Evaluating the Utility of Fresh Tissue in Molecular Diagnostics of Colorectal Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Material Acquisition
2.2. DNA Isolation and Next-Generation Sequencing
2.3. Bioinformatics and Variant Classification
3. Results
3.1. Patient Characteristics
3.2. NGS Results
3.3. Clinical Characteristics
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keum, N.; Giovannucci, E. Global Burden of Colorectal Cancer: Emerging Trends, Risk Factors and Prevention Strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Didkowska, J.; Barańska, K.; Miklewska, M.J.; Wojciechowska, U. Cancer Incidence and Mortality in Poland in 2023. Nowotw. J. Oncol. 2024, 74, 75–93. [Google Scholar] [CrossRef]
- DeStefanis, R.A.; Kratz, J.D.; Emmerich, P.B.; Deming, D.A. Targeted Therapy in Metastatic Colorectal Cancer: Current Standards and Novel Agents in Review. Curr. Color. Cancer Rep. 2019, 15, 61–69. [Google Scholar] [CrossRef]
- Fleming, M.; Ravula, S.; Tatishchev, S.F.; Wang, H.L. Colorectal Carcinoma: Pathologic Aspects. J. Gastrointest. Oncol. 2012, 3, 153–173. [Google Scholar] [CrossRef]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef]
- Goel, G. Molecular Characterization and Biomarker Identification in Colorectal Cancer: Toward Realization of the Precision Medicine Dream. Cancer Manag. Res. 2018, 10, 5895–5908. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; Rachiglio, A.M.; Roma, C.; Fenizia, F.; Esposito, C.; Pasquale, R.; La Porta, M.L.; Iannaccone, A.; Micheli, F.; Santangelo, M.; et al. Molecular Diagnostics and Personalized Medicine in Oncology: Challenges and Opportunities. J. Cell. Biochem. 2013, 114, 514–524. [Google Scholar] [CrossRef]
- Rey, J.-M.; Ducros, V.; Pujol, P.; Wang, Q.; Buisine, M.-P.; Aissaoui, H.; Maudelonde, T.; Olschwang, S. Improving Mutation Screening in Patients with Colorectal Cancer Predisposition Using Next-Generation Sequencing. J. Mol. Diagn. 2017, 19, 589–601. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Steiert, T.A.; Parra, G.; Gut, M.; Arnold, N.; Trotta, J.-R.; Tonda, R.; Moussy, A.; Gerber, Z.; Abuja, P.M.; Zatloukal, K.; et al. A Critical Spotlight on the Paradigms of FFPE-DNA Sequencing. Nucleic Acids Res. 2023, 51, 7143–7162. [Google Scholar] [CrossRef]
- Cho, M.; Ahn, S.; Hong, M.; Bang, H.; Van Vrancken, M.; Kim, S.; Lee, J.; Park, S.H.; Park, J.O.; Park, Y.S.; et al. Tissue Recommendations for Precision Cancer Therapy Using next Generation Sequencing: A Comprehensive Single Cancer Center’s Experiences. Oncotarget 2017, 8, 42478–42486. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Codina, A.; Arpi, O.; Martinez-García, M.; Pineda, E.; Mallo, M.; Gut, M.; Carrato, C.; Rovira, A.; Lopez, R.; Tortosa, A.; et al. A Comparison of RNA-Seq Results from Paired Formalin-Fixed Paraffin-Embedded and Fresh-Frozen Glioblastoma Tissue Samples. PLoS ONE 2017, 12, e0170632. [Google Scholar] [CrossRef]
- Suciu, B.A.; Pap, Z.; Dénes, L.; Brînzaniuc, K.; Copotoiu, C.; Pávai, Z. Allele-Specific PCR Method for Identification of EGFR Mutations in Non-Small Cell Lung Cancer: Formalin-Fixed Paraffin-Embedded Tissue Versus Fresh Tissue. Rom. J. Morphol. Embryol. 2016, 57, 495–500. [Google Scholar] [PubMed]
- Lehmann, U.; Kreipe, H. Real-Time PCR Analysis of DNA and RNA Extracted from Formalin-Fixed and Paraffin-Embedded Biopsies. Methods 2001, 25, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Gallegos Ruiz, M.I.; Floor, K.; Rijmen, F.; Grünberg, K.; Rodriguez, J.A.; Giaccone, G. EGFR and K-Ras Mutation Analysis in Non-Small Cell Lung Cancer: Comparison of Paraffin Embedded versus Frozen Specimens. Anal. Cell. Oncol. 2007, 29, 257–264. [Google Scholar] [CrossRef]
- Seiler, C.; Sharpe, A.; Barrett, J.C.; Harrington, E.A.; Jones, E.V.; Marshall, G.B. Nucleic Acid Extraction from Formalin-Fixed Paraffin-Embedded Cancer Cell Line Samples: A Trade off between Quantity and Quality? BMC Clin. Pathol. 2016, 16, 17. [Google Scholar] [CrossRef]
- Hedegaard, J.; Thorsen, K.; Lund, M.K.; Hein, A.-M.K.; Hamilton-Dutoit, S.J.; Vang, S.; Nordentoft, I.; Birkenkamp-Demtröder, K.; Kruhøffer, M.; Hager, H.; et al. Next-Generation Sequencing of RNA and DNA Isolated from Paired Fresh-Frozen and Formalin-Fixed Paraffin-Embedded Samples of Human Cancer and Normal Tissue. PLoS ONE 2014, 9, e98187. [Google Scholar] [CrossRef]
- Robbe, P.; Popitsch, N.; Knight, S.J.L.; Antoniou, P.; Becq, J.; He, M.; Kanapin, A.; Samsonova, A.; Vavoulis, D.V.; Ross, M.T.; et al. Clinical Whole-Genome Sequencing from Routine Formalin-Fixed, Paraffin-Embedded Specimens: Pilot Study for the 100,000 Genomes Project. Genet. Med. 2018, 20, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.H.; Li, J.; Gong, H.F.; Yu, G.Y.; Liu, P.; Hao, L.Q.; Liu, L.J.; Bai, C.G.; Zhang, W. Comparison of Fresh Frozen Tissue with Formalin-Fixed Paraffin-Embedded Tissue for Mutation Analysis Using a Multi-Gene Panel in Patients with Colorectal Cancer. Front. Oncol. 2020, 10, 310. [Google Scholar] [CrossRef]
- Sepulveda, A.R.; Hamilton, S.R.; Allegra, C.J.; Grody, W.; Cushman-Vokoun, A.M.; Funkhouser, W.K.; Kopetz, S.E.; Lieu, C.; Lindor, N.M.; Minsky, B.D.; et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J. Mol. Diagn. 2017, 19, 187–225. [Google Scholar] [CrossRef]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.E.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- van der Auwera, G.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2020; ISBN 978-1-4919-7519-0. [Google Scholar]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Ewing, C.M.; Ray, A.M.; Lange, E.M.; Zuhlke, K.A.; Robbins, C.M.; Tembe, W.D.; Wiley, K.E.; Isaacs, S.D.; Johng, D.; Wang, Y.; et al. Germline Mutations in HOXB13 and Prostate-Cancer Risk. N. Engl. J. Med. 2012, 366, 141–149. [Google Scholar] [CrossRef]
- Laitinen, V.H.; Wahlfors, T.; Saaristo, L.; Rantapero, T.; Pelttari, L.M.; Kilpivaara, O.; Laasanen, S.-L.; Kallioniemi, A.; Nevanlinna, H.; Aaltonen, L.; et al. HOXB13 G84E Mutation in Finland: Population-Based Analysis of Prostate, Breast, and Colorectal Cancer Risk. Cancer Epidemiol. Biomark. Prev. 2013, 22, 452–460. [Google Scholar] [CrossRef]
- Armaghany, T.; Wilson, J.D.; Chu, Q.; Mills, G. Genetic Alterations in Colorectal Cancer. Gastrointest. Cancer Res. 2012, 5, 19–27. [Google Scholar]
- Kinzler, K.W.; Vogelstein, B. Lessons from Hereditary Colorectal Cancer. Cell 1996, 87, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Traverso, G.; Shuber, A.; Levin, B.; Johnson, C.; Olsson, L.; Schoetz, D.J.; Hamilton, S.R.; Boynton, K.; Kinzler, K.W.; Vogelstein, B. Detection of APC Mutations in Fecal DNA from Patients with Colorectal Tumors. N. Engl. J. Med. 2002, 346, 311–320. [Google Scholar] [CrossRef]
- Lea, I.A.; Jackson, M.A.; Dunnick, J.K. Genetic Pathways to Colorectal Cancer. Mutat. Res. 2009, 670, 96–98. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gertych, A.; Zurek, N.; Piaseczna, N.; Szkaradnik, K.; Cui, Y.; Zhang, Y.; Nurzynska, K.; Pyciński, B.; Paul, P.; Bartczak, A.; et al. Tumor Cellularity Assessment Using Artificial Intelligence Trained on Immunohistochemistry-Restained Slides Improves Selection of Lung Adenocarcinoma Samples for Molecular Testing. Am. J. Pathol. 2025, 195, 907–922. [Google Scholar] [CrossRef] [PubMed]
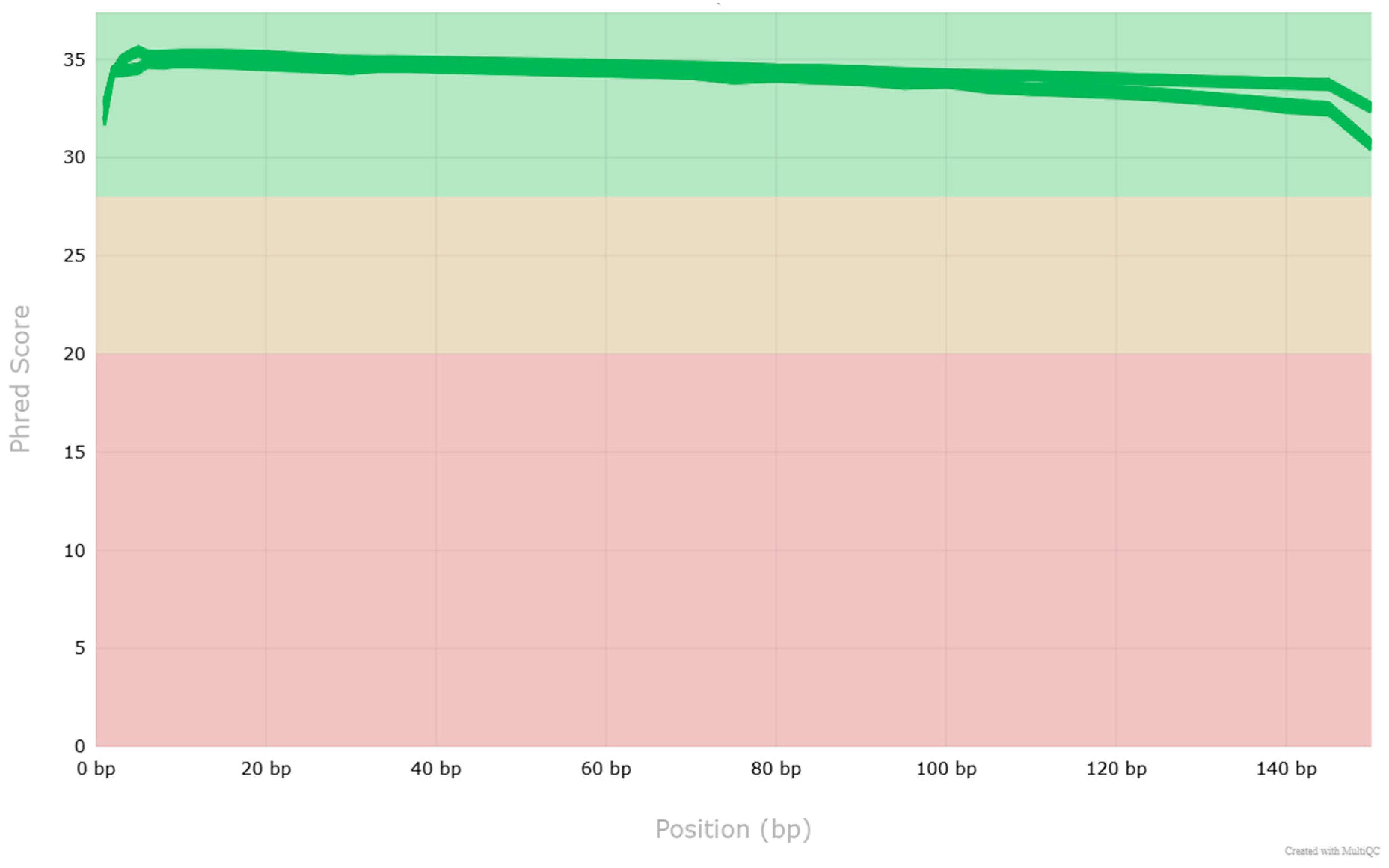
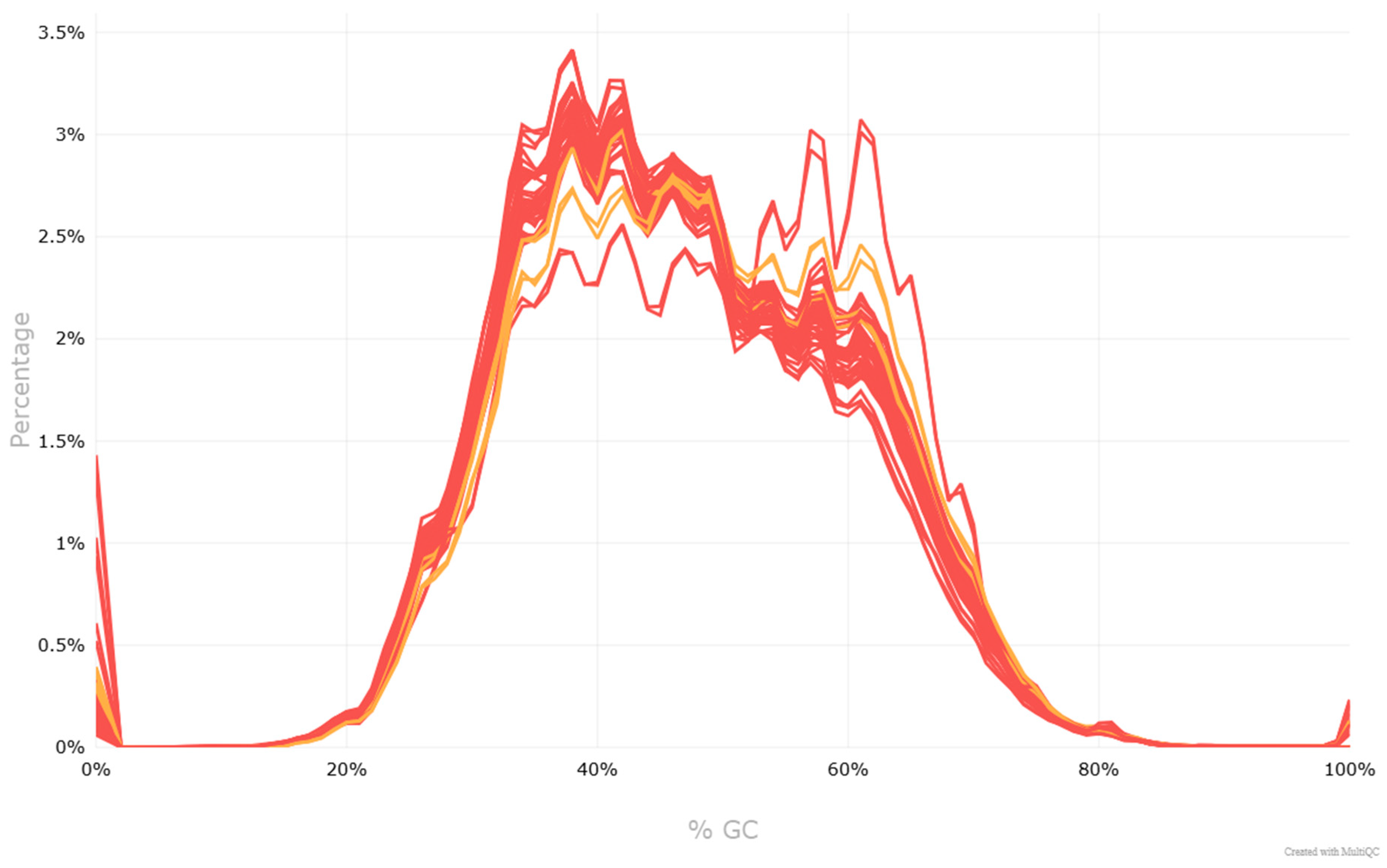

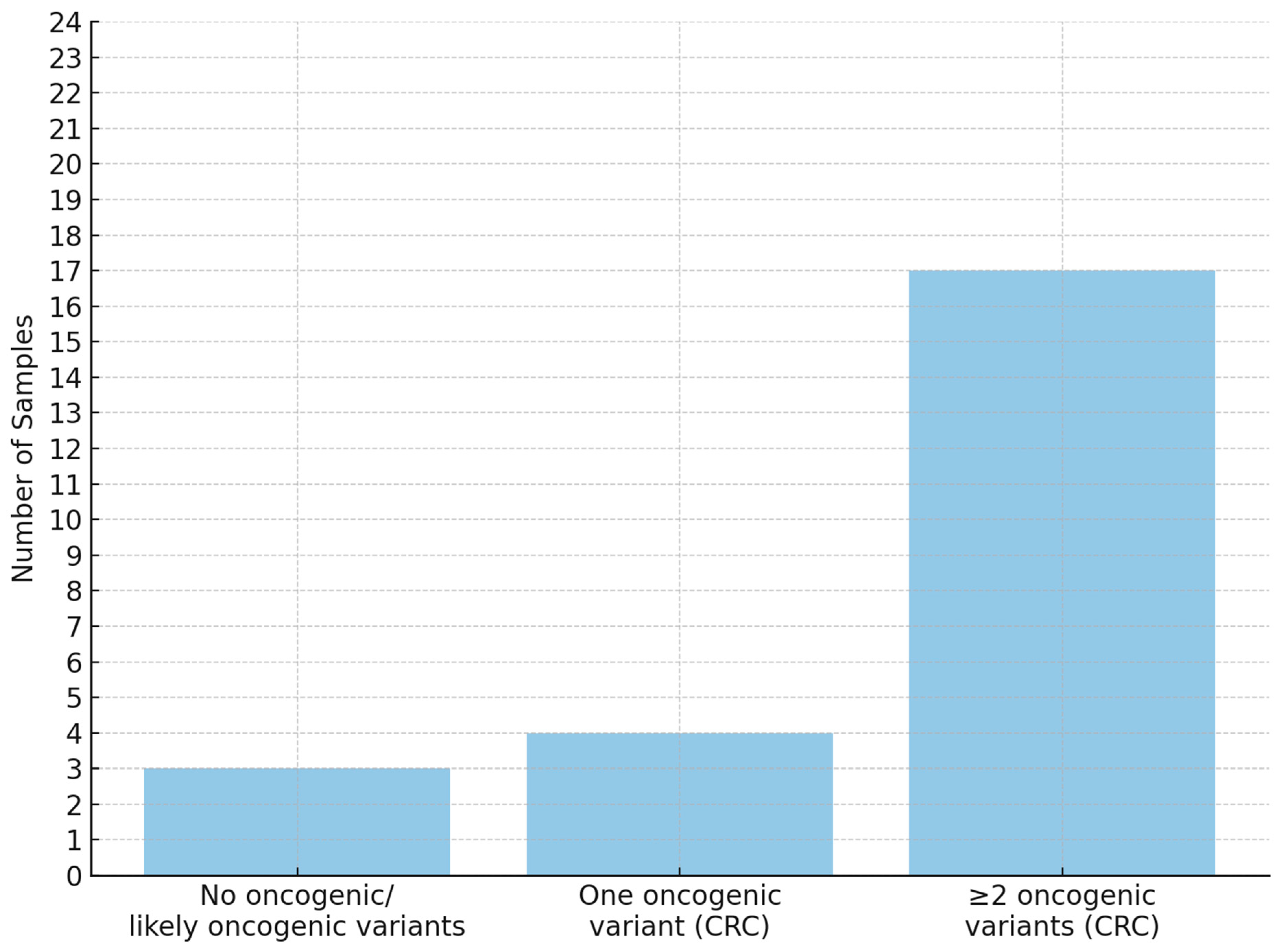
| Sample | Sex | Age | BMI | Smoking (Pack-Year) | Localization | Grade | pTNM | Size (cm) | Lymph Nodes | Metastases |
|---|---|---|---|---|---|---|---|---|---|---|
| 640 | F | 71 | 17.92 | 0 | rectum | GX | T3N0M0 | 4.0 × 3.0 × 1.5 | 0/6 | 0 |
| 638 | F | 77 | 23.44 | 0 | cecum | - | T3N0M0 | 5.0 × 4.0 × 1.5 | 0/18 | 0 |
| 631 | F | 69 | 26.85 | 20 | rectum | G2 | T3N0M0 | 5.5 × 6.0 × 0.7 | 0/22 | 0 |
| 625 | M | 79 | 26.12 | 0 | rectum | G2 | T3N2aM0 | 4.0 × 4.0 × 1.0 | 5/12 | 0 |
| 618 | M | 70 | 24.69 | 50 | ascending colon | G3 | T3N2aM0 | 6.5 × 4.5 × 2.0 | 5/15 | 0 |
| 616 | F | 79 | 25.39 | 6 | ascending colon | G2 | T2N2bM1a | 5.5 × 6.0 × 3.0 | 9/20 | liver |
| 609 | F | 82 | 21.64 | 1 | rectum | G2 | T4bN2bM1a | 6.0 × 3.0 × 2.1 | 7/13 | spine |
| 599 | F | 71 | 33.46 | 40 | transverse colon | G2 | T3N0M0 | 5.0 × 6.5 × 1.5 | 0/14 | 0 |
| 593 | F | 69 | 25.71 | 0 | ascending colon | G3 | T3N2aM0 | 6.0 × 3.5 × 1.5 | 4/7 | 0 |
| 577 | M | 77 | 25.35 | 30 | rectum | G2 | T2N0M0 | 5.0 × 7.5 × 2.0 | 0/28 | 0 |
| 570 | F | 78 | 25.97 | 0 | cecum | - | T2N0M0 | 2.5 × 3.0 × 0.7 | 0/19 | 0 |
| 568 | M | 70 | 24.57 | 15 | descending colon | G2 | T3N1bM1a | 2.5 × 3.5 × 2.5 | 3/13 | liver |
| 565 | F | 68 | 35.08 | 8 | sigmoid colon | - | TisN0M0 | 3.0 × 2.5 × 2.0 | 0/0 | 0 |
| 560 | F | 63 | 31.25 | 0 | ascending colon | G2 | T3N0M0 | 4.5 × 4.5 × 2.5 | 0/14 | 0 |
| 555 | F | 82 | 24.22 | 0 | sigmoid colon | G2 | T2N0M0 | 7.0 × 4.5 × 0.8 | 0/13 | 0 |
| 554 | M | 77 | 26.20 | 20 | ascending colon | - | T3N0M0 | 7.0 × 5.0 × 2.0 | 0/13 | 0 |
| 532 | F | 50 | 30.07 | 0 | rectum | G2 | T3N1bM0 | 4.5 × 3.0 × 1.0 | 2/19 | 0 |
| 526 | F | 33 | 22.15 | 0 | rectum | G2 | T2N0M0 | 2.0 × 2.0 × 1.0 | 0/13 | 0 |
| 507 | M | 73 | 37.02 | 0 | sigmoid colon | G2 | T3N0M0 | 3.2 × 3.0 × 1.0 | 0/14 | 0 |
| 504 | F | 74 | 30.86 | 10 | ascending colon | G2 | T4N0M0 | 8.0 × 4.0 × 1.0 | 0/14 | 0 |
| 505 | F | 81 | 27.11 | 0 | rectum | G2 | T3N2aM0 | 3.0 × 3.5 × 1.0 | 4/13 | 0 |
| 493 | F | 73 | 26.14 | 0 | sigmoid colon | G2 | T3N0M0 | 3.7 × 4.0 × 1.0 | 0/8 | 0 |
| 486 | F | 75 | 31.65 | 0 | sigmoid colon | - | T2N0M0 | 3.5 × 2.0 × 1.0 | 0/13 | 0 |
| 484 | F | 65 | 27.64 | 0 | rectum | G2 | T2N0M0 | 6.2 × 5.0 × 2.0 | 0/16 | 0 |
| Sample | Nucleotide Variant | Predicted Protein Variant | ClinGen-CGC-VICC Pathogenicity | Classification Criteria |
|---|---|---|---|---|
| 618 | c.543_546del | p.(Thr182Ilefs*2) | Likely Oncogenic | OVS1 + 8, OP4 + 1 |
| 532 | c.646C>T | p.(Arg216*) | Oncogenic | OVS1 + 8, OS1 + 4, OS3 + 4, OP4 + 1 |
| 505 | c.847C>T | p.(Arg283*) | Oncogenic | OVS1 + 8, OS1 + 4, OS3 + 4, OP4 + 1 |
| 526 | c.1495C>T | p.(Arg499*) | Oncogenic | OVS1 + 8, OS1 + 4, OS3 + 4, OP4 + 1 |
| 577 | c.1690C>T | p.(Arg564*) | Oncogenic | OVS1 + 8, OS3 + 4, OP4 + 1 |
| 631 | c.2336del | p.(Leu779*) | Likely Oncogenic | OVS1 + 8, OP4 + 1 |
| 599 | c.2413C>T | p.(Arg805*) | Oncogenic | OVS1 + 8, OS1 + 4, OS3 + 4, OP4 + 1 |
| 484 | c.2804dup | p.(Tyr935*) | Oncogenic | OVS1 + 8, OM3 + 2, OP4 + 1 |
| 616 | c.2928_2929del | p.(Gly977Serfs*7) | Likely Oncogenic | OVS1 + 8, OP4 + 1 |
| 555 | c.3340C>T | p.(Arg1114*) | Oncogenic | OVS1 + 8, OS3 + 4, OP4 + 1 |
| 486 | c.3454C>T | p.(Gln1152*) | Likely Oncogenic | OVS1 + 8, OP4 + 1 |
| 625 | c.3852del | p.(Asp1285Metfs*3) | Likely Oncogenic | OVS1 + 8, OP4 + 1 |
| 532 | c.3859del | p.(Ile1287*) | Oncogenic | OVS1 + 8, OM3 + 2, OP4 + 1 |
| 555 | c.3907C>T | p.(Gln1303*) | Oncogenic | OVS1 + 8, OS3 + 4, OP4 + 1 |
| 609, 526, 609 | c.3927_3931del | p.(Glu1309Aspfs*4) | Oncogenic | OVS1 + 8, OS1 + 4, OS3 + 4, OP4 + 1 |
| 493, 593 | c.4033G>T | p.(Glu1345*) | Oncogenic | OVS1 + 8, OS3 + 4, OP4 + 1 |
| 565 | c.4129_4130del | p.(Val1377Serfs*8) | Likely Oncogenic | OVS1 + 8, OP4 + 1 |
| 505 | c.4135G>T | p.(Glu1379*) | Oncogenic | OVS1 + 8, OS3 + 4, OP4 + 1 |
| 484 | c.4391_4394del | p.(Glu1464Valfs*8) | Oncogenic | OVS1 + 8, OS1 + 4, OS3 + 4, OP4 + 1 |
| 507 | c.4473dup | p.(Ala1492Cysfs*22) | Likely Oncogenic | OVS1 + 8, OP4 + 1 |
| 616 | c.4666dup | p.(Thr1556Asnfs*3) | Oncogenic | OVS1 + 8, OS3 + 4, OP4 + 1 |
| 554 | c.4741del | p.(Ser1581Leufs*69) | Oncogenic | OVS1 + 8, OM3 + 2, OP4 + 1 |
| Sample | Nucleotide Variant | Predicted Protein Variant | ClinGen-CGC-VICC Pathogenicity | Classification Criteria |
|---|---|---|---|---|
| 493, 593 | c.378C>A | p.(Tyr126*) | Oncogenic | OVS1 + 8, OS1 + 4, OM1 + 2, OP4 + 1 |
| 631 | c.389T>C | p.(Leu130Pro) | Oncogenic | OS2 + 4, OM1 + 2, OP1 + 1, OP3 + 1, OP4 + 1 |
| 577 | c.396G>C | p.(Lys132Asn) | Oncogenic | OS1 + 4, OS2 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
| 526 | c.475G>C | p.(Ala159Pro) | Oncogenic | OS2 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
| 554 | c.487T>C | p.(Tyr163His) | Oncogenic | OS2 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
| 618 | c.524G>A | p.(Arg175His) | Oncogenic | OS1 + 4, OS2 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
| 599 | c.527G>T | p.(Cys176Phe) | Oncogenic | OS1 + 4, OS2 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
| 505, 532 | c.743G>A | p.(Arg248Gln) | Oncogenic | OS1 + 4, OS2 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
| 486 | c.809T>G | p.(Phe270Cys) | Likely Oncogenic | OS2 + 4, OM3 + 4, OP1 + 1, OP4 + 1 |
| 616 | c.818G>A | p.(Arg273His) | Oncogenic | OS1 + 4, OS2 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
| 507 | c.844C>T | p.(Arg282Trp) | Oncogenic | OS1 + 4, OS2 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
| 625 | c.1024C>T | p.(Arg342*) | Oncogenic | OVS1 + 8, OS1 + 4, OP4 + 1 |
| Sample | Nucleotide Variant | Predicted Protein Variant | ClinGen-CGC-VICC Pathogenicity | Classification Criteria |
|---|---|---|---|---|
| 505 | c.35G>T | p.Gly12Val | Oncogenic | OS1 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
| 507, 526, 616, 599 | c.35G>A | p.Gly12Asp | Oncogenic | OS1 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
| 484, 631 | c.38G>A | p.Gly13Asp | Oncogenic | OS1 + 4, OS3 + 4, OP1 + 1, OP4 + 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kałużewski, T.; Wcisło, S.; Sałacińska, K.; Kępczyński, Ł.; Kubiak, I.; Grabiec, M.; Kalinka, E.; Kałużewski, B.; Gach, A. Evaluating the Utility of Fresh Tissue in Molecular Diagnostics of Colorectal Cancer. Cancers 2025, 17, 3709. https://doi.org/10.3390/cancers17223709
Kałużewski T, Wcisło S, Sałacińska K, Kępczyński Ł, Kubiak I, Grabiec M, Kalinka E, Kałużewski B, Gach A. Evaluating the Utility of Fresh Tissue in Molecular Diagnostics of Colorectal Cancer. Cancers. 2025; 17(22):3709. https://doi.org/10.3390/cancers17223709
Chicago/Turabian StyleKałużewski, Tadeusz, Szymon Wcisło, Kinga Sałacińska, Łukasz Kępczyński, Izabela Kubiak, Magdalena Grabiec, Ewa Kalinka, Bogdan Kałużewski, and Agnieszka Gach. 2025. "Evaluating the Utility of Fresh Tissue in Molecular Diagnostics of Colorectal Cancer" Cancers 17, no. 22: 3709. https://doi.org/10.3390/cancers17223709
APA StyleKałużewski, T., Wcisło, S., Sałacińska, K., Kępczyński, Ł., Kubiak, I., Grabiec, M., Kalinka, E., Kałużewski, B., & Gach, A. (2025). Evaluating the Utility of Fresh Tissue in Molecular Diagnostics of Colorectal Cancer. Cancers, 17(22), 3709. https://doi.org/10.3390/cancers17223709







