KIF11 and KIF14 Are a Novel Potential Prognostic Biomarker in Patients with Endometrioid Carcinoma
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Specimens
2.2. Tissue Samples
2.3. Immunohistochemistry
2.4. Expression Analysis
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics of the Study Cohort
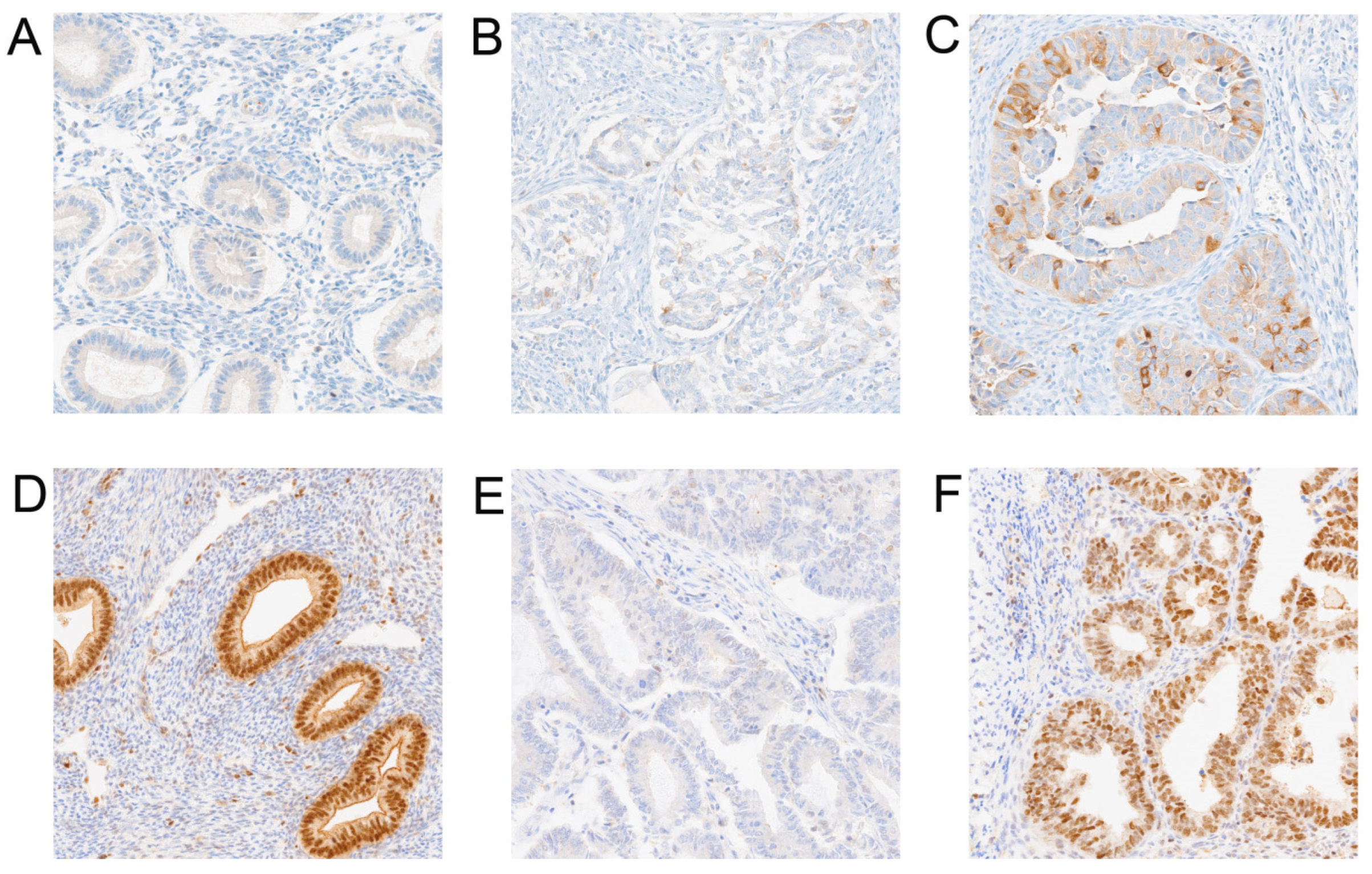
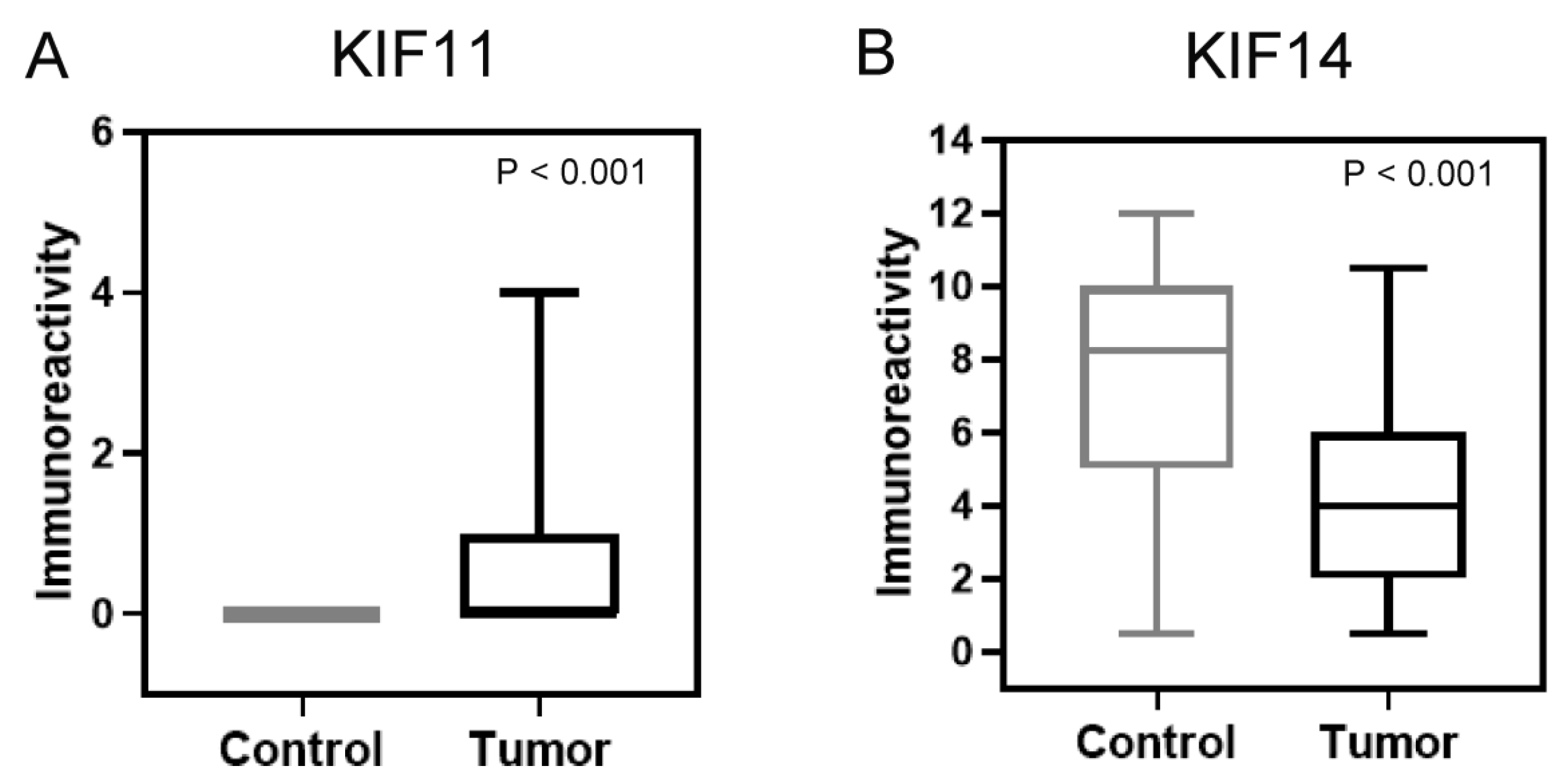
3.2. Comparison of Protein Expression Levels Between Cancer and Normal Tissues in Our Own Cohort and Evaluation of Its Relationship with Clinical-Pathological Features
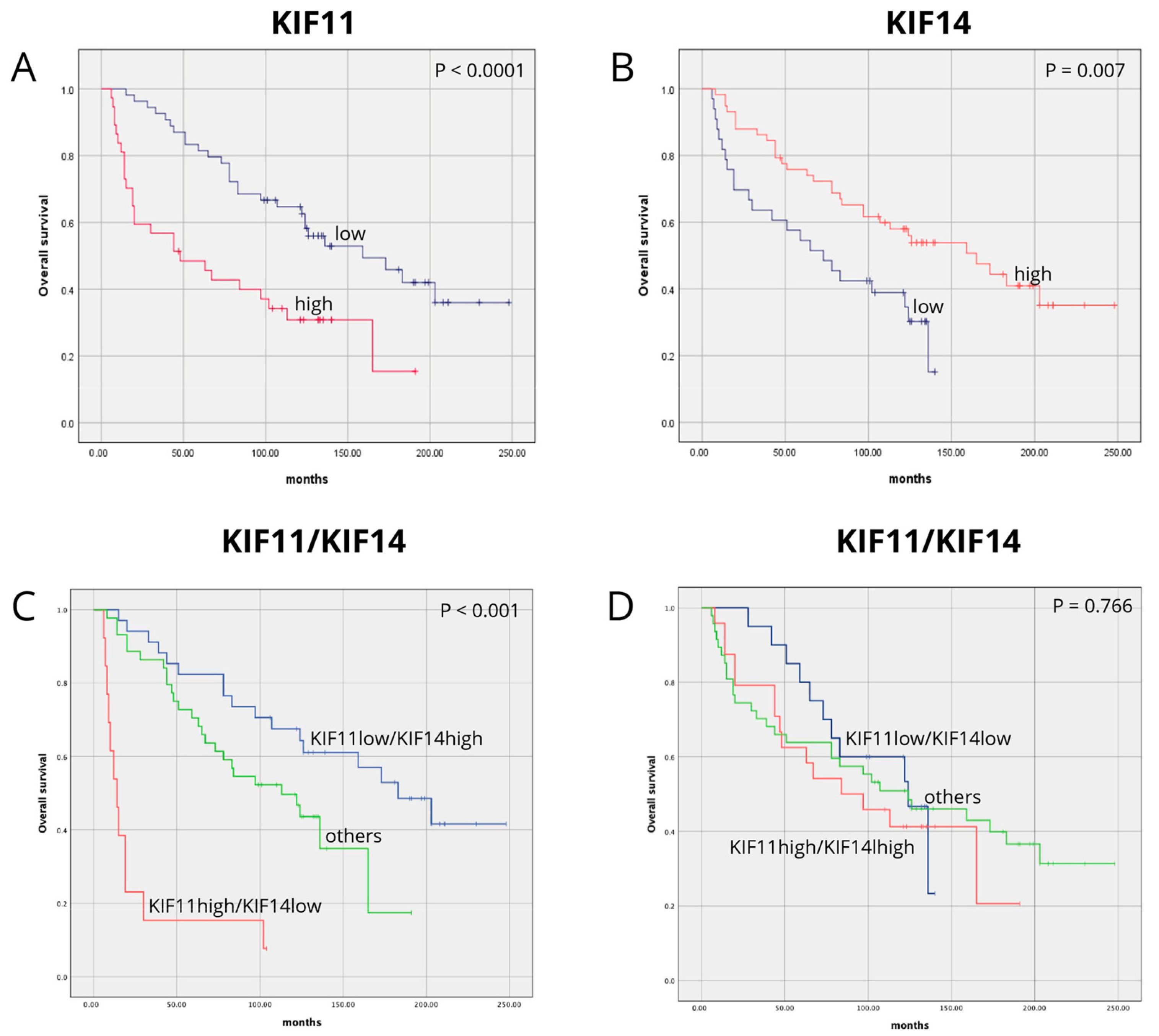
3.3. Association Between KIF11, KIF14, and Combined KIF11/KIF14 Expression and Patient Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Doherty, M.T.; Sanni, O.B.; Coleman, H.G.; Cardwell, C.R.; Glenn McCluggage, W.; Quinn, D.; Wylie, J.; McMenamin, Ú.C. Concurrent and Future Risk of Endometrial Cancer in Women with Endometrial Hyperplasia: A Systematic Review and Meta-Analysis. PLoS ONE 2020, 15, e0232231. [Google Scholar] [CrossRef] [PubMed]
- Jonusiene, V.; Sasnauskiene, A. Notch and Endometrial Cancer. Adv. Exp. Med. Biol. 2021, 1287, 47–57. [Google Scholar] [CrossRef]
- Arciuolo, D.; Travaglino, A.; Raffone, A.; Raimondo, D.; Santoro, A.; Russo, D.; Varricchio, S.; Casadio, P.; Inzani, F.; Seracchioli, R.; et al. TCGA Molecular Prognostic Groups of Endometrial Carcinoma: Current Knowledge and Future Perspectives. Int. J. Mol. Sci. 2022, 23, 11684. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.E.; González-Montiel, A.; Allos-Villalva, J.C.C.; Cantú, D.; Barquet, S.; Olivares-Mundo, A.; Herrera, L.A.; Prada, D. Prognostic Molecular Biomarkers in Endometrial Cancer: A Review. J. Cancer Res. Ther. 2019, 7, 17. [Google Scholar] [CrossRef]
- Devis-Jauregui, L.; Eritja, N.; Davis, M.L.; Matias-Guiu, X.; Llobet-Navàs, D. Autophagy in the Physiological Endometrium and Cancer. Autophagy 2021, 17, 1077–1095. [Google Scholar] [CrossRef] [PubMed]
- Sorosky, J.I. Endometrial Cancer. Obstet. Gynecol. 2012, 120, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.H.; Broaddus, R.R. Endometrial Cancer. N. Engl. J. Med. 2020, 383, 2053–2064. [Google Scholar] [CrossRef] [PubMed]
- IARC—International Agency for Research on Cancer. Available online: https://www.iarc.who.int/ (accessed on 10 February 2025).
- Green, A.K.; Feinberg, J.; Makker, V. A Review of Immune Checkpoint Blockade Therapy in Endometrial Cancer. In American Society of Clinical Oncology Educational Book; American Society of Clinical Oncology: Alexandria, VA, USA, 2020; pp. 238–244. [Google Scholar] [CrossRef]
- Van Den Heerik, A.S.V.M.; Horeweg, N.; De Boer, S.M.; Bosse, T.; Creutzberg, C.L. Adjuvant Therapy for Endometrial Cancer in the Era of Molecular Classification: Radiotherapy, Chemoradiation and Novel Targets for Therapy. Int. J. Gynecol. Cancer 2021, 31, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Münstedt, K.; Grant, P.; Woenckhaus, J.; Roth, G.; Tinneberg, H.R. Cancer of the Endometrium: Current Aspects of Diagnostics and Treatment. World J. Surg. Oncol. 2004, 2, 24. [Google Scholar] [CrossRef] [PubMed]
- Deligdisch, L. Hormonal Pathology of the Endometrium. Mod. Pathol. 2000, 13, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Miki, H.; Okada, Y.; Hirokawa, N. Analysis of the Kinesin Superfamily: Insights into Structure and Function. Trends Cell Biol. 2005, 15, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Feng, Y.M. The Role of Kinesin Family Proteins in Tumorigenesis and Progression: Potential Biomarkers and Molecular Targets for Cancer Therapy. Cancer 2010, 116, 5150–5160. [Google Scholar] [CrossRef] [PubMed]
- Castillo, A.; Morse, H.C.; Godfrey, V.L.; Naeem, R.; Justice, M.J. Overexpression of Eg5 Causes Genomic Instability and Tumor Formation in Mice. Cancer Res. 2007, 67, 10138–10147. [Google Scholar] [CrossRef]
- Shi, B.; Bao, J.; Liu, Y.; Shi, J. Death Receptor 6 Promotes Ovarian Cancer Cell Migration through KIF11. FEBS Open Bio 2018, 8, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Corson, T.W.; Gallie, B.L. KIF14 MRNA Expression Is a Predictor of Grade and Outcome in Breast Cancer. Int. J. Cancer 2006, 119, 1088–1094. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, W.R.; Yang, L.C.; Wang, J.; Sun, J.Y.; Zhang, W.W.; He, Z.Y.; Wu, S.G. KIF11 Functions as an Oncogene and Is Associated with Poor Outcomes from Breast Cancer. Cancer Res. Treat. 2019, 51, 1207–1221. [Google Scholar] [CrossRef] [PubMed]
- OSF Preprints|Over-Expression of Kinesin Family Member 11 in Human Endometrial Cancer. Available online: https://osf.io/preprints/osf/k39gx_v1 (accessed on 10 February 2025).
- Ogłuszka, M.; Orzechowska, M.; Jędroszka, D.; Witas, P.; Bednarek, A.K. Evaluate Cutpoints: Adaptable Continuous Data Distribution System for Determining Survival in Kaplan-Meier Estimator. Comput. Methods Programs Biomed. 2019, 177, 133–139. [Google Scholar] [CrossRef]
- Gao, W.; Lu, J.; Yang, Z.; Li, E.; Cao, Y.; Xie, L. Mitotic Functions and Characters of KIF11 in Cancers. Biomolecules 2024, 14, 386. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.Y.; Li, G.C.; Ran, J.; Wei, F.X. Kinesin Family Member 11 Contributes to the Progression and Prognosis of Human Breast Cancer. Oncol. Lett. 2017, 14, 6618. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, P.; Cao, X.; Liu, Y.; Jia, L.; Sun, P.; Cao, X.; Liu, Y.; Jia, L. Eg5 High Expression Predicts Dismal Prognosis in Epithelial Ovarian Cancer. Blood Genom. 2019, 3, 145–151. [Google Scholar] [CrossRef]
- Jin, Q.; Dai, Y.; Wang, Y.; Zhang, S.; Liu, G. High Kinesin Family Member 11 Expression Predicts Poor Prognosis in Patients with Clear Cell Renal Cell Carcinoma. J. Clin. Pathol. 2019, 72, 354–362. [Google Scholar] [CrossRef]
- Neska-Długosz, I.; Buchholz, K.; Durślewicz, J.; Gagat, M.; Grzanka, D.; Tojek, K.; Klimaszewska-Wiśniewska, A. Prognostic Impact and Functional Annotations of KIF11 and KIF14 Expression in Patients with Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 9732. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhou, N.; Li, J.; Kong, J.; Guan, X.; Wang, X. Eg5 Overexpression Is Predictive of Poor Prognosis in Hepatocellular Carcinoma Patients. Dis. Markers 2017, 2017, 2176460. [Google Scholar] [CrossRef] [PubMed]
- Klimaszewska-Wiśniewska, A.; Neska-Długosz, I.; Buchholz, K.; Durślewicz, J.; Grzanka, D.; Kasperska, A.; Antosik, P.; Zabrzyński, J.; Grzanka, A.; Gagat, M. Prognostic Significance of KIF11 and KIF14 Expression in Pancreatic Adenocarcinoma. Cancers 2021, 13, 3017. [Google Scholar] [CrossRef]
- Daigo, K.; Takano, A.; Thang, P.M.; Yoshitake, Y.; Shinohara, M.; Tohnai, I.; Murakami, Y.; Maegawa, J.; Daigo, Y. Characterization of KIF11 as a Novel Prognostic Biomarker and Therapeutic Target for Oral Cancer. Int. J. Oncol. 2018, 52, 155–165. [Google Scholar] [CrossRef]
- Liu, J.; Tian, Y.; Yi, L.; Gao, Z.; Lou, M.; Yuan, K. High KIF11 Expression Is Associated with Poor Outcome of NSCLC. Tumori 2022, 108, 40–46. [Google Scholar] [CrossRef]
- Guo, X.; Zhou, L.; Wu, Y.; Li, J. KIF11 As a Potential Pan-Cancer Immunological Biomarker Encompassing the Disease Staging, Prognoses, Tumor Microenvironment, and Therapeutic Responses. Oxid. Med. Cell. Longev. 2022, 2022, 2764940. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, Y.; Kamakura, S.; Hayase, J.; Sumimoto, H. Interaction of NuMA Protein with the Kinesin Eg5: Its Possible Role in Bipolar Spindle Assembly and Chromosome Alignment. Biochem. J. 2013, 451, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Bao, L.; Li, X.; Sun, G.; Yang, W.; Xie, N.; Lei, L.; Chen, W.; Zhang, H.; Chen, M.; et al. Identification and Validation of the Important Role of KIF11 in the Development and Progression of Endometrial Cancer. J. Transl. Med. 2025, 23, 48. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.; Dong, M.; Zhang, K.; Tian, W.; Wang, Y.; Xue, F. Bioinformatics Analysis of Key Differentially Expressed Genes in Well and Poorly Differentiated Endometrial Carcinoma. Mol. Med. Rep. 2018, 18, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhuang, H.; Xia, R.; Gan, L.; Wu, Y.; Ma, J.; Sun, Y.; Zhuang, Z. KIF11 Is Required for Proliferation and Self-Renewal of Docetaxel Resistant Triple Negative Breast Cancer Cells. Oncotarget 2017, 8, 92106. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, L.; Xiong, L.; Wang, K.; Hou, X.; Li, Q.; Kong, F.; Liu, X.; He, J. KIF11 Is Upregulated in Colorectal Cancer and Silencing of It Impairs Tumor Growth and Sensitizes Colorectal Cancer Cells to Oxaliplatin via P53/GSK3β Signaling. J. Cancer 2021, 12, 3741–3753. [Google Scholar] [CrossRef]
- Hagan, I.; Yanagida, M. Novel Potential Mitotic Motor Protein Encoded by the Fission Yeast Cut7+ Gene. Nature 1990, 347, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Slangy, A.; Lane, H.A.; d’Hérin, P.; Harper, M.; Kress, M.; Niggt, E.A. Phosphorylation by P34cdc2 Regulates Spindle Association of Human Eg5, a Kinesin-Related Motor Essential for Bipolar Spindle Formation in Vivo. Cell 1995, 83, 1159–1169. [Google Scholar] [CrossRef]
- Weil, D.; Garçon, L.; Harper, M.; Duménil, D.; Dautry, F.; Kress, M. Targeting the Kinesin Eg5 to Monitor SiRNA Transfection in Mammalian Cells. Biotechniques 2002, 33, 1244–1248. [Google Scholar] [CrossRef] [PubMed]
- Singel, S.M.; Cornelius, C.; Zaganjor, E.; Batten, K.; Sarode, V.R.; Buckley, D.L.; Peng, Y.; John, G.B.; Li, H.C.; Sadeghi, N.; et al. KIF14 Promotes AKT Phosphorylation and Contributes to Chemoresistance in Triple-Negative Breast Cancer. Neoplasia 2014, 16, 247–256.e2. [Google Scholar] [CrossRef] [PubMed]
- Hung, P.F.; Hong, T.M.; Hsu, Y.C.; Chen, H.Y.; Chang, Y.L.; Wu, C.T.; Chang, G.C.; Jou, Y.S.; Pan, S.H.; Yang, P.C. The Motor Protein KIF14 Inhibits Tumor Growth and Cancer Metastasis in Lung Adenocarcinoma. PLoS ONE 2013, 8, e61664. [Google Scholar] [CrossRef] [PubMed]
- Gerashchenko, T.S.; Zolotaryova, S.Y.; Kiselev, A.M.; Tashireva, L.A.; Novikov, N.M.; Krakhmal, N.V.; Cherdyntseva, N.V.; Zavyalova, M.V.; Perelmuter, V.M.; Denisov, E.V. The Activity of KIF14, Mieap, and EZR in a New Type of the Invasive Component, Torpedo-Like Structures, Predetermines the Metastatic Potential of Breast Cancer. Cancers 2020, 12, 1909. [Google Scholar] [CrossRef]
- Yang, Z.; Li, C.; Yan, C.; Li, J.; Yan, M.; Liu, B.; Zhu, Z.; Wu, Y.; Gu, Q. KIF14 Promotes Tumor Progression and Metastasis and Is an Independent Predictor of Poor Prognosis in Human Gastric Cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhang, X.B.; Zheng, Z.M. Suppression of KIF14 Expression Inhibits Hepatocellular Carcinoma Progression and Predicts Favorable Outcome. Cancer Sci. 2013, 104, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, L.; Li, D.; Deng, J.; Zhao, Z.; He, S.; Zhang, Y.; Tu, Y. Kinesin Family Member 14 Is a Candidate Prognostic Marker for Outcome of Glioma Patients. Cancer Epidemiol. 2013, 37, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Li, K.K.W.; Qi, Y.; Xia, T.; Chan, A.K.Y.; Zhang, Z.Y.; Aibaidula, A.; Zhang, R.; Zhou, L.; Yao, Y.; Ng, H.K. The Kinesin KIF14 Is Overexpressed in Medulloblastoma and Downregulation of KIF14 Suppressed Tumor Proliferation and Induced Apoptosis. Lab. Investig. 2017, 97, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Corson, T.W.; Chang, Q.Z.; Lau, S.K.; Shepherd, F.A.; Tsao, M.S.; Gallie, B.L. KIF14 Messenger RNA Expression Is Independently Prognostic for Outcome in Lung Cancer. Clin. Cancer Res. 2007, 13, 3229–3234. [Google Scholar] [CrossRef] [PubMed]
- Li, T.F.; Zeng, H.J.; Shan, Z.; Ye, R.Y.; Cheang, T.Y.; Zhang, Y.J.; Lu, S.H.; Zhang, Q.; Shao, N.; Lin, Y. Overexpression of Kinesin Superfamily Members as Prognostic Biomarkers of Breast Cancer. Cancer Cell Int. 2020, 20, 123. [Google Scholar] [CrossRef]
| KIF11 | KIF14 | ||||||
|---|---|---|---|---|---|---|---|
| Variables | Number | High | Low | p Value | High | Low | p Value |
| n = 38 | n = 54 | n = 58 | n = 34 | ||||
| Age | |||||||
| ≤60 | 30 | 10 | 20 | 0.3673 | 21 | 9 | 0.3669 |
| >60 | 62 | 28 | 34 | 37 | 25 | ||
| Histologic grade | |||||||
| G1 | 7 | 1 | 6 | 0.1026 | 4 | 3 | 0.7181 |
| G2 | 60 | 23 | 37 | 36 | 24 | ||
| G3 | 25 | 14 | 11 | 18 | 17 | ||
| pT status | |||||||
| T1 | 54 | 22 | 32 | 0.7133 | 34 | 20 | 0.6209 |
| T2 | 22 | 11 | 11 | 14 | 8 | ||
| T3 | 13 | 4 | 9 | 10 | 3 | ||
| T4 | 3 | 1 | 2 | 0 | 3 | ||
| pN status | |||||||
| N0 | 79 | 31 | 48 | 0.3713 | 50 | 29 | >0.9999 |
| N1 | 13 | 7 | 6 | 8 | 5 | ||
| pM status | |||||||
| M0 | 84 | 33 | 51 | 0.2675 | 55 | 29 | 0.1403 |
| M1 | 8 | 5 | 3 | 3 | 5 | ||
| Lymphovascular invasion | |||||||
| Present | 16 | 9 | 7 | 0.7800 | 6 | 10 | 0.0253 |
| Absent | 76 | 47 | 29 | 52 | 24 | ||
| FIGO | |||||||
| I | 48 | 19 | 29 | 0.3259 | 30 | 18 | 0.3918 |
| II | 18 | 9 | 9 | 12 | 6 | ||
| III | 18 | 5 | 13 | 13 | 5 | ||
| IV | 8 | 5 | 3 | 3 | 5 | ||
| Variable | Univariate Analysis of Own Cohort | Multivariate Analysis of Own Cohort: KIF11 | Multivariate Analysis of Own Cohort: KIF14 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p Value | HR | 95% CI | p Value | HR | 95% CI | p Value | ||||
| Lower | Upper | Lower | Upper | Lower | Upper | |||||||
| KIF11 | 2.645 | 1.540 | 4.542 | <0.0001 | 2.327 | 1.346 | 4.023 | 0.002 | - | - | - | - |
| KIF14 | 2.106 | 1.210 | 3.665 | 0.008 | - | - | - | - | 2.127 | 1.150 | 3.937 | 0.016 |
| age | 3.561 | 1.721 | 7.367 | 0.001 | 3.227 | 1.473 | 7.068 | 0.003 | 3.167 | 1.472 | 6.813 | 0.003 |
| grade | 2.656 | 1.538 | 4.585 | <0.0001 | 2.478 | 1.300 | 4.725 | 0.006 | 2.963 | 1.482 | 5.921 | 0.002 |
| pT | 1.273 | 0.747 | 2.170 | 0.374 | 0.704 | 0.366 | 1.353 | 0.292 | 0.704 | 0.362 | 1.369 | 0.300 |
| pN | 1.974 | 1.017 | 3.832 | 0.044 | 1.624 | 0.686 | 3.845 | 0.270 | 1.858 | 0.739 | 4.670 | 0.188 |
| pM | 1.489 | 0.588 | 3.768 | 0.401 | 0.593 | 0.184 | 1.915 | 0.383 | 0.286 | 0.074 | 1.104 | 0.069 |
| LVSI | 5.419 | 2.873 | 10.221 | <0.0001 | 3.529 | 1.785 | 6.979 | <0.0001 | 3.419 | 1.637 | 7.140 | 0.001 |
| Variable | Univariate Analysis of Own Cohort | |||
|---|---|---|---|---|
| HR | 95% CI | p Value | ||
| Lower | Upper | |||
| Others | ||||
| KIF11lowKIF14high | 0.515 | 0.267 | 0.996 | 0.048 |
| KIF11highKIF14low | 4.663 | 2.339 | 9.296 | <0.0001 |
| Variable | Multivariate Analysis of Own Cohort: KIF11/KIF14 | |||
|---|---|---|---|---|
| HR | 95% CI | p Value | ||
| Lower | Upper | |||
| Others | ||||
| KIF11lowKIF14high | 0.570 | 0.284 | 1.143 | 0.113 |
| KIF11highKIF14low | 3.769 | 1.806 | 7.869 | <0.0001 |
| age | 2.780 | 1.263 | 6.119 | 0.011 |
| grading | 2.636 | 1.343 | 5.172 | 0.005 |
| pT | 0.675 | 0.347 | 1.313 | 0.247 |
| pN | 2.102 | 0.840 | 5.259 | 0.112 |
| pM | 0.365 | 0.103 | 1.292 | 0.118 |
| LVSI | 3.506 | 1.702 | 7.220 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antosik, P.; Durślewicz, J.; Smolińska-Świtała, M.; Podemski, J.; Podemska, E.; Neska-Długosz, I.; Jóźwicki, J.; Grzanka, D. KIF11 and KIF14 Are a Novel Potential Prognostic Biomarker in Patients with Endometrioid Carcinoma. Cancers 2025, 17, 804. https://doi.org/10.3390/cancers17050804
Antosik P, Durślewicz J, Smolińska-Świtała M, Podemski J, Podemska E, Neska-Długosz I, Jóźwicki J, Grzanka D. KIF11 and KIF14 Are a Novel Potential Prognostic Biomarker in Patients with Endometrioid Carcinoma. Cancers. 2025; 17(5):804. https://doi.org/10.3390/cancers17050804
Chicago/Turabian StyleAntosik, Paulina, Justyna Durślewicz, Marta Smolińska-Świtała, Jonasz Podemski, Edyta Podemska, Izabela Neska-Długosz, Jakub Jóźwicki, and Dariusz Grzanka. 2025. "KIF11 and KIF14 Are a Novel Potential Prognostic Biomarker in Patients with Endometrioid Carcinoma" Cancers 17, no. 5: 804. https://doi.org/10.3390/cancers17050804
APA StyleAntosik, P., Durślewicz, J., Smolińska-Świtała, M., Podemski, J., Podemska, E., Neska-Długosz, I., Jóźwicki, J., & Grzanka, D. (2025). KIF11 and KIF14 Are a Novel Potential Prognostic Biomarker in Patients with Endometrioid Carcinoma. Cancers, 17(5), 804. https://doi.org/10.3390/cancers17050804







