Comparative Oncology: Cross-Sectional Single-Cell Transcriptomic Profiling of the Tumor Microenvironment Across Seven Human Cancers
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Single-Cell RNA-seq Data Processing
2.2. Cell Type Annotation
2.3. Cell–Cell Communication Analysis
2.4. Survival Analysis Using Gene Expression Data
2.5. Statistical Analysis
2.6. Declaration of Generative AI and AI-Assisted Technologies in the Writing Process
3. Results
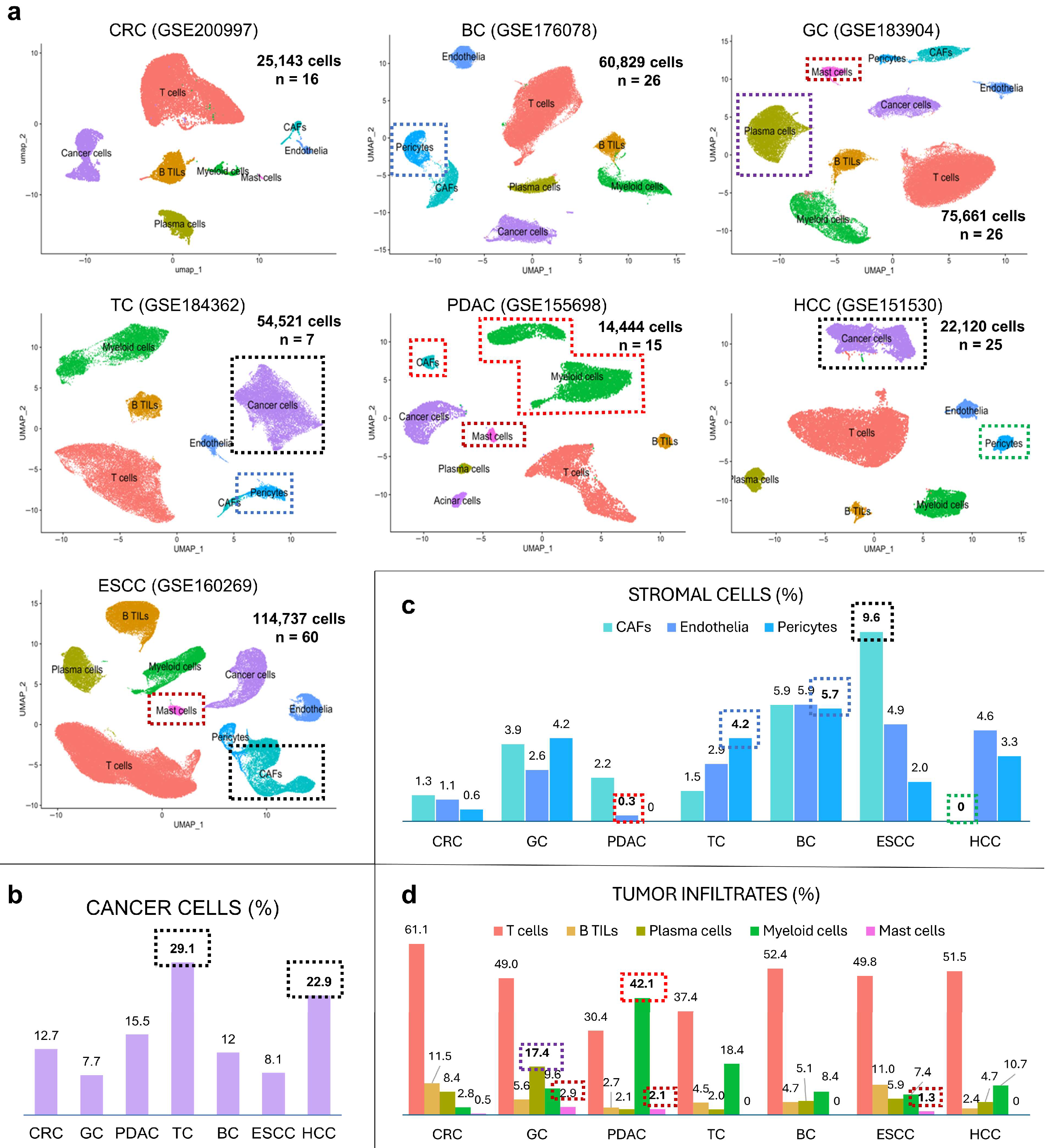
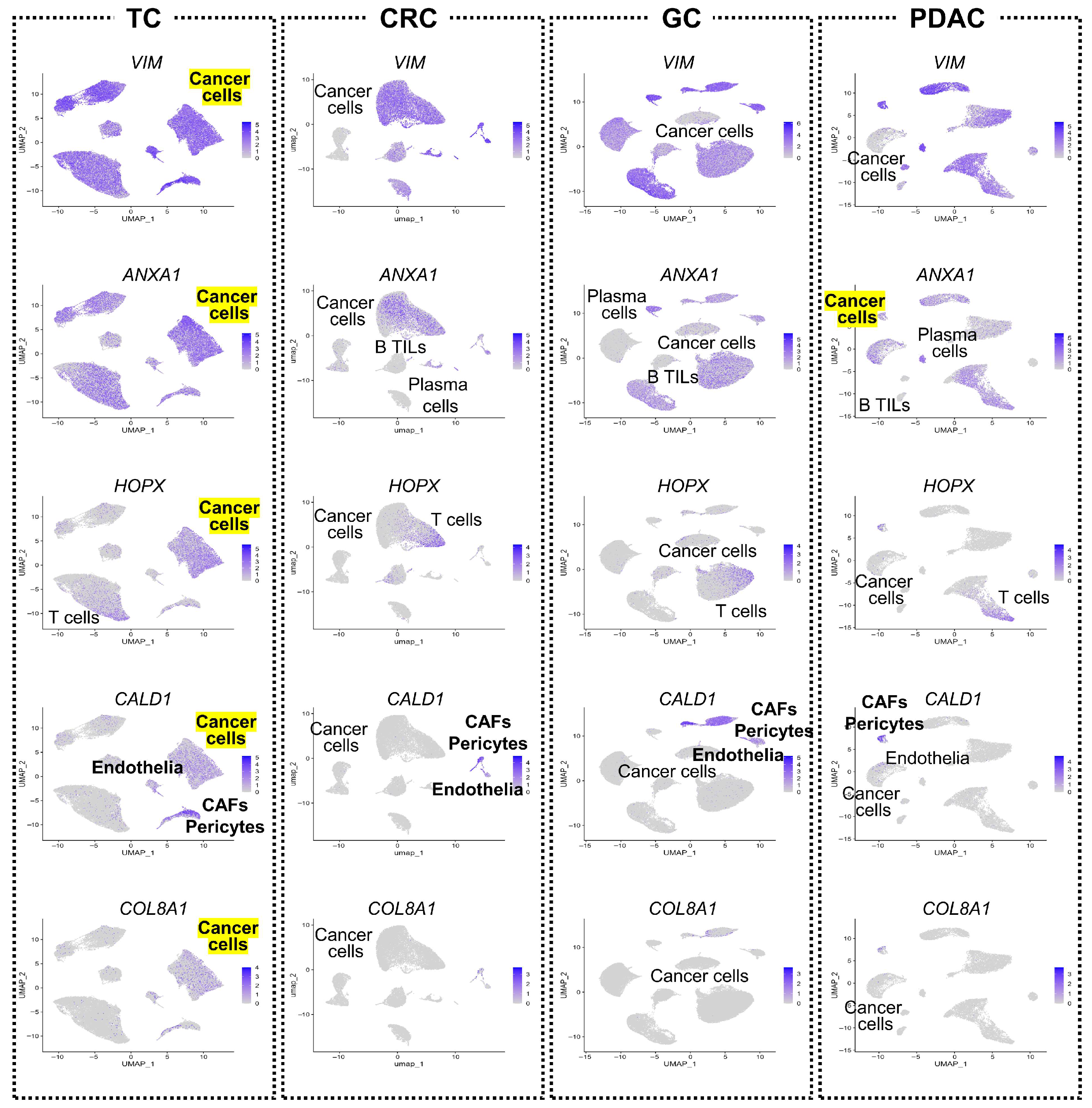
3.1. UMAP-Based Comparison of Seven Human Cancers
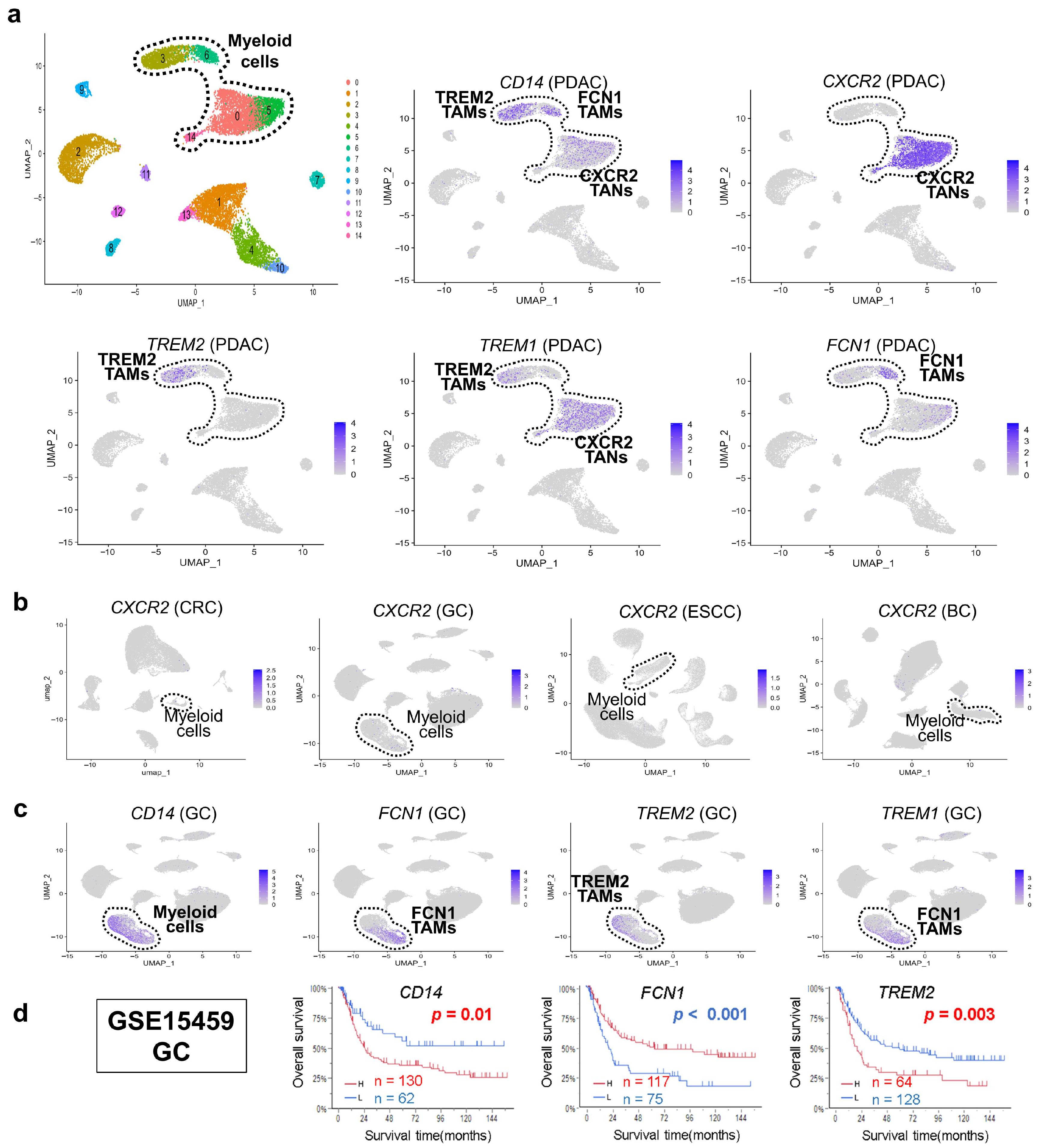
3.2. Discovery of CXCR2/CXCR1 Myeloid Cells Unique to PDAC
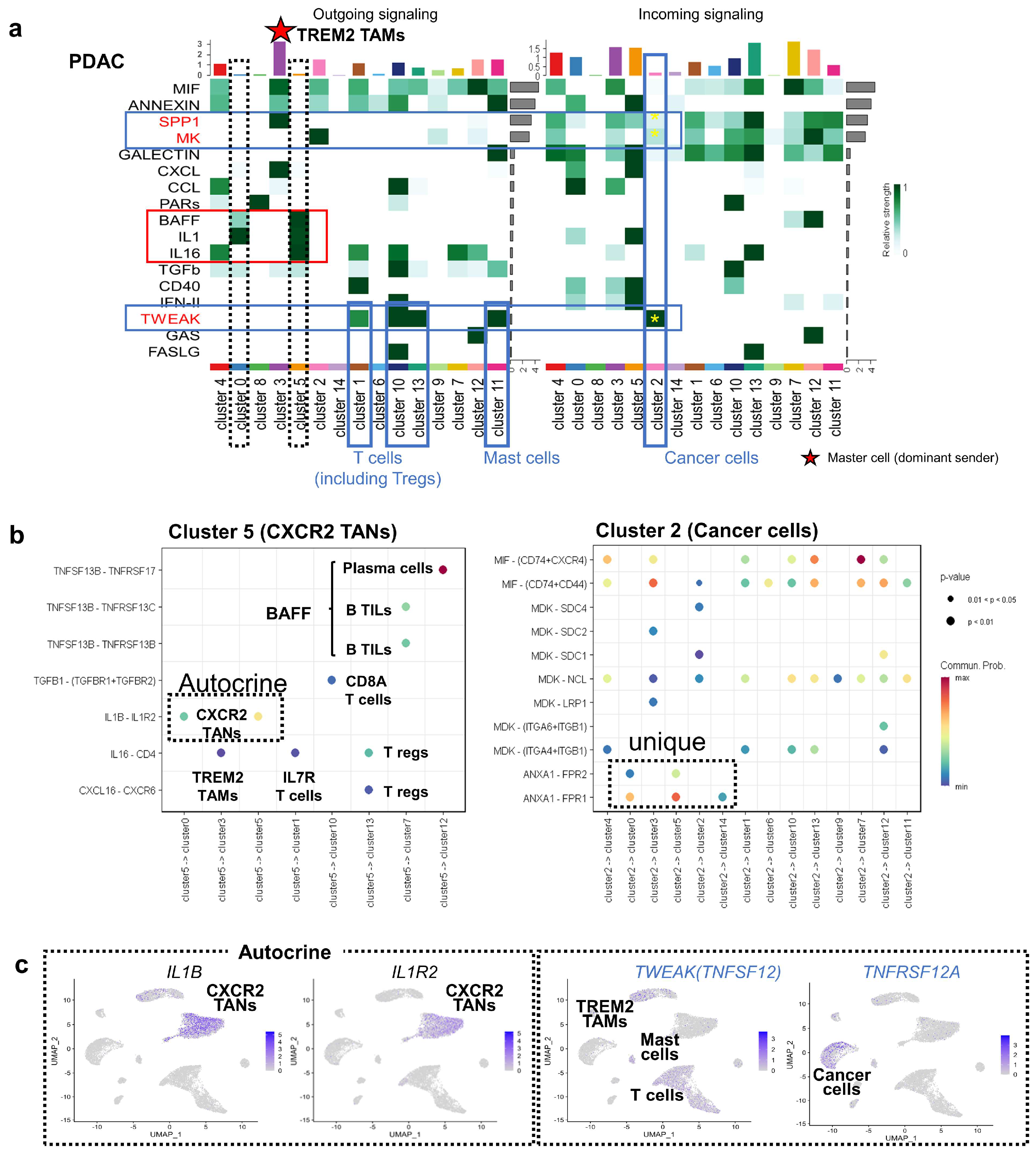
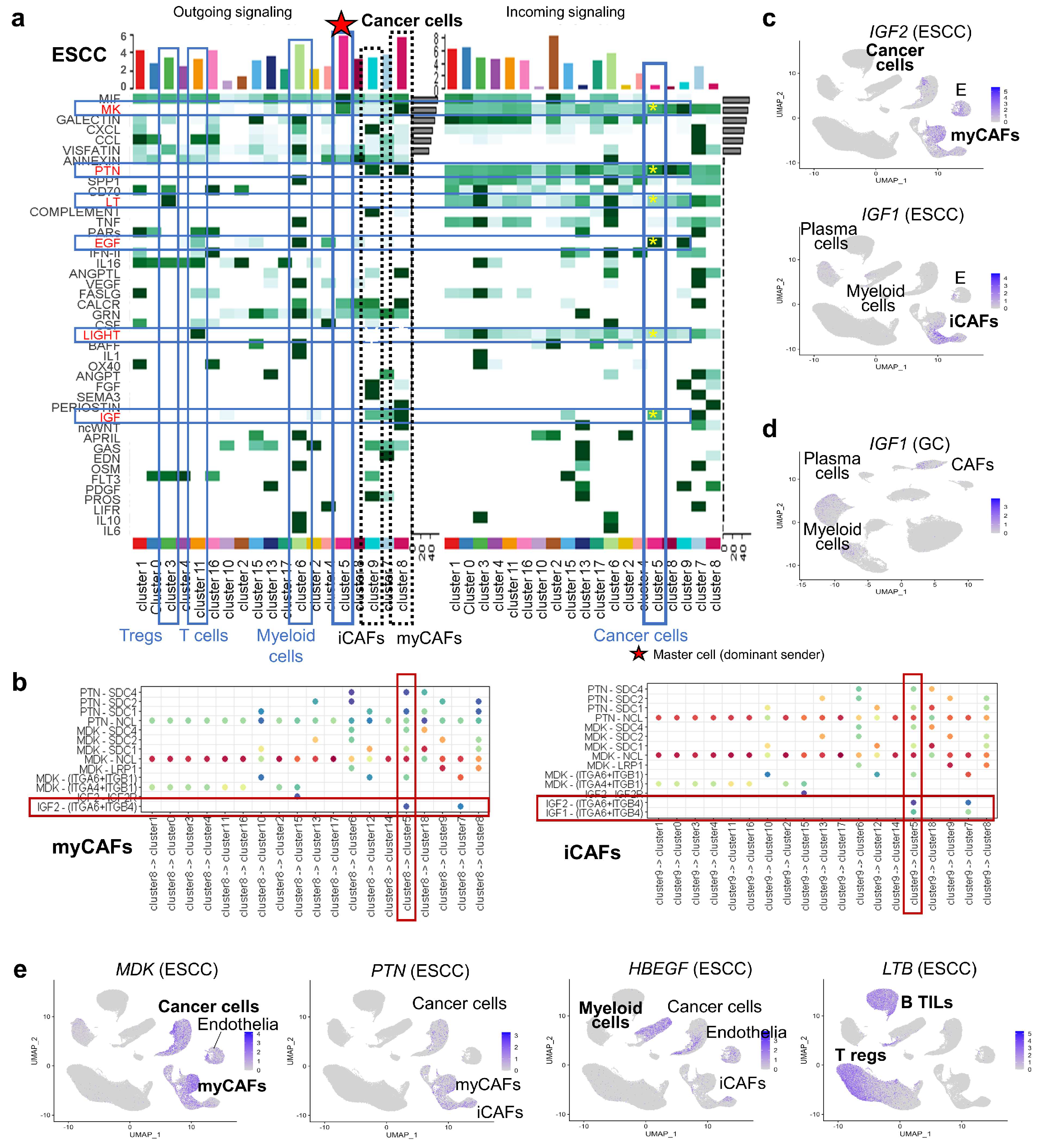
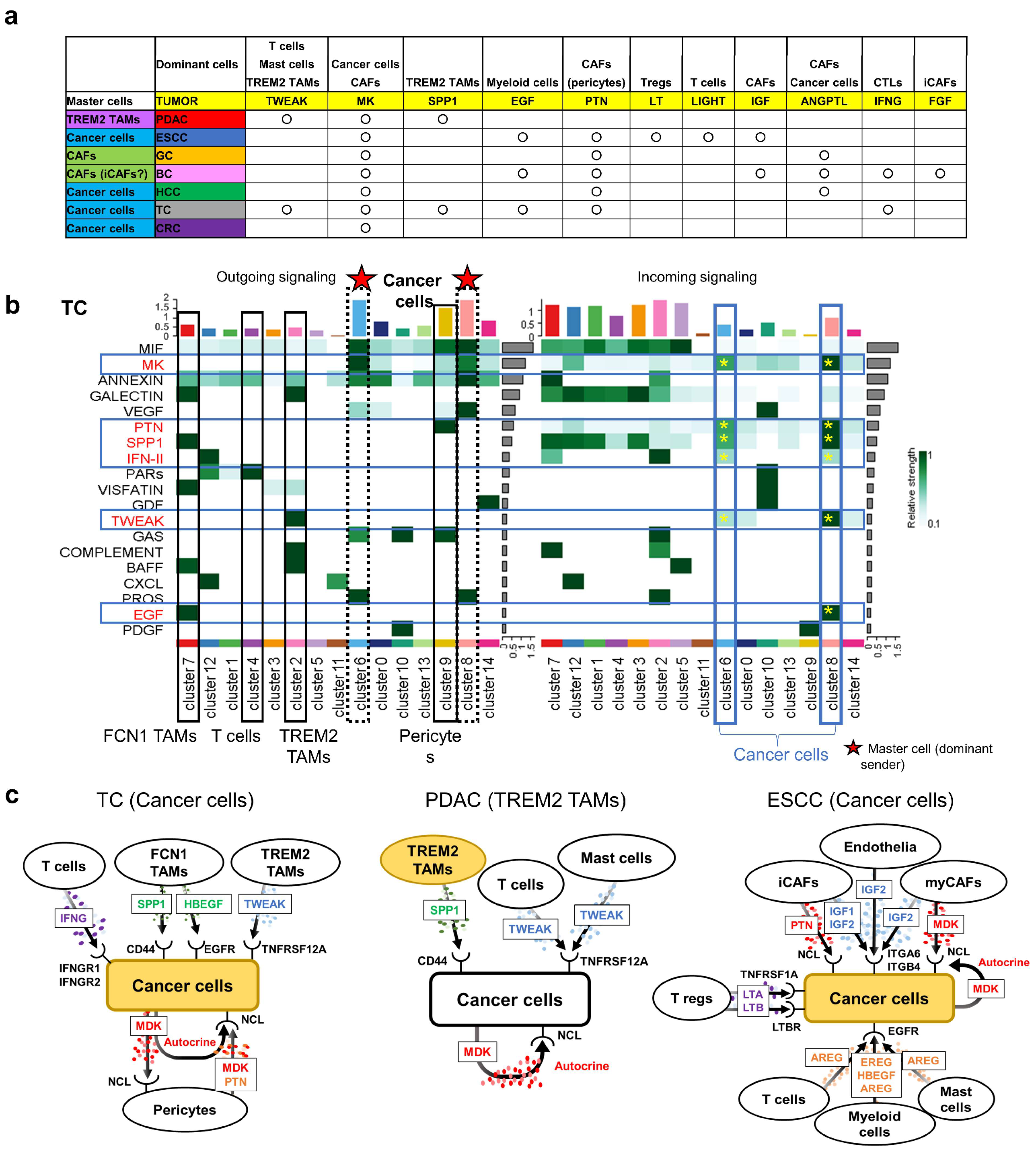
3.3. Intercellular Interaction Analysis in PDAC
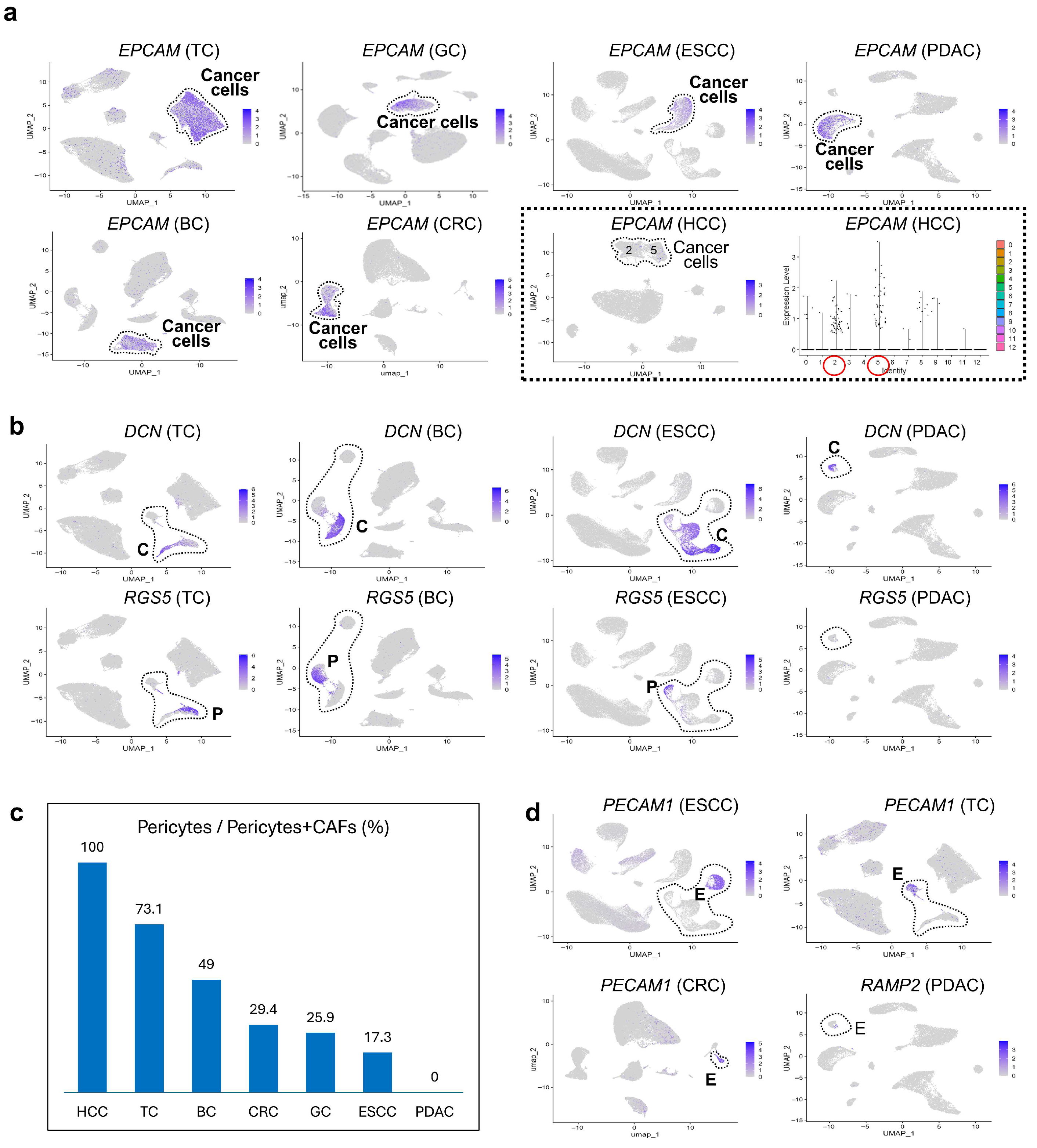
3.4. Comparative scRNA-seq Reveals Cancer-Specific Molecular Features
3.5. Unique Characteristics of CAFs Revealed by Comparative Oncology
3.6. CellChat Reveals Distinct Sender Cell Populations Across Cancers
3.7. Retention of Tumor-Suppressor Genes in TC Tumor Cells
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | Breast cancer |
| B TILs | B-type tumor-infiltrating lymphocytes. |
| CAFs | Cancer-associated fibroblast |
| CAFGs | Cancer-associated fibroblast genes |
| CRC | Colorectal cancer |
| ESCC | Esophageal squamous cell carcinoma |
| GC | Gastric cancer |
| HCC | Hepatocellular carcinoma |
| PDAC | Pancreatic ductal adenocarcinoma |
| scRNA-seq | Single-cell RNA sequencing |
| TAN | Tumor-associated neutrophil |
| TC | Thyroid cancer |
| TME | Tumor microenvironment |
| TNBC | Triple-Negative Breast Cancer |
| UMAP | Uniform manifold approximation and projection |
References
- Seferbekova, Z.; Lomakin, A.; Yates, L.R.; Gerstung, M. Spatial biology of cancer evolution. Nat. Rev. Genet. 2023, 24, 295–313. [Google Scholar] [CrossRef]
- Li, Y.; Cai, L.; Wang, H.; Wu, P.; Gu, W.; Chen, Y.; Hao, H.; Tang, K.; Yi, P.; Liu, M.; et al. Pleiotropic regulation of macrophage polarization and tumorigenesis by formyl peptide receptor-2. Oncogene 2011, 30, 3887–3899. [Google Scholar] [CrossRef]
- Lee, H.O.; Hong, Y.; Etlioglu, H.E.; Cho, Y.B.; Pomella, V.; Van den Bosch, B.; Vanhecke, J.; Verbandt, S.; Hong, H.; Min, J.W.; et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 2020, 52, 594–603. [Google Scholar] [CrossRef]
- Xie, Y.; Zhou, T.; Li, X.; Zhao, K.; Bai, W.; Hou, X.; Liu, Z.; Ni, B.; Zhang, Z.; Yan, J.; et al. Targeting ESE3/EHF With Nifurtimox Inhibits CXCR2(+) Neutrophil Infiltration and Overcomes Pancreatic Cancer Resistance to Chemotherapy and Immunotherapy. Gastroenterology 2024, 167, 281–297. [Google Scholar] [CrossRef]
- Coulouarn, C.; Corlu, A.; Glaise, D.; Guénon, I.; Thorgeirsson, S.S.; Clément, B. Hepatocyte-stellate cell cross-talk in the liver engenders a permissive inflammatory microenvironment that drives progression in hepatocellular carcinoma. Cancer Res. 2012, 72, 2533–2542. [Google Scholar] [CrossRef] [PubMed]
- Croizer, H.; Mhaidly, R.; Kieffer, Y.; Gentric, G.; Djerroudi, L.; Leclere, R.; Pelon, F.; Robley, C.; Bohec, M.; Meng, A.; et al. Deciphering the spatial landscape and plasticity of immunosuppressive fibroblasts in breast cancer. Nat. Commun. 2024, 15, 2806. [Google Scholar] [CrossRef]
- Khaliq, A.M.; Erdogan, C.; Kurt, Z.; Turgut, S.S.; Grunvald, M.W.; Rand, T.; Khare, S.; Borgia, J.A.; Hayden, D.M.; Pappas, S.G.; et al. Refining colorectal cancer classification and clinical stratification through a single-cell atlas. Genome Biol. 2022, 23, 113. [Google Scholar] [CrossRef]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef]
- Kumar, V.; Ramnarayanan, K.; Sundar, R.; Padmanabhan, N.; Srivastava, S.; Koiwa, M.; Yasuda, T.; Koh, V.; Huang, K.K.; Tay, S.T.; et al. Single-Cell Atlas of Lineage States, Tumor Microenvironment, and Subtype-Specific Expression Programs in Gastric Cancer. Cancer Discov. 2022, 12, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Pu, W.; Shi, X.; Yu, P.; Zhang, M.; Liu, Z.; Tan, L.; Han, P.; Wang, Y.; Ji, D.; Gan, H.; et al. Single-cell transcriptomic analysis of the tumor ecosystems underlying initiation and progression of papillary thyroid carcinoma. Nat. Commun. 2021, 12, 6058. [Google Scholar] [CrossRef] [PubMed]
- Steele, N.G.; Carpenter, E.S.; Kemp, S.B.; Sirihorachai, V.R.; The, S.; Delrosario, L.; Lazarus, J.; Amir, E.D.; Gunchick, V.; Espinoza, C.; et al. Multimodal Mapping of the Tumor and Peripheral Blood Immune Landscape in Human Pancreatic Cancer. Nat. Cancer. 2020, 1, 1097–1112. [Google Scholar] [CrossRef]
- Ma, L.; Wang, L.; Khatib, S.A.; Chang, C.W.; Heinrich, S.; Dominguez, D.A.; Forgues, M.; Candia, J.; Hernandez, M.O.; Kelly, M.; et al. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Hepatol. 2021, 75, 1397–1408. [Google Scholar] [CrossRef]
- Zhang, X.; Peng, L.; Luo, Y.; Zhang, S.; Pu, Y.; Chen, Y.; Guo, W.; Yao, J.; Shao, M.; Fan, W.; et al. Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat. Commun. 2021, 12, 5291. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, C.S.; Murrow, L.M.; Gartner, Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019, 8, 329–337.e4. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Ooi, C.H.; Ivanova, T.; Wu, J.; Lee, M.; Tan, I.B.; Tao, J.; Ward, L.; Koo, J.H.; Gopalakrishnan, V.; Zhu, Y.; et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009, 5, e1000676. [Google Scholar] [CrossRef]
- Hornburg, M.; Desbois, M.; Lu, S.; Guan, Y.; Lo, A.A.; Kaufman, S.; Elrod, A.; Lotstein, A.; DesRochers, T.M.; Munoz-Rodriguez, J.L.; et al. Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell. 2021, 39, 928–944.e6. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Sun, D.; Tu, C.; Wang, J.; Fu, R.; Gong, R.; Xiao, Y.; Liu, Q.; Li, X. Defining gastric cancer ecology: The crucial roles of TREM2(+) macrophages and fibroblasts in tumor microenvironments. Commun. Biol. 2025, 8, 514. [Google Scholar] [CrossRef] [PubMed]
- Mayer, S.; Milo, T.; Isaacson, A.; Halperin, C.; Miyara, S.; Stein, Y.; Lior, C. The tumor microenvironment shows a hierarchy of cell-cell interactions dominated by fibroblasts. Nat. Commun. 2023, 14, 5810. [Google Scholar] [CrossRef]
- Li, H.; Courtois, E.T.; Sengupta, D.; Tan, Y.; Chen, K.H.; Goh, J.J.L.; Kong, S.L.; Chua, C.; Hon, L.K.; Tan, W.S.; et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 2017, 49, 708–718. [Google Scholar] [CrossRef]
- Yamashita, T.; Budhu, A.; Forgues, M.; Wang, X.W. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007, 67, 10831–10839. [Google Scholar] [CrossRef] [PubMed]
- Arpinati, L.; Carradori, G.; Scherz-Shouval, R. CAF-induced physical constraints controlling T cell state and localization in solid tumours. Nat. Rev. Cancer 2024, 24, 676–693. [Google Scholar] [CrossRef]
- Thorlacius-Ussing, J.; Jensen, C.; Nissen, N.I.; Cox, T.R.; Kalluri, R.; Karsdal, M.; Willumsen, N. The collagen landscape in cancer: Profiling collagens in tumors and in circulation reveals novel markers of cancer-associated fibroblast subtypes. J. Pathol. 2024, 262, 22–36. [Google Scholar] [CrossRef]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef]
- Aldred, M.A.; Huang, Y.; Liyanarachchi, S.; Pellegata, N.S.; Gimm, O.; Jhiang, S.; Davuluri, R.V.; de la Chapelle, A.; Eng, C. Papillary and follicular thyroid carcinomas show distinctly different microarray expression profiles and can be distinguished by a minimum of five genes. J. Clin. Oncol. 2004, 22, 3531–3539. [Google Scholar] [CrossRef]
- Wang, Q.S.; Shen, S.Q.; Sun, H.W.; Xing, Z.X.; Yang, H.L. Interferon-gamma induces autophagy-associated apoptosis through induction of cPLA2-dependent mitochondrial ROS generation in colorectal cancer cells. Biochem. Biophys. Res. Commun. 2018, 498, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Okuno, K.; Ikemura, K.; Okamoto, R.; Oki, K.; Watanabe, A.; Kuroda, Y.; Kidachi, M.; Fujino, S.; Nie, Y.; Higuchi, T.; et al. CAF-associated genes putatively representing distinct prognosis by in silico landscape of stromal components of colon cancer. PLoS ONE 2024, 19, e0299827. [Google Scholar] [CrossRef]
- Nie, Y.; Fujiyama, Y.; Shibaki, S.; Okamoto, R.; Okuno, K.; Oki, K.; Watanabe, A.; Kuroda, Y.; Goto, T.; Yokota, K.; et al. SPARC Induces COL1A1/COL3A1 Expressions Representing Aggressive Molecular Cancer-Associated Fibroblasts Signatures and CSF1-Mediated Cancer Invasion in Colorectal Cancer. Ann. Surg. Oncol. 2025, 32, 9354–9365. [Google Scholar] [CrossRef]
- Ren, X.; Yang, X.; Cheng, B.; Chen, X.; Zhang, T.; He, Q.; Li, B.; Li, Y.; Tang, X.; Wen, X.; et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat. Commun. 2017, 8, 14053. [Google Scholar] [CrossRef]
- Cheung, W.K.; Zhao, M.; Liu, Z.; Stevens, L.E.; Cao, P.D.; Fang, J.E.; Westbrook, T.F.; Nguyen, D.X. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell 2013, 23, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Shapiro, B.; Vucic, E.A.; Vogt, S.; Bar-Sagi, D. Tumor Cell-Derived IL1β Promotes Desmoplasia and Immune Suppression in Pancreatic Cancer. Cancer Res. 2020, 80, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, K.; Yoshimitsu, K.; Irie, H.; Shinozaki, K.; Nagata, S.; Yamaguchi, K.; Honda, H. Imaging of intraductal tubular tumors of the pancreas. AJR Am. J. Roentgenol. 2008, 191, 1836–1840. [Google Scholar] [CrossRef]
- Xu, L.; Ashkenazi, A.; Chaudhuri, A. Duffy antigen/receptor for chemokines (DARC) attenuates angiogenesis by causing senescence in endothelial cells. Angiogenesis 2007, 10, 307–318. [Google Scholar] [CrossRef]
- Devi, S.; Wang, Y.; Chew, W.K.; Lima, R.; N, A.G.; Mattar, C.N.; Chong, S.Z.; Schlitzer, A.; Bakocevic, N.; Chew, S.; et al. Neutrophil mobilization via plerixafor-mediated CXCR4 inhibition arises from lung demargination and blockade of neutrophil homing to the bone marrow. J. Exp. Med. 2013, 210, 2321–2336. [Google Scholar] [CrossRef]
- Fabre, T.; Barron, A.M.S.; Christensen, S.M.; Asano, S.; Bound, K.; Lech, M.P.; Wadsworth, M.H., 2nd; Chen, X.; Wang, C.; Wang, J.; et al. Identification of a broadly fibrogenic macrophage subset induced by type 3 inflammation. Sci. Immunol. 2023, 8, eadd8945. [Google Scholar] [CrossRef] [PubMed]
- Murakata, A.; Tanaka, S.; Mogushi, K.; Yasen, M.; Noguchi, N.; Irie, T.; Kudo, A.; Nakamura, N.; Tanaka, H.; Arii, S. Gene expression signature of the gross morphology in hepatocellular carcinoma. Ann. Surg. 2011, 253, 94–100. [Google Scholar] [CrossRef]
- Ogawa, K.; Tanaka, S.; Matsumura, S.; Murakata, A.; Ban, D.; Ochiai, T.; Irie, T.; Kudo, A.; Nakamura, N.; Tanabe, M.; et al. EpCAM-targeted therapy for human hepatocellular carcinoma. Ann. Surg. Oncol. 2014, 21, 1314–1322. [Google Scholar] [CrossRef]
- Ramadori, G.; Heinz, H.P.; Martin, H.; Meyer zum Büschenfelde, K.H.; Loos, M. Biosynthesis of the subcomponents C1q, C1r and C1s of the first component of complement (C1) by guinea pig hepatocyte primary cultures. Eur. J. Immunol. 1986, 16, 1137–1141. [Google Scholar] [CrossRef]
- Song, D.; Wu, Y.; Li, J.; Liu, J.; Yi, Z.; Wang, X.; Sun, J.; Li, L.; Wu, Q.; Chen, Y.; et al. Insulin-like growth factor 2 drives fibroblast-mediated tumor immunoevasion and confers resistance to immunotherapy. J. Clin. Investig. 2024, 134, e183366. [Google Scholar] [CrossRef]
- Duesberg, M.; LeVee, A.; Chang, H.; Tsai, K.; Crossman, B.; Tadi, M.; Xu, S.; Wheeler, D.; Kang, I. Breast Cancer Immunotherapy: A Team Science Approach. Cancer Treat. Res. 2025, 129, 67–82. [Google Scholar] [PubMed]
- Ooizumi, Y.; Katoh, H.; Yokota, M.; Watanabe, M.; Yamashita, K. Epigenetic silencing of HOPX is critically involved in aggressive phenotypes and patient prognosis in papillary thyroid cancer. Oncotarget 2019, 10, 5906–5918. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okamoto, R.; Okuno, K.; Watanabe, A.; Naito, K.; Minoura, H.; Shibaki, S.; Ikemura, K.; Oki, K.; Kuroda, Y.; Fujino, S.; et al. Comparative Oncology: Cross-Sectional Single-Cell Transcriptomic Profiling of the Tumor Microenvironment Across Seven Human Cancers. Cancers 2025, 17, 3527. https://doi.org/10.3390/cancers17213527
Okamoto R, Okuno K, Watanabe A, Naito K, Minoura H, Shibaki S, Ikemura K, Oki K, Kuroda Y, Fujino S, et al. Comparative Oncology: Cross-Sectional Single-Cell Transcriptomic Profiling of the Tumor Microenvironment Across Seven Human Cancers. Cancers. 2025; 17(21):3527. https://doi.org/10.3390/cancers17213527
Chicago/Turabian StyleOkamoto, Riku, Kota Okuno, Akiko Watanabe, Kanako Naito, Hiroyuki Minoura, Shumpei Shibaki, Kyonosuke Ikemura, Keiko Oki, Yu Kuroda, Shiori Fujino, and et al. 2025. "Comparative Oncology: Cross-Sectional Single-Cell Transcriptomic Profiling of the Tumor Microenvironment Across Seven Human Cancers" Cancers 17, no. 21: 3527. https://doi.org/10.3390/cancers17213527
APA StyleOkamoto, R., Okuno, K., Watanabe, A., Naito, K., Minoura, H., Shibaki, S., Ikemura, K., Oki, K., Kuroda, Y., Fujino, S., Nie, Y., Nishizawa, N., Watanabe, E., Kikuchi, M., Kumagai, K., Yamanashi, T., Katoh, H., Takayasu, H., Sato, T., ... Yamashita, K. (2025). Comparative Oncology: Cross-Sectional Single-Cell Transcriptomic Profiling of the Tumor Microenvironment Across Seven Human Cancers. Cancers, 17(21), 3527. https://doi.org/10.3390/cancers17213527






