Identification of the Interacting Domains Between Tissue Factor and β1-Integrin and the Signalling Properties of the Two Fibronectin-like Domains of Tissue Factor
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
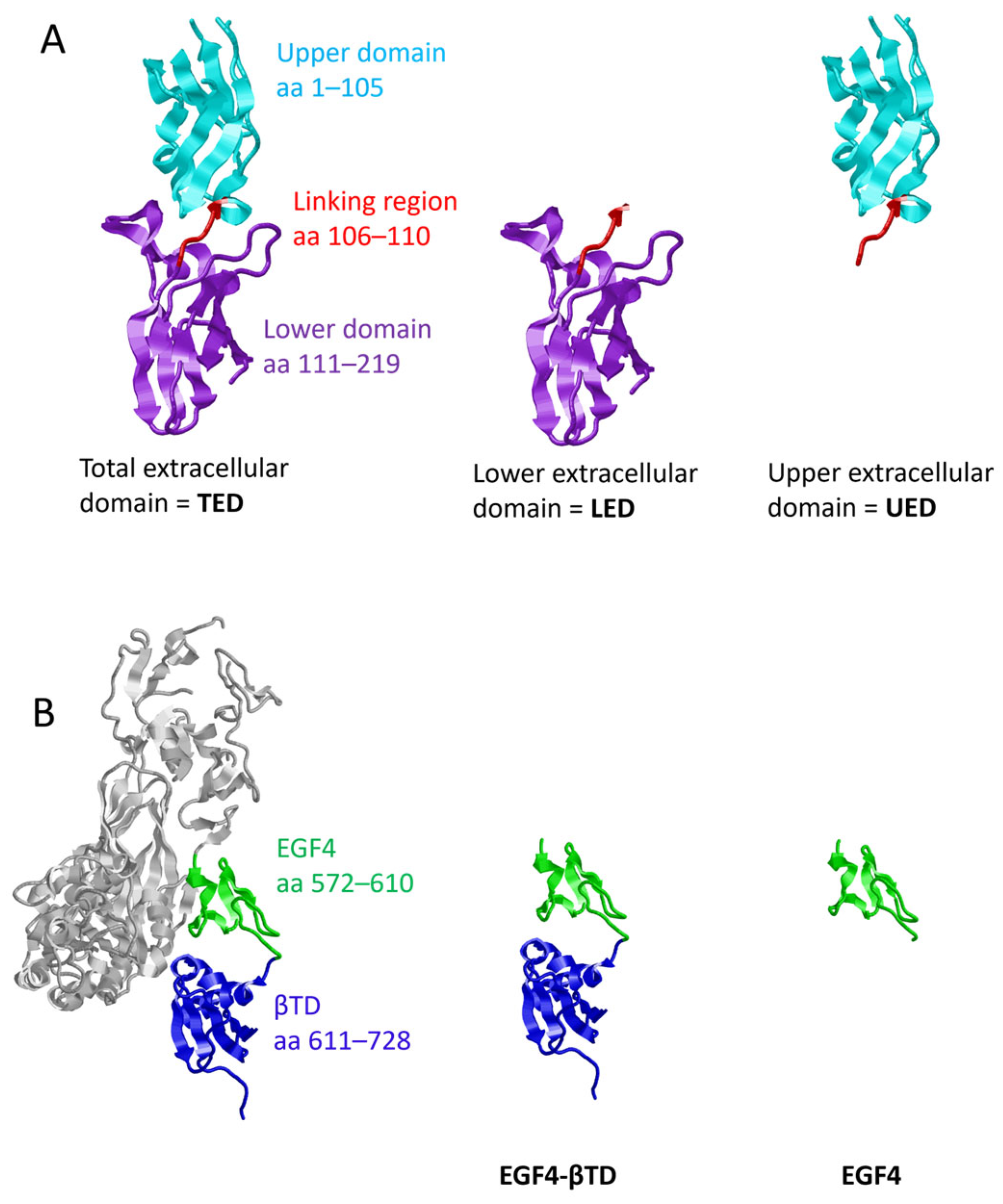
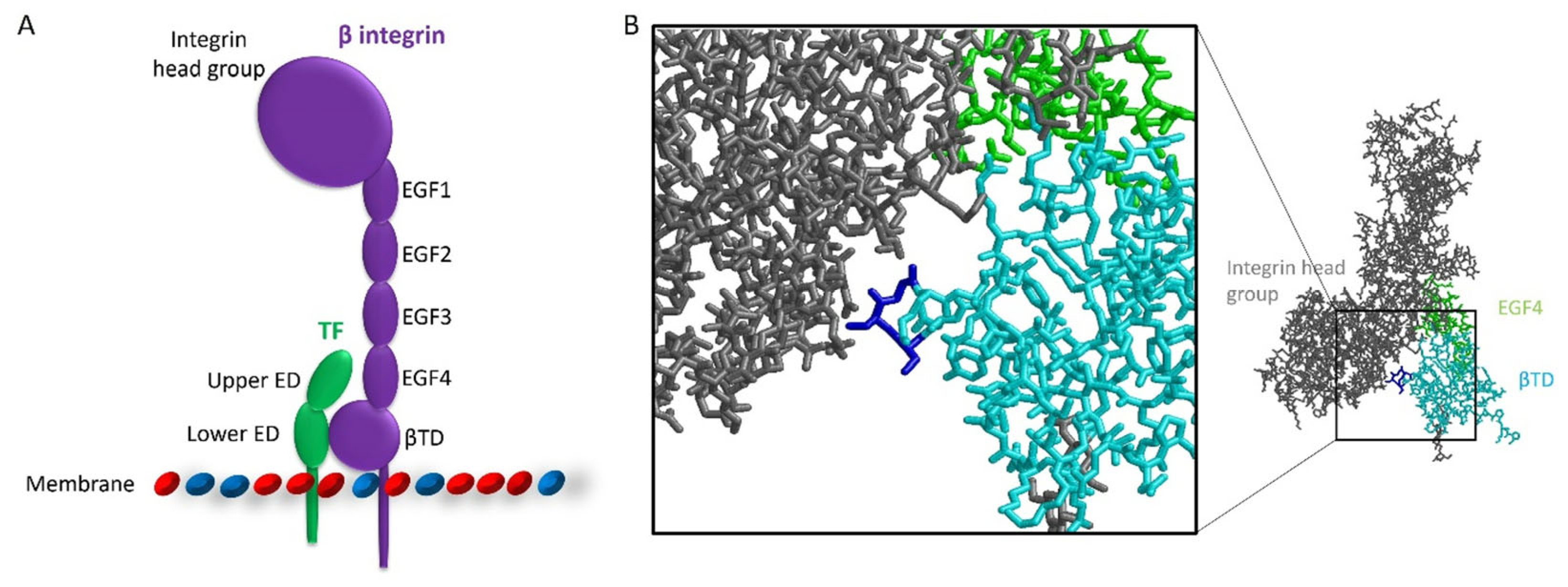
2.1. Preparation of Plasmid Constructs for the Expression of TF and β1-Integrin Domains
2.2. Cell Culture and Transfection of Plasmid Constructs
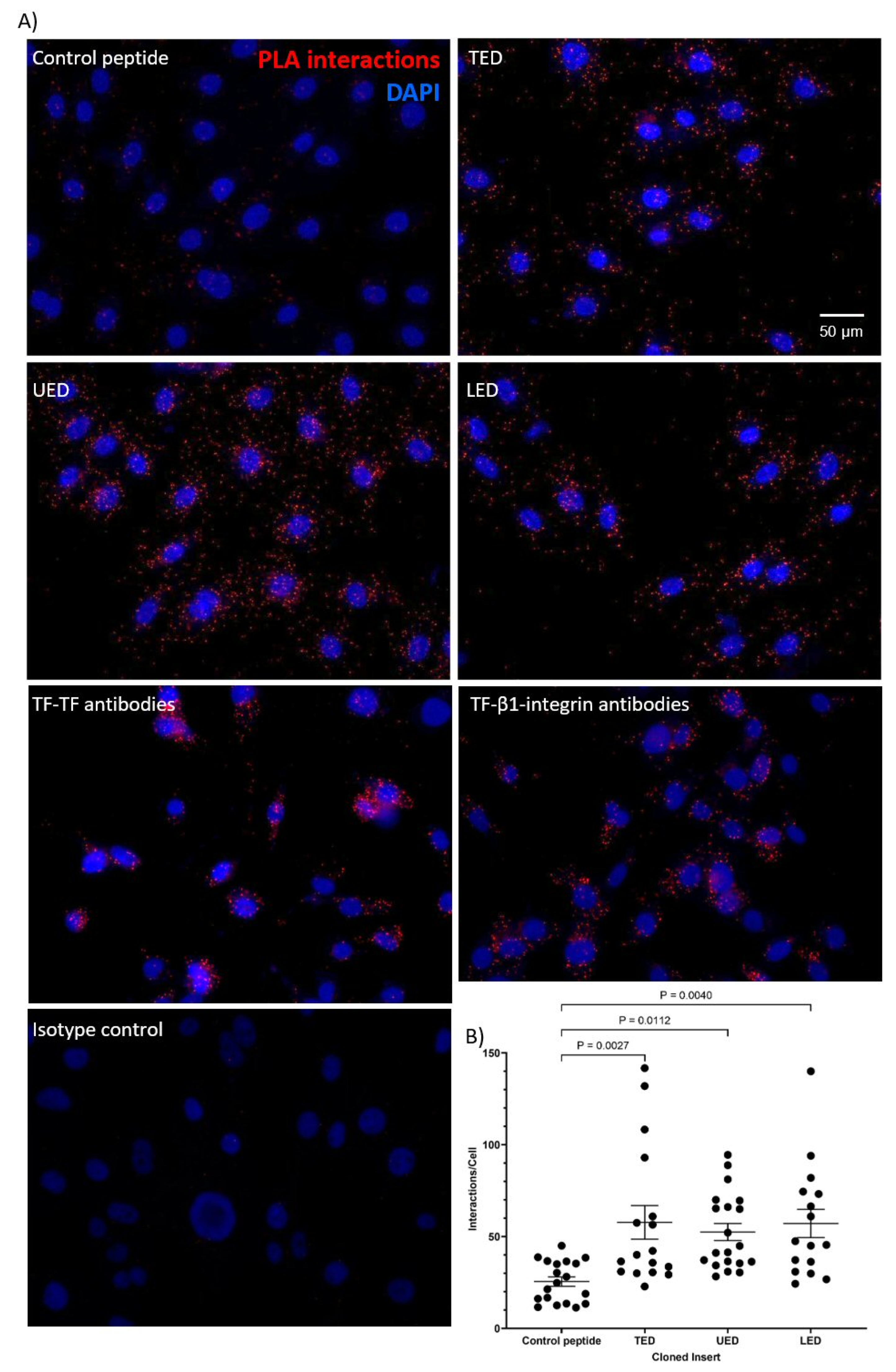
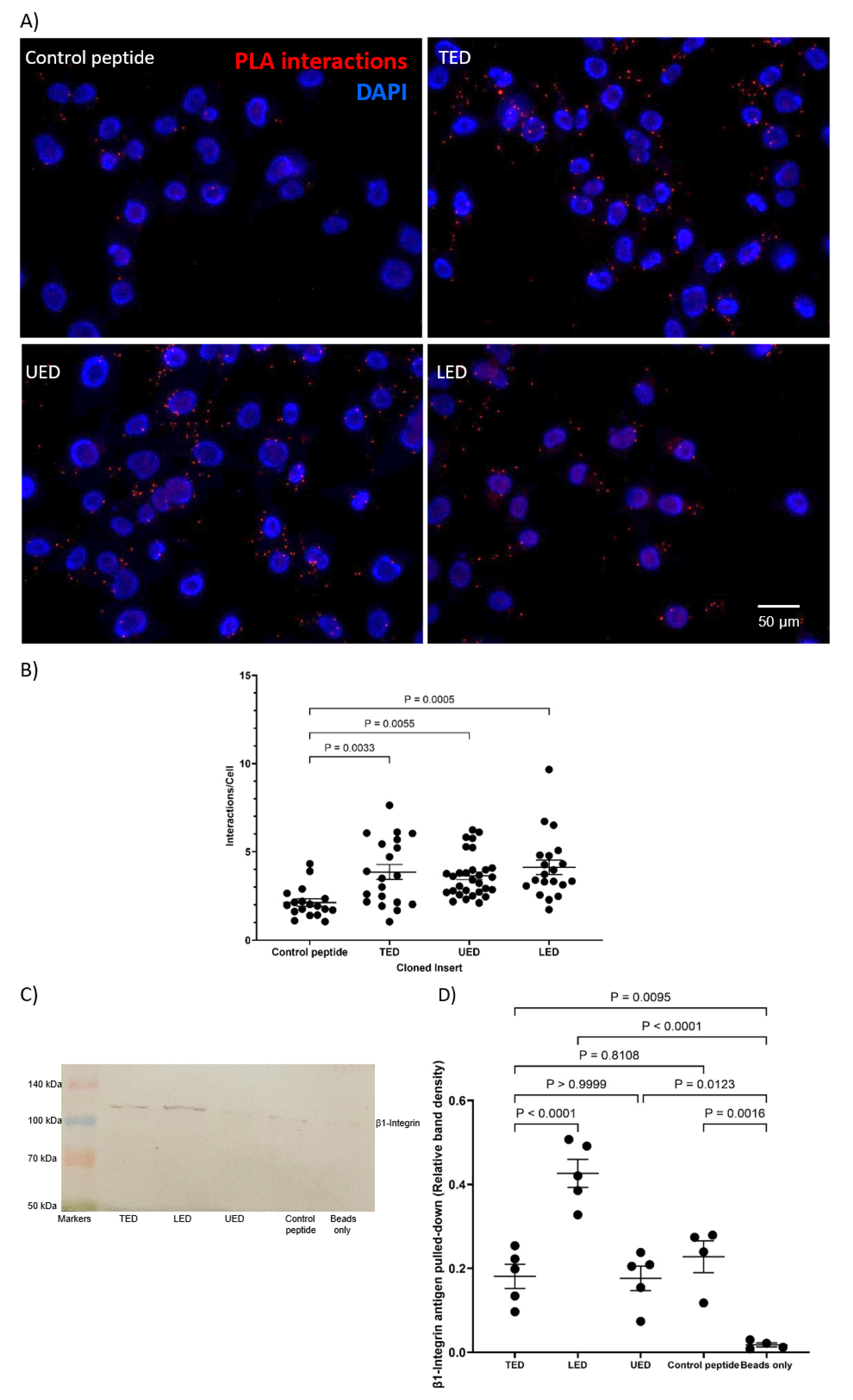
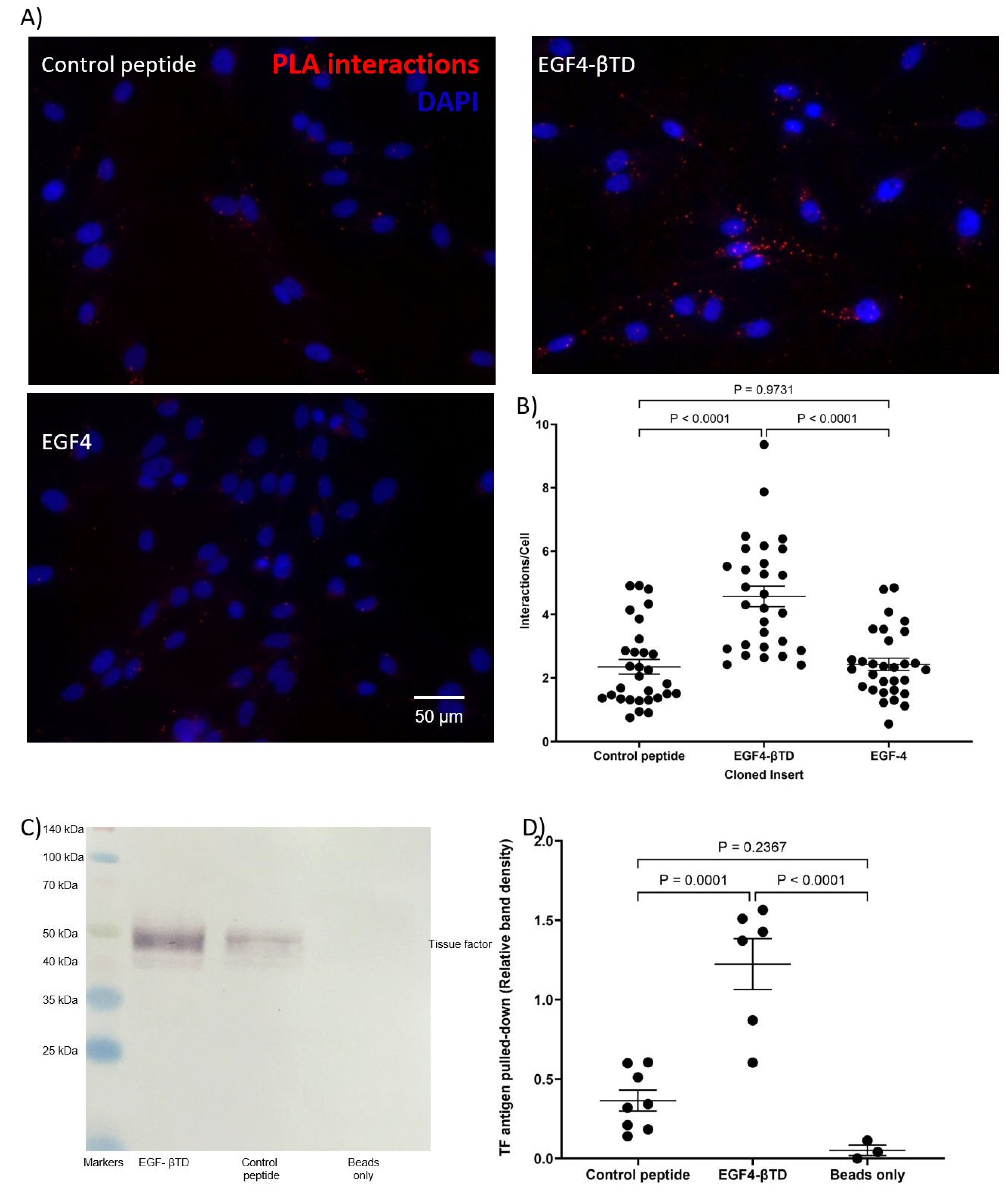
2.3. Analysis of Protein Interaction by Proximity Ligation Assay (PLA)
2.4. Analysis of Protein Interaction by Co-Immunoprecipitation
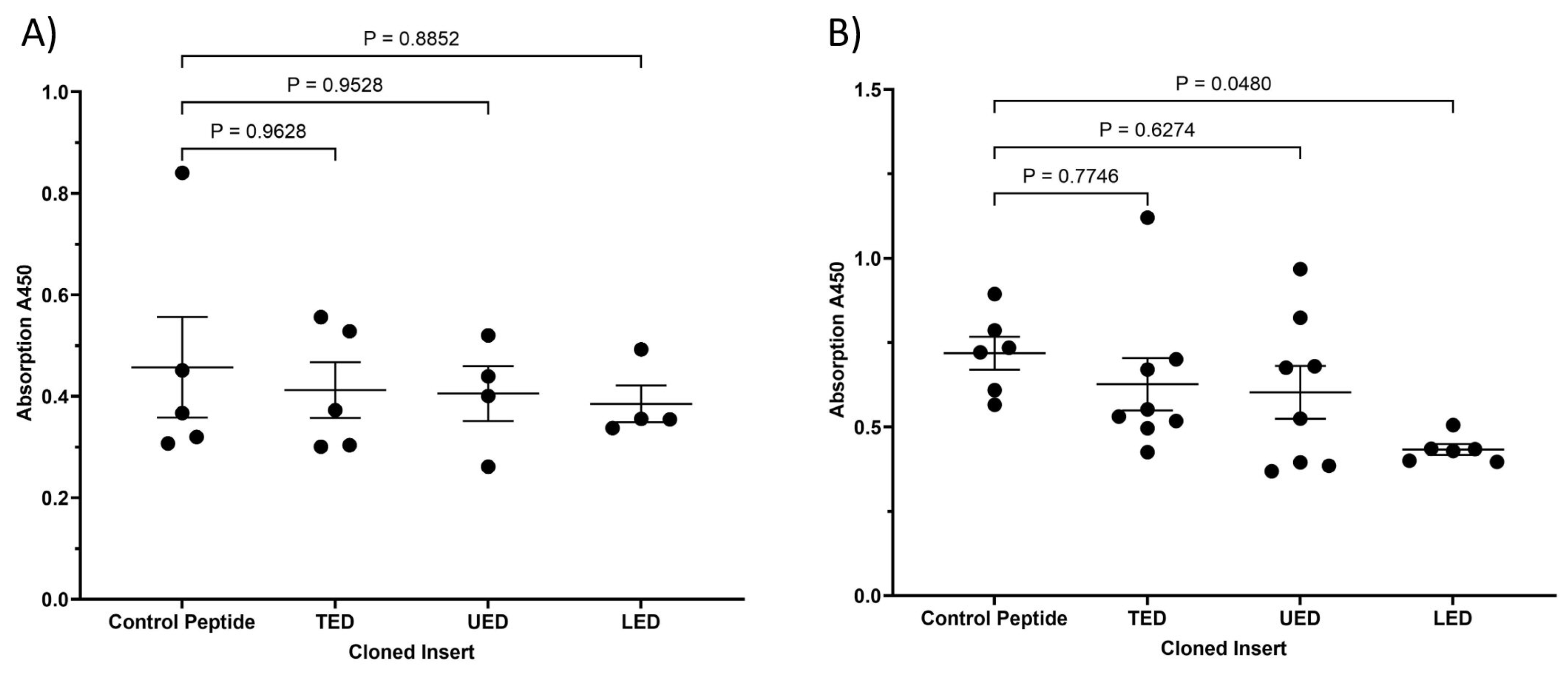
2.5. Assessment of the Conformation of β1-Integrin Using Antibody Binding Assay
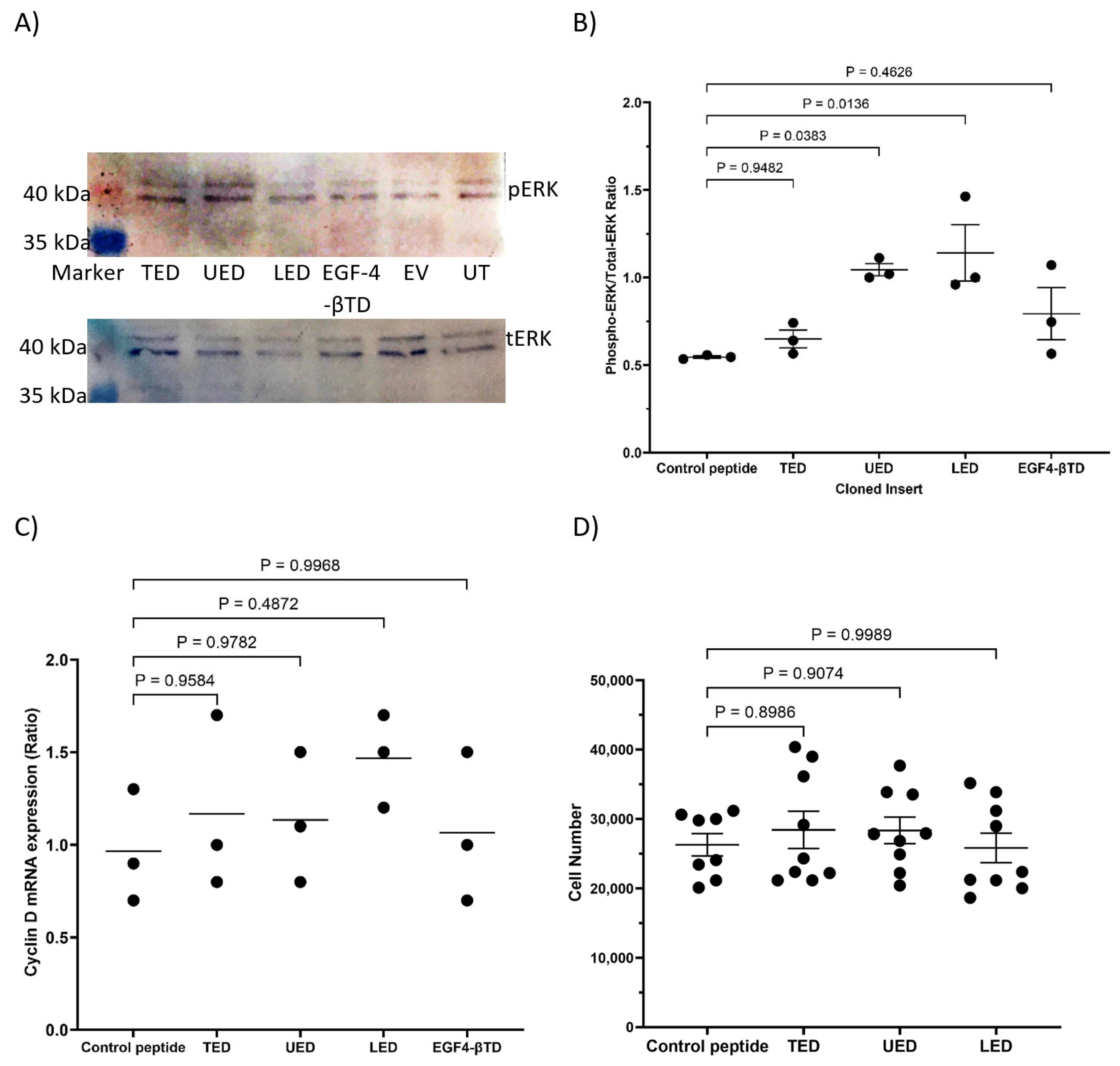
2.6. Measurement of ERK1/2 Phosphorylation by SDS-PAGE and Western Blot Analysis
2.7. Preparation of cDNA and Quantification of Cyclin D1 Expression by qPCR
2.8. Determination of Cell Numbers by Crystal Violet Assay
2.9. Statistical Analysis
3. Results
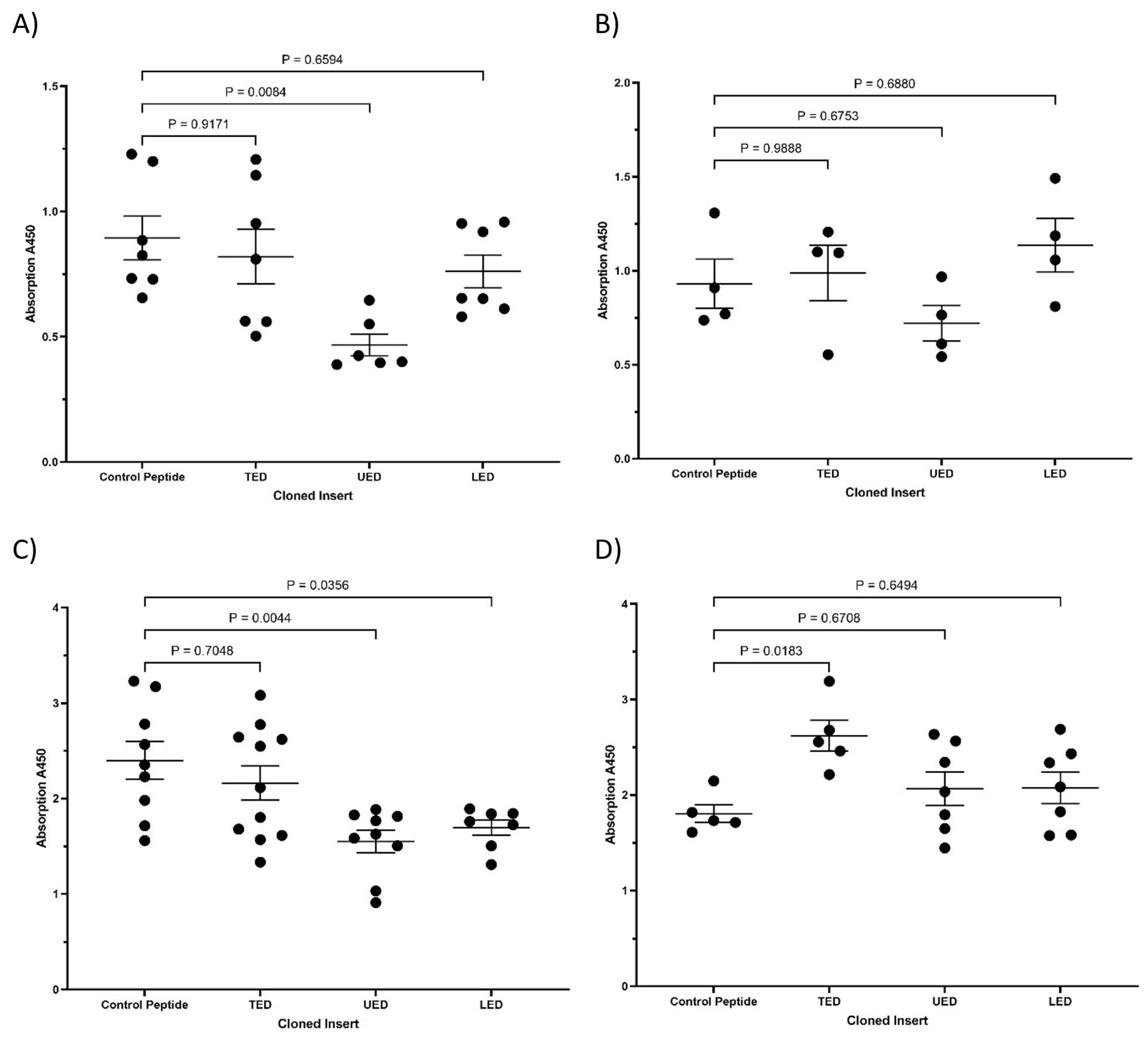
3.1. Identification of the Interacting Domains Between TF and β1-Integrin by PLA and Co-Immunoprecipitation
3.2. Characterisation of the Influence of Expressed TF Domains on the Conformation of β1-Integrin
3.3. Analysis of the Outcome of the Expression of the Construct Peptides on Cellular Proliferation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Butenas, S. Tissue factor structure and function. Scientifica 2012, 2012, 964862. [Google Scholar] [CrossRef]
- Morrissey, J.H.; Fakhrai, H.; Edgington, T.S. Molecular cloning of the cDNA for tissue factor, the cellular receptor for the initiation of the coagulation protease cascade. Cell 1987, 50, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Spicer, E.K.; Horton, R.; Bloem, L.; Bach, R.; Williams, K.R.; Guha, A.; Kraus, J.; Lin, T.C.; Nemerson, Y.; Konigsberg, W.H. Isolation of cDNA clones coding for human tissue factor: Primary structure of the protein and cDNA. Proc. Natl. Acad. Sci. USA 1987, 84, 5148–5152. [Google Scholar] [CrossRef]
- Mackman, N. The role of tissue factor and factor VIIa in hemostasis. Anesth. Analg. 2009, 108, 1447–1452. [Google Scholar] [CrossRef] [PubMed]
- Drake, T.A.; Morrissey, J.H.; Edgington, T.S. Selective cellular expression of tissue factor in human tissues. Implications for disorders of hemostasis and thrombosis. Am. J. Pathol. 1989, 134, 1087–1097. [Google Scholar] [PubMed]
- Unruh, D.; Horbinski, C. Beyond thrombosis: The impact of tissue factor signaling in cancer. J. Hematol. Oncol. 2020, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, Y.W.; Osanto, S.; Reitsma, P.H.; Versteeg, H.H. The relationship between tissue factor and cancer progression: Insights from bench and bedside. Blood 2012, 119, 924–932. [Google Scholar] [CrossRef]
- Kocaturk, B.; Versteeg, H.H. Tissue factor-integrin interactions in cancer and thrombosis: Every Jack has his Jill. J. Thromb. Haemost. 2013, 11 (Suppl. 1), 285–293. [Google Scholar] [CrossRef] [PubMed]
- Campbell, I.D.; Humphries, M.J. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a004994. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhou, C.; Ma, R.; Guo, Q.; Huang, H.; Hao, J.; Liu, H.; Shi, R.; Liu, B. Prognostic value of increased integrin-beta 1 expression in solid cancers: A meta-analysis. OncoTargets Ther. 2018, 11, 1787–1799. [Google Scholar] [CrossRef]
- Dorfleutner, A.; Hintermann, E.; Tarui, T.; Takada, Y.; Ruf, W. Cross-talk of integrin alpha3beta1 and tissue factor in cell migration. Mol. Biol. Cell 2004, 15, 4416–4425. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; Albrecht, S.; Golfert, F.; Hofer, A.; Funk, R.H.; Magdolen, V.; Flossel, C.; Luther, T. Localization of tissue factor in actin-filament-rich membrane areas of epithelial cells. Exp. Cell Res. 1999, 248, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Collier, M.E.; Li, C.; Ettelaie, C. Influence of exogenous tissue factor on estrogen receptor alpha expression in breast cancer cells: Involvement of beta1-integrin, PAR2, and mitogen-activated protein kinase activation. Mol. Cancer Res. 2008, 6, 1807–1818. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, H.H.; Schaffner, F.; Kerver, M.; Petersen, H.H.; Ahamed, J.; Felding-Habermann, B.; Takada, Y.; Mueller, B.M.; Ruf, W. Inhibition of tissue factor signaling suppresses tumor growth. Blood 2008, 111, 190–199. [Google Scholar] [CrossRef]
- Collier, M.E.; Ettelaie, C. Induction of endothelial cell proliferation by recombinant and microparticle-tissue factor involves beta1-integrin and extracellular signal regulated kinase activation. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1810–1817. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pradier, A.; Ettelaie, C. The influence of exogenous tissue factor on the regulators of proliferation and apoptosis in endothelial cells. J. Vasc. Res. 2008, 45, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Kocaturk, B.; Van den Berg, Y.W.; Tiekena, C.; Mieog, J.S.D.; de Kruijf, E.M.; Engels, C.C.; van der Ent, M.A.; Kuppen, P.J.; Van De Velde, C.J.; Ruf, W.; et al. Alternatively spliced tissue factor promotes breast cancer growth in a beta 1 integrin-dependent manner. Proc. Natl. Acad. Sci. USA 2013, 110, 11517–11522. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, Y.W.; van den Hengel, L.G.; Myers, H.R.; Ayachi, O.; Jordanova, E.; Ruf, W.; Spek, C.A.; Reitsma, P.H.; Bogdanov, V.Y.; Versteeg, H.H. Alternatively spliced tissue factor induces angiogenesis through integrin ligation. Proc. Natl. Acad. Sci. USA 2009, 106, 19497–19502. [Google Scholar] [CrossRef] [PubMed]
- Rothmeier, A.S.; Liu, E.; Chakrabarty, S.; Disse, J.; Mueller, B.M.; Ostergaard, H.; Ruf, W. Identification of the integrin-binding site on coagulation factor VIIa required for proangiogenic PAR2 signaling. Blood 2018, 131, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, J.E.; Zakarija, A.; Cundiff, D.L.; Doll, J.A.; Hymen, E.; Cornwell, M.; Crawford, S.E.; Liu, N.; Signaevsky, M.; Soff, G.A. Alternatively spliced human tissue factor promotes tumor growth and angiogenesis in a pancreatic cancer tumor model. Thromb. Res. 2007, 120 (Suppl. 2), S13–S21. [Google Scholar] [CrossRef]
- Teplyakov, A.; Obmolova, G.; Malia, T.J.; Wu, B.; Zhao, Y.; Taudte, S.; Anderson, G.M.; Gilliland, G.L. Crystal structure of tissue factor in complex with antibody 10H10 reveals the signaling epitope. Cell. Signal. 2017, 36, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.H.; Carman, C.V.; Springer, T.A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007, 25, 619–647. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.; Giancotti, F.G. Integrin signaling in cancer: Mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 2019, 35, 347–367. [Google Scholar] [CrossRef] [PubMed]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Collier, M.E.W.; Ettelaie, C.; Goult, B.T.; Maraveyas, A.; Goodall, A.H. Investigation of the filamin A-dependent mechanisms of tissue factor incorporation into microvesicles. Thromb. Haemost. 2017, 117, 2034–2044. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mohammad, M.A.; Greenman, J.; Maraveyas, A.; Ettelaie, C. Activation of PAR2 by tissue factor induces the release of the PTEN from MAGI proteins and regulates PTEN and Akt activities. Sci. Rep. 2020, 10, 20908. [Google Scholar] [CrossRef] [PubMed]
- Madkhali, Y.; Rondon, A.M.R.; Featherby, S.; Maraveyas, A.; Greenman, J.; Ettelaie, C. Factor VIIa Regulates the Level of Cell-Surface Tissue Factor through Separate but Cooperative Mechanisms. Cancers 2021, 13, 3718. [Google Scholar] [CrossRef]
- Bazzoni, G.; Shih, D.T.; Buck, C.A.; Hemler, M.E. Monoclonal-antibody 9EG7 defines a novel beta(1) integrin epitope induced by soluble ligand and manganese, but inhibited by calcium. J. Biol. Chem. 1995, 270, 25570–25577. [Google Scholar] [CrossRef] [PubMed]
- Park, C.C.; Zhang, H.; Pallavicini, M.; Gray, J.W.; Baehner, F.; Park, C.J.; Bissell, M.J. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Res. 2006, 66, 1526–1535. [Google Scholar] [CrossRef]
- Spiess, M.; Hernandez-Varas, P.; Oddone, A.; Olofsson, H.; Blom, H.; Waithe, D.; Lock, J.G.; Lakadamyali, M.; Stromblad, S. Active and inactive beta1 integrins segregate into distinct nanoclusters in focal adhesions. J. Cell Biol. 2018, 217, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Bijwaard, K.E.; Aguilera, N.S.; Monczak, Y.; Trudel, M.; Taubenberger, J.K.; Lichy, J.H. Quantitative real-time reverse transcription-PCR assay for cyclin D1 expression: Utility in the diagnosis of mantle cell lymphoma. Clin. Chem. 2001, 47, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- ElKeeb, A.M.; Collier, M.E.; Maraveyas, A.; Ettelaie, C. Accumulation of tissue factor in endothelial cells induces cell apoptosis, mediated through p38 and p53 activation. Thromb. Haemost. 2015, 114, 364–378. [Google Scholar] [CrossRef]
- Ettelaie, C.; Collier, M.E.; Mei, M.P.; Xiao, Y.P.; Maraveyas, A. Enhanced binding of tissue factor-microparticles to collagen-IV and fibronectin leads to increased tissue factor activity in vitro. Thromb. Haemost. 2013, 109, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, H.H.; Ruf, W. Emerging insights in tissue factor-dependent signaling events. Semin. Thromb. Hemost. 2006, 32, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Kakkar, A.K.; Lemoine, N.R.; Scully, M.F.; Tebbutt, S.; Williamson, R.C. Tissue factor expression correlates with histological grade in human pancreatic cancer. Br. J. Surg. 1995, 82, 1101–1104. [Google Scholar] [CrossRef]
- Seto, S.; Onodera, H.; Kaido, T.; Yoshikawa, A.; Ishigami, S.; Arii, S.; Imamura, M. Tissue factor expression in human colorectal carcinoma: Correlation with hepatic metastasis and impact on prognosis. Cancer 2000, 88, 295–301. [Google Scholar] [CrossRef]
- Rothmeier, A.S.; Versteeg, H.H.; Ruf, W. Factor VIIa-induced interaction with integrin controls the release of tissue factor on extracellular vesicles from endothelial cells. J. Thromb. Haemost. 2019, 17, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Gylling, A.; Alonso, J.L.; Sugimori, T.; Ianakiev, P.; Xiong, J.P.; Arnaout, M.A. The beta-tail domain (beta TD) regulates physiologic ligand binding to integrin CD11b/CD18. Blood 2007, 109, 3513–3520. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.P.; Stehle, T.; Goodman, S.L.; Arnaout, M.A. New insights into the structural basis of integrin activation. Blood 2003, 102, 1155–1159. [Google Scholar] [CrossRef]
- Ünlü, B.; Kocaturk, B.; Rondon, A.M.R.; Lewis, C.S.; Swier, N.; van den Akker, R.F.P.; Krijgsman, D.; Noordhoek, I.; Blok, E.J.; Bogdanov, V.Y.; et al. Integrin regulation by tissue factor promotes cancer stemness and metastatic dissemination in breast cancer. Oncogene 2022, 41, 5176–5185. [Google Scholar] [CrossRef] [PubMed]
- Kariya, Y.; Oyama, M.; Hashimoto, Y.; Gu, J.; Kariya, Y. beta4-Integrin/PI3K Signaling Promotes Tumor Progression through the Galectin-3-N-Glycan Complex. Mol. Cancer Res. 2018, 16, 1024–1034. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.S.; Kang, C.W.; Kang, S.; Jang, Y.; Chae, Y.C.; Kim, B.G.; Cho, N.H. ITGB4-mediated metabolic reprogramming of cancer-associated fibroblasts. Oncogene 2020, 39, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Desgrosellier, J.S.; Barnes, L.A.; Shields, D.J.; Huang, M.; Lau, S.K.; Prevost, N.; Tarin, D.; Shattil, S.J.; Cheresh, D.A. An integrin alpha(v)beta(3)-c-Src oncogenic unit promotes anchorage-independence and tumor progression. Nat. Med. 2009, 15, 1163–1169. [Google Scholar] [CrossRef]
- Parvani, J.G.; Galliher-Beckley, A.J.; Schiemann, B.J.; Schiemann, W.P. Targeted inactivation of beta1 integrin induces beta3 integrin switching, which drives breast cancer metastasis by TGF-beta. Mol. Biol. Cell 2013, 24, 3449–3459. [Google Scholar] [CrossRef] [PubMed]
- Vachon, P.H. Integrin signaling, cell survival, and anoikis: Distinctions, differences, and differentiation. J. Signal Transduct. 2011, 2011, 738137. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, H.H.; Spek, C.A.; Richel, D.J.; Peppelenbosch, M.P. Coagulation factors VIIa and Xa inhibit apoptosis and anoikis. Oncogene 2004, 23, 410–417. [Google Scholar] [CrossRef]


| MDA-MB-231 (TF+) | HDBEC (TF−) | |
|---|---|---|
| PLA | TED, LED, UED interact with β1-integrin. EGF4-βTD interacts with TF. | TED, LED, UED interact with β1-integrin. |
| IP | LED IP with β1-integrin. EGF4-βTD IP with TF. | N/A |
| ERK phosphorylation | UED and EGF4-βTD reduce ERK phosphorylation. | UED and LED increase ERK phosphorylation. |
| Cyclin D1 expression | UED and EGF4-βTD reduce cyclin D expression. | No significant changes. |
| Proliferation | UED reduces cell proliferation. | No significant changes. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Featherby, S.J.; Faulkner, E.C.; Maraveyas, A.; Ettelaie, C. Identification of the Interacting Domains Between Tissue Factor and β1-Integrin and the Signalling Properties of the Two Fibronectin-like Domains of Tissue Factor. Cancers 2025, 17, 644. https://doi.org/10.3390/cancers17040644
Featherby SJ, Faulkner EC, Maraveyas A, Ettelaie C. Identification of the Interacting Domains Between Tissue Factor and β1-Integrin and the Signalling Properties of the Two Fibronectin-like Domains of Tissue Factor. Cancers. 2025; 17(4):644. https://doi.org/10.3390/cancers17040644
Chicago/Turabian StyleFeatherby, Sophie J., Eamon C. Faulkner, Anthony Maraveyas, and Camille Ettelaie. 2025. "Identification of the Interacting Domains Between Tissue Factor and β1-Integrin and the Signalling Properties of the Two Fibronectin-like Domains of Tissue Factor" Cancers 17, no. 4: 644. https://doi.org/10.3390/cancers17040644
APA StyleFeatherby, S. J., Faulkner, E. C., Maraveyas, A., & Ettelaie, C. (2025). Identification of the Interacting Domains Between Tissue Factor and β1-Integrin and the Signalling Properties of the Two Fibronectin-like Domains of Tissue Factor. Cancers, 17(4), 644. https://doi.org/10.3390/cancers17040644






