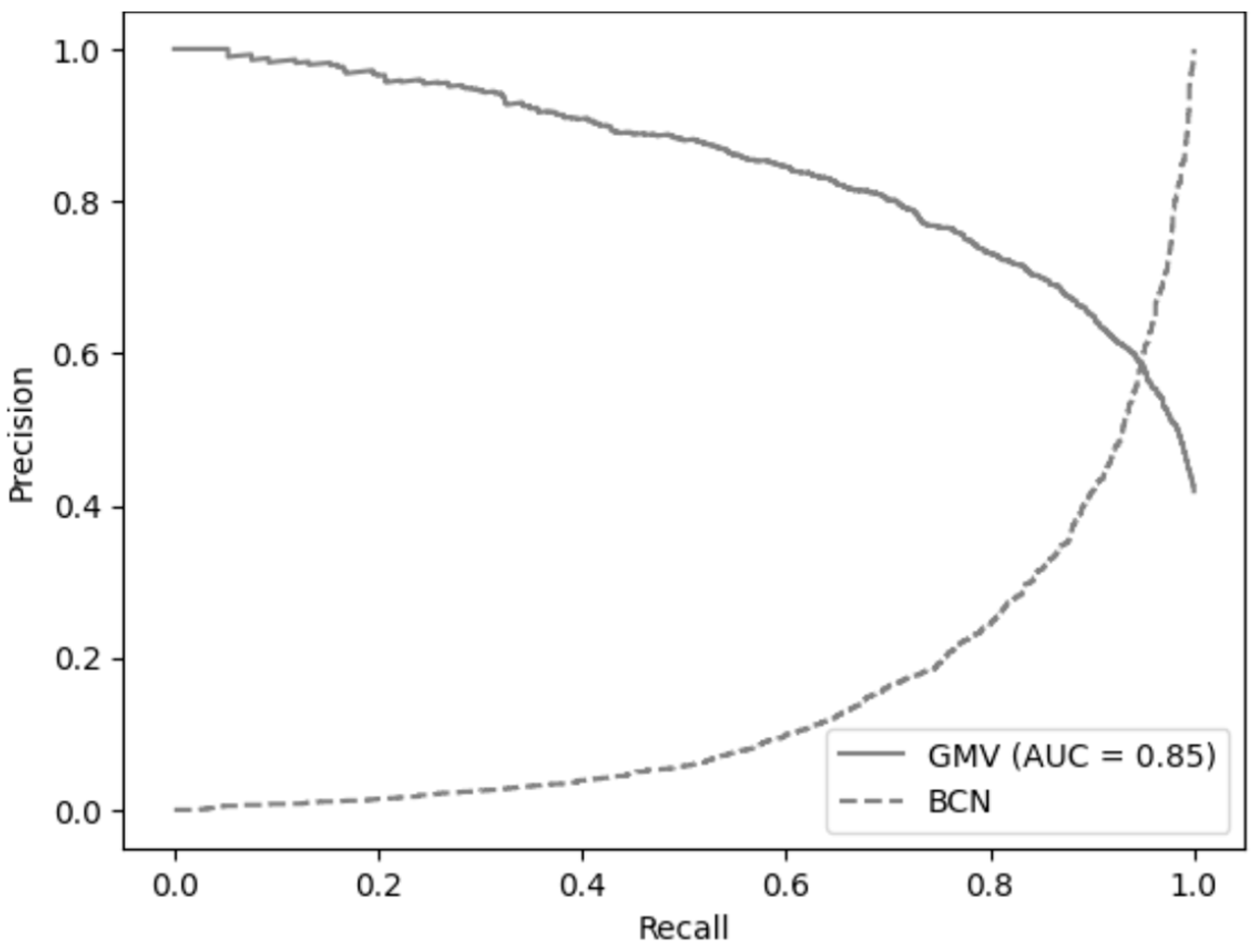
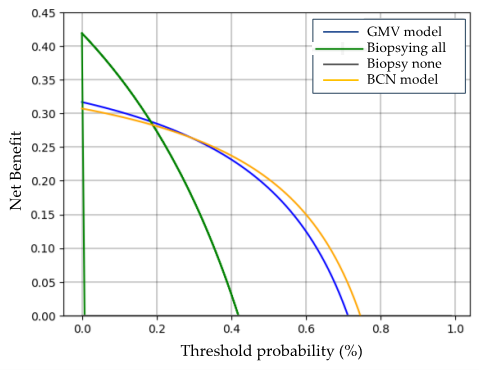
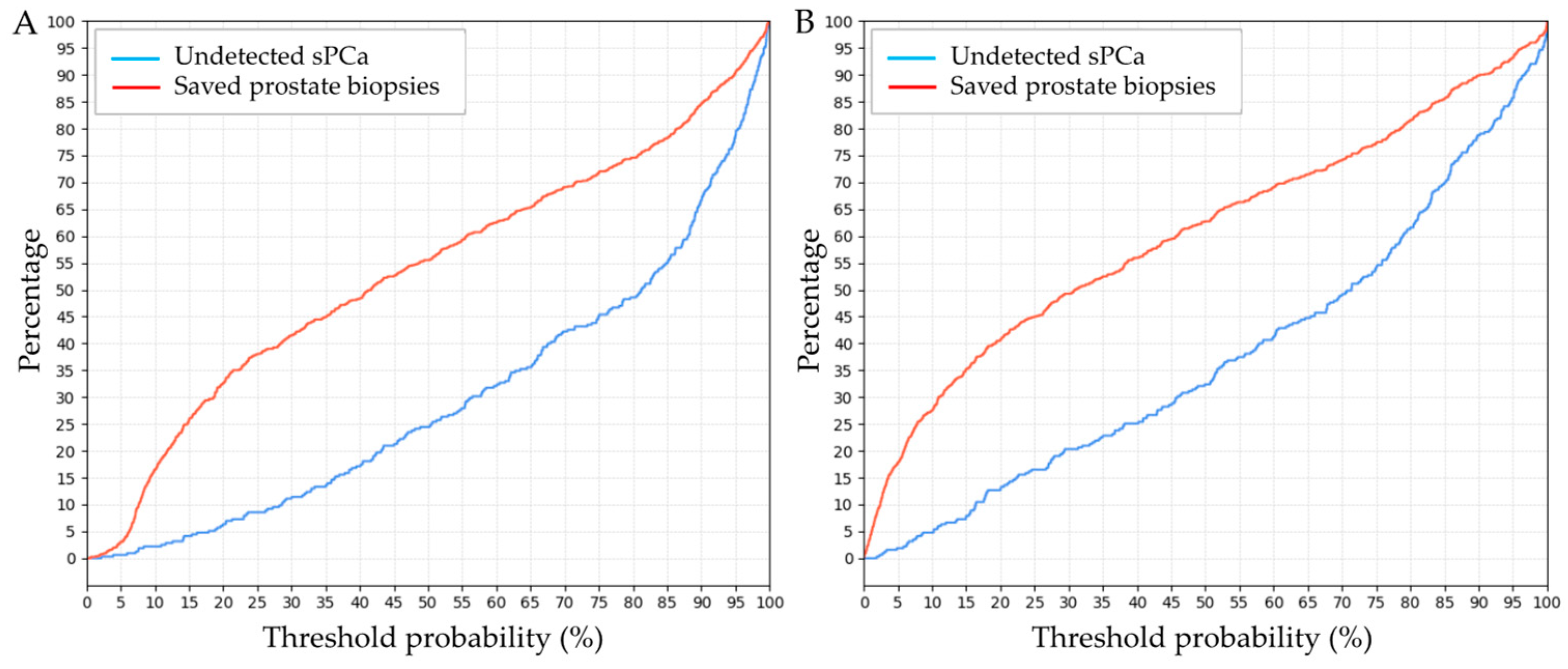
3.4. Clinical Comparison of GMV and BCN Predictive Models for sPCa Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Characteristic | Development Cohort | Validation Cohort | Test Cohort | p-Value |
|---|---|---|---|---|
| Number of men | 4254 | 639 | 751 | |
| Mean age at biopsy, years (SD) | 68 (8.3) | 68 (8.3) | 68 (8.2) | 1 |
| Mean serum PSA, ng/mL (SD) | 13.4 (75.0) | 12.7 (35.5) | 12.8 (47.6) | 0.36 |
| Suspicious DRE, n (%) | 1210 (28.5) | 192 (30.1) | 217 (28.9) | 0.11 |
| PCa family history, n (%) | 304 (7.2) | 51 (8.0) | 48 (6.4) | 0.2 |
| Mean prostate volume, mL (SD) | 61.5 (32.3) | 62.0 (33.1) | 64.6 (35.8) | 0.65 |
| Previous negative prostate biopsy, n (%) | 1281 (30.2) | 216 (33.9) | 224 (29.9) | 0.41 |
| PI-RADS version used | 2 | 2.1 | 2 | |
| Mean number of suspicious lesions | 2 | 2 | 2 | |
| PI-RADS score of index lesion, n (%) | ||||
| 1 | 470 (11.1) | 71 (11.2) | 104 (13.9) | 0.33 |
| 2 | 148 (3.5) | 15 (2.4) | 27 (3.6) | 0.09 |
| 3 | 1053 (24.8) | 161 (25.2) | 197 (26.3) | 0.27 |
| 4 | 1743 (41.0) | 261 (40.9) | 272 (36.3) | 0.07 |
| 5 | 840 (19.8) | 131 (20.6) | 151 (20.2) | 0.71 |
| sPCa detection, n (%) | 1782 (41.9) | 268 (42.0) | 315 (42.0) | 1 |
References
- Van Poppel, H.; Albreht, T.; Basu, P.; Hogenhout, R.; Collen, S.; Roobol, M. Serum PSA-based early detection of prostate cancer in Europe and globally: Past, present and future. Nat. Rev. Urol. 2022, 19, 562–572. [Google Scholar]
- Schroder, F.H.; Hugosson, J.; Roobol, M.J.; Tammela, T.L.; Ciatto, S.; Nelen, V.; Kwiatkowski, M.; Lujan, M.; Lilja, H.; Zappa, M.; et al. Screening and prostate-cancer mortality in a randomized European study. N. Engl. J. Med. 2009, 360, 1320–1328. [Google Scholar] [PubMed]
- Frånlund, M.; Månsson, M.; Godtman, R.A.; Aus, G.; Holmberg, E.; Kollberg, K.S.; Lodding, P.; Pihl, C.G.; Stranne, J.; Lilja, H.; et al. Results from 22 years of Followup in the Göteborg Randomized Population-Based Prostate Cancer Screening Trial. J. Urol. 2022, 208, 292–300. [Google Scholar] [PubMed]
- Kasivisvanathan, V.; Rannikko, A.S.; Borghi, M.; Panebianco, V.; Mynderse, L.A.; Vaarala, M.H.; Briganti, A.; Budäus, L.; Hellawell, G.; Hindley, R.G.; et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N. Engl. J. Med. 2018, 378, 1767–1777. [Google Scholar] [PubMed]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Tempany, C.M.; Choyke, P.L.; Cornud, F.; Margolis, D.J.; et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar]
- Westphalen, A.C.; McCulloch, C.E.; Anaokar, J.M.; Arora, S.; Barashi, N.S.; Barentsz, J.O.; Bathala, T.K.; Bittencourt, L.K.; Booker, M.T.; Braxton, V.G.; et al. Variability of the Positive Predictive Value of PI-RADS for Prostate MRI across 26 Centers: Experience of the Society of Abdominal Radiology Prostate Cancer Disease-focused Panel. Radiology 2020, 296, 76–84. [Google Scholar]
- Girometti, R.; Peruzzi, V.; Polizzi, P.; De Martino, M.; Cereser, L.; Casarotto, L.; Pizzolitto, S.; Isola, M.; Crestani, A.; Giannarini, G.; et al. Case-by-case combination of the prostate imaging reporting and data system version 2.1 with the Likert score to reduce the false-positives of prostate MRI: A proof-of-concept study. Abdom. Radiol. 2024, 49, 4273–4285. [Google Scholar]
- Morote, J.; Borque-Fernando, A.; Triquell, M.; Celma, A.; Regis, L.; Mast, R.; de Torres, I.M.; Semidey, M.E.; Abascal, J.M.; Servian, P.; et al. Comparative Analysis of PSA Density and an MRI-Based Predictive Model to Improve the Selection of Candidates for Prostate Biopsy. Cancers 2022, 11, 2374. [Google Scholar] [CrossRef]
- Haj-Mirzaian, A.; Burk, K.S.; Lacson, R.; Glazer, D.I.; Saini, S.; Kibel, A.S.; Khorasani, R. Magnetic Resonance Imaging, Clinical, and Biopsy Findings in Suspected Prostate Cancer: A Systematic Review and Meta-Analysis. JAMA Netw. Open 2024, 7, e244258. [Google Scholar]
- Eyrich, N.W.; Morgan, T.M.; Tosoian, J.J. Biomarkers for detection of clinically significant prostate cancer: Contemporary clinical data and future directions. Transl. Androl. Urol. 2021, 10, 3091–3103. [Google Scholar]
- Li, K.; Wang, Q.; Tang, X.; Akakuru, O.U.; Li, R.; Wang, Y.; Zhang, R.; Jiang, Z.; Yang, Z. Advances in Prostate Cancer Biomarkers and Probes. Cyborg Bionic Syst. 2024, 5, 0129. [Google Scholar]
- Morote, J.; Borque-Fernando, A.; Triquell, M.; Celma, A.; Regis, L.; Escobar, M.; Mast, R.; de Torres, I.M.; Semidey, M.E.; Abascal, J.M.; et al. The Barcelona Predictive Model of Clinically Significant Prostate Cancer. Cancers 2022, 14, 1589. [Google Scholar] [CrossRef]
- Davis, S.E.; Greevy, R.A.; Lasko, T.A.; Walsh, C.G.; Matheny, M.E. Comparison of Prediction Model Performance Updating Protocols: Using a Data-Driven Testing Procedure to Guide Updating. AMIA Ann. Symp. Proc. 2020, 2019, 1002–1010. [Google Scholar]
- Aladwani, M.; Lophatananon, A.; Ollier, W.; Muir, K. Prediction models for prostate cancer to be used in the primary care setting: A systematic review. BMJ Open 2020, 10, e034661. [Google Scholar] [PubMed]
- Riaz, I.B.; Harmon, S.; Chen, Z.; Naqvi, S.A.A.; Cheng, L. Applications of Artificial Intelligence in Prostate Cancer Care: A Path to Enhanced Efficiency and Outcomes. Am. Soc. Clin. Oncol. Educ. Book. 2024, 44, 3. [Google Scholar]
- Nandi, A.; Xhafa, F. A federated learning method for real-time emotion state classification from multi-modal streaming. Methods 2022, 204, 340–347. [Google Scholar] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Available online: http://www.deeplearningbook.org (accessed on 23 December 2024).
- Jui, J.J.; Molla, M.M.I.; Alam, M.K.; Ferdowsi, A. Prostate Cancer Prediction Using Feedforward Neural Network Trained with Particle Swarm Optimizer. In Proceedings of the 6th International Conference on Electrical, Control and Computer Engineering, Kuantan, Malaysia, 23 August 2021; Lecture Notes in Electrical Engineering. Zain, M.D., Sulaiman, M.H., Mohamed, A.I., Bakar, M.S., Ramli, M.S., Eds.; Springer: Singapore, 2022; Volume 842. [Google Scholar] [CrossRef]
- Wu, H.; Liao, B.; Ji, T.; Ma, K.; Luo, Y.; Zhang, S. Comparison between traditional logistic regression and machine learning for predicting mortality in adult sepsis patients. Front. Med. 2024, 11, 1496869. [Google Scholar]
- Afifi, J.; Ahmad, T.; Guida, A.; Vincer, M.J.; Stewart, S.A. Prediction of Neurodevelopmental Outcomes in Very Preterm Infants: Comparing Machine Learning Methods to Logistic Regression. Children 2024, 11, 1512. [Google Scholar] [CrossRef]
- Austin, P.C.; Lee, D.S.; Wang, B. The relative data hungriness of unpenalized and penalized logistic regression and ensemble-based machine learning methods: The case of calibration. Diagn. Progn. Res. 2024, 8, 15. [Google Scholar]
- Hong, S.; Lu, B.; Wang, S.; Jiang, Y. Comparison of logistic regression and machine learning methods for predicting depression risks among disabled elderly individuals: Results from the China Health and Retirement Longitudinal Study. BMC Psychiatry 2025, 25, 128. [Google Scholar]
- Gui, Q.; Wang, X.; Wu, D.; Guo, Y. Constructing and Validating Models for Predicting Gleason Grade Group Upgrading following Radical Prostatectomy in Localized Prostate Cancer: A Comparison between Machine Learning Algorithms and Conventional Logistic Regression. Oncology 2025, 24, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Wang, S.; Wan, D.; Zhang, Q.; Shen, W.; Liu, X.; Zhang, Y. Relative Fat Mass and Physical Indices as Predictors of Gallstone Formation: Insights From Machine Learning and Logistic Regression. Int. J. Gen. Med. 2025, 18, 509–527. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, A.; Yanagisawa, T.; Bekku, K.; Kardoust Parizi, M.; Laukhtina, E.; Klemm, J.; Chiujdea, S.; Mori, K.; Kimura, S.; Fazekas, T.; et al. Comparing the Performance of Digital Rectal Examination and Prostate-specific Antigen as a Screening Test for Prostate Cancer: A Systematic Review and Meta-analysis. Eur. Urol. Oncol. 2024, 7, 697–704. [Google Scholar] [CrossRef]
- Barentsz, J.O.; Richenberg, J.; Clements, R.; Choyke, P.; Verma, S.; Villeirs, G.; Rouviere, O.; Logager, V.; Futterer, J.J. ESUR prostate MR guidelines 2012. Eur. Radiol. 2012, 22, 746–757. [Google Scholar] [CrossRef]
- Weinreb, J.C.; Barentsz, J.O.; Choyke, P.L.; Cornud, F.; Haider, M.A.; Macura, K.J.; Margolis, D.; Schnall, M.D.; Shtern, F.; Tempany, C.M.; et al. PI-RADS Prostate Imaging—Reporting and Data System: 2015, Version 2. Eur. Urol. 2016, 69, 16–40. [Google Scholar] [CrossRef]
- Khoo, C.C.; Eldred-Evans, D.; Peters, M.; van Son, M.; van Rossum, P.S.N.; Connor, M.J.; Hosking-Jervis, F.; Tanaka, M.B.; Reddy, D.; Bass, E.; et al. A Comparison of Prostate Cancer Detection between Visual Estimation (Cognitive Registration) and Image Fusion (Software Registration) Targeted Transperineal Prostate Biopsy. J. Urol. 2021, 205, 1075–1081. [Google Scholar] [CrossRef]
- Wu, Q.; Tu, X.; Zhang, C.; Ye, J.; Lin, T.; Liu, Z.; Yang, L.; Qiu, S.; Bao, Y.; Wei, Q. Transperineal magnetic resonance imaging targeted biopsy versus transrectal route in the detection of prostate cancer: A systematic review and meta-analysis. Prostate Cancer Prostatic Dis. 2024, 27, 212–221. [Google Scholar] [CrossRef]
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A.; Grading, C. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am. J. Surg. Pathol. 2016, 40, 244–252. [Google Scholar]
- Moore, C.M.; Kasivisvanathan, V.; Eggener, S.; Emberton, M.; Fütterer, J.J.; Gill, I.S.; Grubb Iii, R.L.; Hadaschik, B.; Klotz, L.; Margolis, D.J.; et al. Standards of reporting for MRI-targeted biopsy studies (START) of the prostate: Recommendations from an International Working Group. Eur. Urol. 2013, 64, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Anjum, M.; Khan, K.; Ahmad, W.; Ahmad, A.; Amin, M.N.; Nafees, A. New SHapley Additive ExPlanations (SHAP) approach to evaluate the raw materials interactions of steel-fiber-reinforced concrete. Materials 2022, 15, 6261. [Google Scholar] [CrossRef]
- Stephan, C.; Xu, C.; Finne, P.; Cammann, H.; Meyer, H.A.; Lein, M.; Jung, K.; Stenman, U.H. Comparison of two different artificial neural networks for prostate biopsy indication in two different patient populations. Urology 2007, 70, 596–601. [Google Scholar]
- Jansen, F.H.; van Schaik, R.H.; Kurstjens, J.; Horninger, W.; Klocker, H.; Bektic, J.; Wildhagen, M.F.; Roobol, M.J.; Bangma, C.H.; Bartsch, G. Prostate-specific antigen (PSA) isoform p2PSA in combination with total PSA and free PSA improves diagnostic accuracy in prostate cancer detection. Eur. Urol. 2010, 57, 921–927. [Google Scholar]
- Ecke, T.H.; Bartel, P.; Hallmann, S.; Koch, S.; Ruttloff, J.; Cammann, H.; Lein, M.; Schrader, M.; Miller, K.; Stephan, C. Outcome prediction for prostate cancer detection rate with artificial neural network (ANN) in daily routine. Urol. Oncol. 2012, 30, 139–144. [Google Scholar]
- Takeuchi, T.; Hattori-Kato, M.; Okuno, Y.; Iwai, S.; Mikami, K. Prediction of prostate cancer by deep learning with multilayer artificial neural network. Can. Urol. Assoc. J. 2019, 13, E145–E150. [Google Scholar] [PubMed]
- Checcucci, E.; Rosati, S.; De Cillis, S.; Giordano, N.; Volpi, G.; Granato, S.; Zamengo, D.; Verri, P.; Amparore, D.; De Luca, S.; et al. Machine-Learning-Based Tool to Predict Target Prostate Biopsy Outcomes: An Internal Validation Study. J. Clin. Med. 2023, 17, 4358. [Google Scholar] [CrossRef]
- Rippa, M.; Schulze, R.; Kenyon, G.; Himstedt, M.; Kwiatkowski, M.; Grobholz, R.; Wyler, S.; Cornelius, A.; Schindera, S.; Burn, F. Evaluation of Machine Learning Classification Models for False-Positive Reduction in Prostate Cancer Detection Using MRI Data. Diagnostics 2024, 14, 1677. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.C.; Nakai, H.; Kuanar, S.; Froemming, A.T.; Bolan, C.W.; Kawashima, A.; Takahashi, H.; Mynderse, L.A.; Dora, C.D.; Humphreys, M.R.; et al. Fully Automated Deep Learning Model to Detect Clinically Significant Prostate Cancer at MRI. Radiology 2024, 312, e232635. [Google Scholar]
- Chen, F.; Esmaili, R.; Khajir, G.; Zeevi, T.; Gross, M.; Leapman, M.; Sprenkle, P.; Justice, A.C.; Arora, S.; Weinreb, J.C.; et al. Comparative Performance of Machine Learning Models in Reducing Unnecessary Targeted Prostate Biopsies. Eur. Urol. Oncol. 2025, in press. [CrossRef]
- Zheng, B.; Mo, F.; Shi, X.; Li, W.; Shen, Q.; Zhang, L.; Liao, Z.; Fan, C.; Liu, Y.; Zhong, J.; et al. An Automatic Deep-Radiomics Framework for Prostate Cancer Diagnosis and Stratification in Patients with Serum Prostate-Specific Antigen of 4.0–10.0 ng/mL: A Multicenter Retrospective Study. Acad. Radiol. 2025; in press. [Google Scholar] [CrossRef]
- Lee, Y.J.; Moon, H.W.; Choi, M.H.; Eun Jung, S.; Park, Y.H.; Lee, J.Y.; Kim, D.H.; Eun Rha, S.; Kim, S.H.; Lee, K.W.; et al. MRI-based Deep Learning Algorithm for Assisting Clinically Significant Prostate Cancer Detection: A Bicenter Prospective Study. Radiology 2025, 314, e232788. [Google Scholar]
- Michaely, H.J.; Aringhieri, G.; Cioni, D.; Neri, E. Current Value of Biparametric Prostate MRI with Machine-Learning or Deep-Learning in the Detection, Grading, and Characterization of Prostate Cancer: A Systematic Review. Diagnostics 2022, 12, 799. [Google Scholar] [CrossRef]
- Huynh, L.M.; Hwang, Y.; Taylor, O.; Baine, M.J. The Use of MRI-Derived Radiomic Models in Prostate Cancer Risk Stratification: A Critical Review of Contemporary Literature. Diagnostics 2023, 13, 1128. [Google Scholar] [CrossRef]
- Antolin, A.; Roson, N.; Mast, R.; Arce, J.; Almodovar, R.; Cortada, R.; Maceda, A.; Escobar, M.; Trilla, E.; Morote, J. The Role of Radiomics in the Prediction of Clinically Significant Prostate Cancer in the PI-RADS v2 and v2.1 Era: A Systematic Review. Cancers 2024, 16, 2951. [Google Scholar] [CrossRef]
- Boesen, L.; Thomsen, F.B.; Nørgaard, N.; Løgager, V.; Balslev, I.; Bisbjerg, R.; Thomsen, H.S.; Jakobsen, H. A predictive model based on biparametric magnetic resonance imaging and clinical parameters for improved risk assessment and selection of biopsy-naïve men for prostate biopsies. Prostate Cancer Prostatic Dis. 2019, 22, 609–616. [Google Scholar] [PubMed]
- Thompson, I.M.; Ankerst, D.P.; Chi, C.; Goodman, P.J.; Tangen, C.M.; Lucia, M.S.; Feng, Z.; Parnes, H.L.; Coltman, C.A., Jr. Assessing prostate cancer risk: Results from the Prostate Cancer Prevention Trial. J. Natl. Cancer Inst. 2006, 98, 529–534. [Google Scholar] [PubMed]
- Rapisarda, S.; Bada, M.; Crocetto, F.; Barone, B.; Arcaniolo, D.; Polara, A.; Imbimbo, C.; Grosso, G. The role of multiparametric resonance and biopsy in prostate cancer detection: Comparison with definitive histological report after laparoscopic/robotic radical prostatectomy. Abdom. Radiol. 2020, 45, 4178–4184. [Google Scholar]
- Morote, J.; Borque-Fernando, Á.; Triquell, M.; Campistol, M.; Servian, P.; Abascal, J.M.; Planas, J.; Méndez, O.; Esteban, L.M.; Trilla, E. Comparison of Rotterdam and Barcelona Magnetic Resonance Imaging Risk Calculators for Predicting Clinically Significant Prostate Cancer. Eur. Urol. Open Sci. 2023, 53, 46–54. [Google Scholar] [PubMed]
- Ogbonnaya, C.N.; Alsaedi, B.S.O.; Alhussaini, A.J.; Hislop, R.; Pratt, N.; Steele, J.D.; Kernohan, N.; Nabi, G. Radiogenomics Map-Based Molecular and Imaging Phenotypical Characterization in Localized Prostate Cancer Using Pre-Biopsy Biparametric MR Imaging. Int. J. Mol. Sci. 2024, 25, 5379. [Google Scholar] [CrossRef]
- Arita, Y.; Roest, C.; Kwee, T.V.; Paudyal, R.; Lema-Dopico, A.; Fransen, S.; Hirahara, D.; Takaya, E.; Ueda, R.; Ruby, R.; et al. Advancements in artificial intelligence for prostate cancer: Optimizing diagnosis, treatment, and prognostic assessment. Asian J. Urol. 2025, 11, 545–554. [Google Scholar] [CrossRef]

| Characteristic | sPCa | nsPCa | Odds Ratio (95% CI) | p-Value |
|---|---|---|---|---|
| Number of men (%) | 2097 (41.9) | 2908 (58.1) | - | - |
| Mean age, years (SD) | 70 (8.2) | 66 (7.6) | 1.07 (1.06–1.08) | <0.001 |
| Mean serum PSA, ng/mL (SD) | 20 (109) | 8.4 (9.6) | 1.04 (1.03–1.05) | <0.001 |
| PCa family history, n (%) | ||||
| No | 1930 (92%) | 2723 (93.6%) | - | Ref. |
| Yes | 167 (8%) | 185 (6.4%) | 1.27 (1.02–1.58) | 0.033 |
| Type of prostate biopsy, n (%) | ||||
| Initial | 1594 (76%) | 1906 (65.5%) | - | Ref. |
| Repeated | 503 (24%) | 1002 (34.5%) | 0.6 (0.53–0.68) | <0.001 |
| DRE, n (%) | ||||
| Normal | 1161 (55.4%) | 2417 (83.1%) | - | Ref. |
| Suspicious | 936 (44.6%) | 491 (16.9%) | 3.97 (3.49–4.52) | <0.001 |
| Prostate volume (mL) | 51.9 (27.6) | 69.1 (34.4) | 0.98 (0.98–0.98) | <0.001 |
| PI-RADS score, n (%) | ||||
| 1 | 60 (2.9%) | 514 (17.7%) | - | Ref. |
| 2 | 23 (1.1%) | 152 (5.2%) | 1.3 (0.76–2.14) | 0.322 |
| 3 | 206 (9.8%) | 1044 (35.9%) | 1.69 (1.25–2.31) | 0.001 |
| 4 | 991 (47.3%) | 1024 (35.2%) | 8.29 (6.31–11.08) | <0.001 |
| 5 | 817 (39%) | 174 (6%) | 40.22 (29.61–55.48) | <0.001 |
| Metric | Training Set (n = 4–254) | Validation Set (n = 631) | Test Set (n = 751) | |||
|---|---|---|---|---|---|---|
| GMV Model | BCN Model | GMV Model | BCN Model | GMV Model | BCN Model | |
| AUC (95% CI) | 0.88 (0.87−0.90) | 0.85 (0.84−0.86) | 0.88 (0.86−0.91) | 0.86 (0.85−0.87) | 0.85 (0.83−0.88) | 0.84 (0.82−0.86) |
| Precision (95% CI) | 0.7171 (0.6973−0.7362) | 0.7435 (0.7228−0.7633) | 0.7184 (0.6659−0.7657) | 0.7426 (0.6876−0.7910) | 0.7126 (0.6618−0.7585) | 0.7607 (0.7074−0.8069) |
| Recall (95% CI) | 0.8266 (0.8083−0.8435) | 0.7435 (0.7228−0.7633) | 0.8284 (0.7787−0.8688) | 0.7537 (0.6988−0.8015) | 0.7556 (0.7052−0.7997) | 0.6762 (0.6227−0.7255) |
| Specificity (95% CI) | 0.765 (0.7478−0.7813) | 0.8151 (0.7993−0.8299) | 0.7655 (0.7198−0.8058) | 0.8113 (0.7684−0.8479) | 0.7798 (0.7386−0.8162) | 0.8463 (0.8095−0.8771) |
| Accuracy (95% CI) | 0.7908 (0.7783−0.8027) | 0.7851 (0.7725−0.7972) | 0.7919 (0.7587−0.8215) | 0.7872 (0.7538−0.8171) | 0.7696 (0.7382−0.7983) | 0.7750 (0.7437−0.8034) |
| F1 score (95% CI) | 0.768 (0.7537−0.7832) | 0.7435 (0.7275−0.7592) | 0.7695 (0.7298−0.8047) | 0.7481 (0.7025−0.7842) | 0.7334 (0.6952−0.7695) | 0.7160 (0.6736−0.7537) |
| Kappa score (95% CI) | 0.5792 (0.5548−0.6037) | 0.5587 (0.5325−0.5847) | 0.5815 (0.5163−0.6389) | 0.5639 (0.4903−0.6276) | 0.5309 (0.4671−0.5851) | 0.5307 (0.4671−0.5911) |
| MCC (95% CI) | 0.5841 (0.5603−0.6085) | 0.5587 (0.5325−0.5850) | 0.5864 (0.5243−0.6441) | 0.5639 (0.4910−0.6287) | 0.5316 (0.4682−0.5857) | 0.5332 (0.4685−0.5948) |
| Threshold (%) | GMV Model | BCN Model | ||
|---|---|---|---|---|
| Saved Biopsies (%) | Undetected sPCa (%) | Saved Biopsies (%) | Undetected sPCa (%) | |
| 5 | 3.5 | 0.5 | 18 | 2 |
| 6 | 5 | 0.75 | 20 | 2.5 |
| 7 | 9.5 | 1 | 23 | 3.5 |
| 8 | 10 | 2 | 26 | 4.75 |
| 9 | 14 | 2 | 27 | 5 |
| 10 | 17 | 2 | 27.5 | 5 |
| 11 | 19 | 2.5 | 30 | 6 |
| 12 | 20 | 2.5 | 32.5 | 6.5 |
| 13 | 22 | 2.6 | 33.5 | 6.5 |
| 14 | 23 | 2.6 | 34 | 8 |
| 15 | 26 | 4.5 | 35 | 8.5 |
| 16 | 27.5 | 5 | 36 | 10.5 |
| 17 | 29 | 5 | 37.5 | 10.5 |
| 18 | 30 | 5.1 | 39.5 | 13 |
| 19 | 30 | 5.1 | 40 | 13 |
| 20 | 32.5 | 6.5 | 41 | 13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morote, J.; Miró, B.; Hernando, P.; Paesano, N.; Picola, N.; Muñoz-Rodriguez, J.; Ruiz-Plazas, X.; Muñoz-Rivero, M.V.; Celma, A.; García-de Manuel, G.; et al. Developing a Predictive Model for Significant Prostate Cancer Detection in Prostatic Biopsies from Seven Clinical Variables: Is Machine Learning Superior to Logistic Regression? Cancers 2025, 17, 1101. https://doi.org/10.3390/cancers17071101
Morote J, Miró B, Hernando P, Paesano N, Picola N, Muñoz-Rodriguez J, Ruiz-Plazas X, Muñoz-Rivero MV, Celma A, García-de Manuel G, et al. Developing a Predictive Model for Significant Prostate Cancer Detection in Prostatic Biopsies from Seven Clinical Variables: Is Machine Learning Superior to Logistic Regression? Cancers. 2025; 17(7):1101. https://doi.org/10.3390/cancers17071101
Chicago/Turabian StyleMorote, Juan, Berta Miró, Patricia Hernando, Nahuel Paesano, Natàlia Picola, Jesús Muñoz-Rodriguez, Xavier Ruiz-Plazas, Marta V. Muñoz-Rivero, Ana Celma, Gemma García-de Manuel, and et al. 2025. "Developing a Predictive Model for Significant Prostate Cancer Detection in Prostatic Biopsies from Seven Clinical Variables: Is Machine Learning Superior to Logistic Regression?" Cancers 17, no. 7: 1101. https://doi.org/10.3390/cancers17071101
APA StyleMorote, J., Miró, B., Hernando, P., Paesano, N., Picola, N., Muñoz-Rodriguez, J., Ruiz-Plazas, X., Muñoz-Rivero, M. V., Celma, A., García-de Manuel, G., Servian, P., Abascal, J. M., Trilla, E., & Méndez, O. (2025). Developing a Predictive Model for Significant Prostate Cancer Detection in Prostatic Biopsies from Seven Clinical Variables: Is Machine Learning Superior to Logistic Regression? Cancers, 17(7), 1101. https://doi.org/10.3390/cancers17071101






