The Impact on the Therapeutic Decision of Massive Gene Sequencing (NGS) in Plasma from Patients with Advanced Non-Small Cell Lung Cancer (NSCLC)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Patients
2.2. Tissue Samples
2.3. Next-Generation Sequencing (NGS)-Based Liquid Biopsy
2.4. Outcomes and Statistical Analyses
2.5. Ethical Considerations
3. Results
3.1. Patient Characteristics
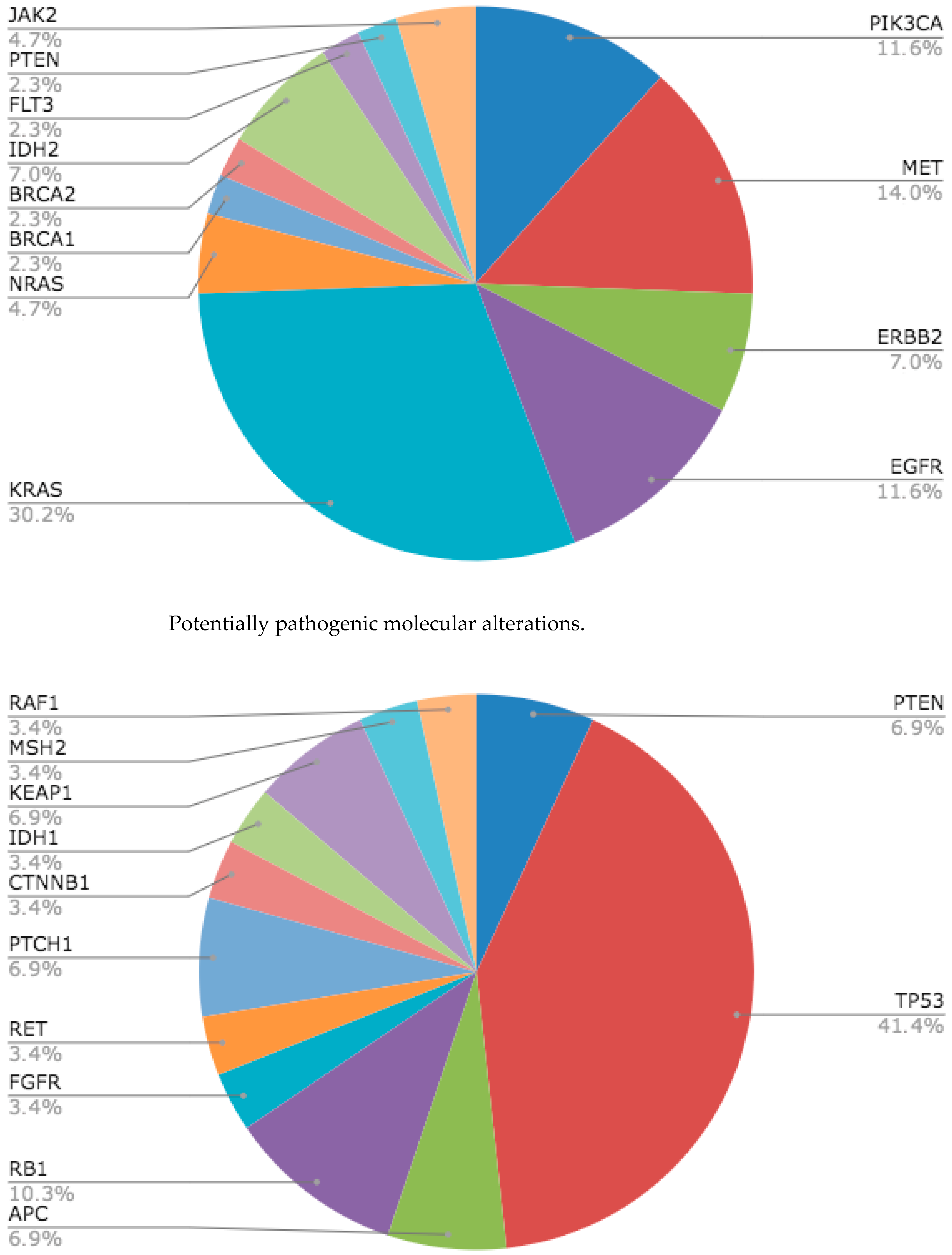
3.2. Molecular Alterations Detected by Massive Gene Sequencing (NGS) in Plasma
3.3. Concordance Between Massive Gene Sequencing (NGS) in Plasma and Tissue
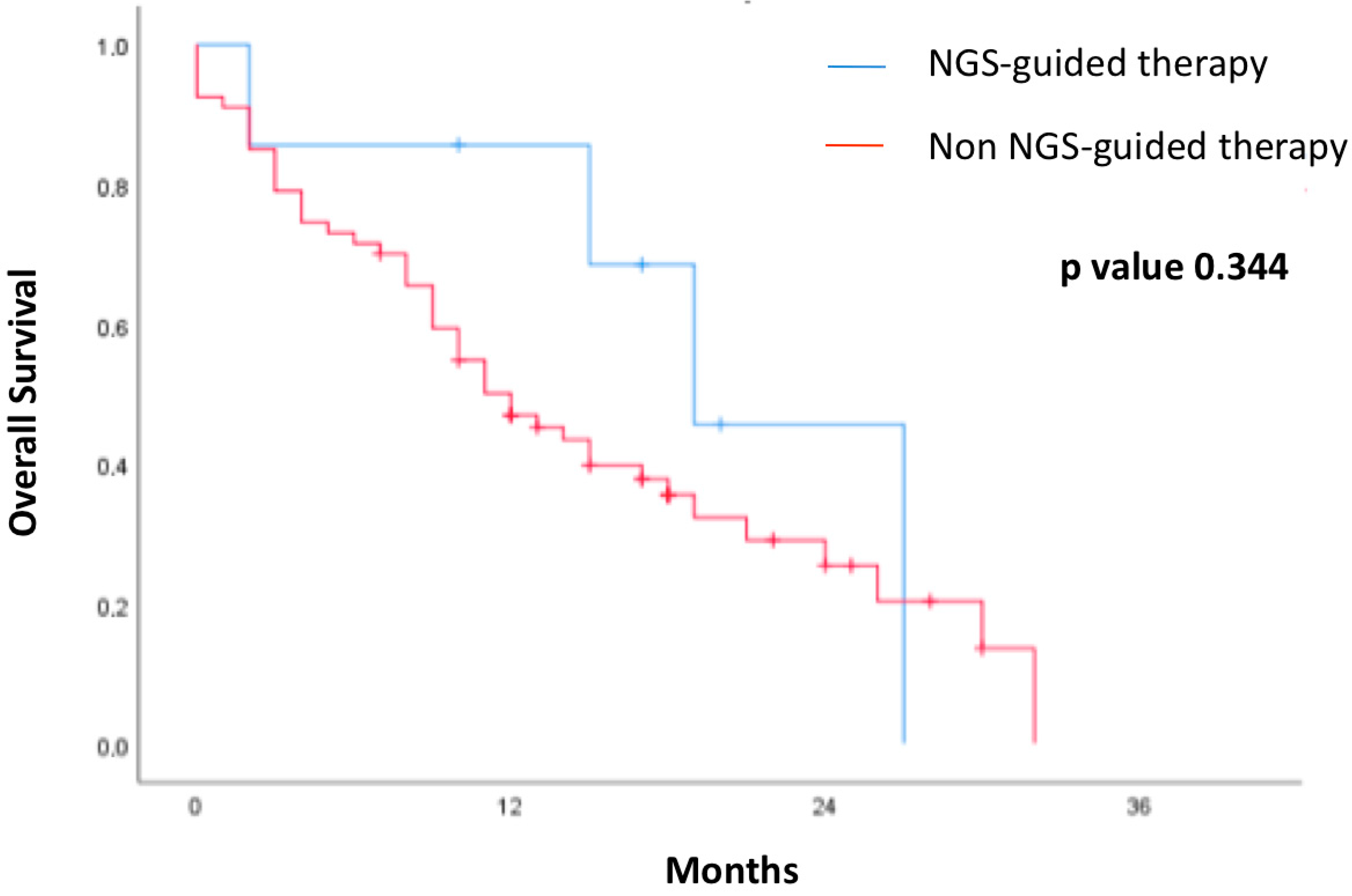
3.4. Survival Outcome in Patients Regarding Treatment Guided by Liquid Biopsy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer Statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacev, S.; Baird, R.; Rodenfeld, N. Liquid Biopsies Come of Age: Towards Implementation of Circulating Tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Cui, W.; Milner-Watts, C.; O’Sullivan, H.; Lyons, H.; Minchom, A.; Bhosle, J.; Davidson, M.; Yousaf, N.; Scott, S.; Faull, I.; et al. Up-front cell-free DNA next generation sequencing improves target identification in UK first line advanced non-small cell lung cancer (NSCLC) patients. Eur. J. Cancer 2022, 171, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, Y.; Tang, S.; Deng, Q.; Jiang, J.; Zhou, C. Comparative analysis of genomic profiles between tissue-based and plasma-based next-generation sequencing in patients with non-small cell lung cancer. Lung Cancer 2023, 182, 107282. [Google Scholar] [CrossRef]
- Park, S.; Olsen, S.; Ku, B.M.; Lee, M.S.; Jung, H.A.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; Park, K.; Choi, Y.L.; et al. High concordance of actionable genomic alterations identified between circulating tumor DNA-based and tissue-based next-generation sequencing testing in advanced non-small cell lung cancer: The Korean Lung Liquid Versus Invasive Biopsy Program. Cancer 2021, 127, 3019–3028. [Google Scholar] [CrossRef]
- Hendriks, L.E.; Kerr, K.M.; Menis, J.; Mok, T.S.; Nestle, U.; Passaro, A.; Peters, S.; Planchard, D.; Smit, E.F.; Solomon, B.J.; et al. Oncogene-Addicted Metastatic Non-Small-Cell Lung Cancer: ESMO Clinical Practice Guideline for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2023, 34, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Riely, G.J.; Wood, D.E.; Ettinger, D.S.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; et al. Non-Small Cell Lung Cancer, Version 4.2024, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2024, 22, 249–274. [Google Scholar] [CrossRef]
- Owen, D.H.; Ismaila, N.; Freeman-Daily, J.; Roof, L.; Singh, N.; Velazquez, A.I.; Leighl, N.B. Therapy for Stage IV Non-Small Cell Lung Cancer with Driver Alterations: ASCO Living Guideline, Version 2024.1. J. Clin. Oncol. 2024, 42, e44–e59. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: Consensus Statement from the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating Liquid Biopsies into the Management of Cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.K.; Oh, M.S. Detection of Minimal Residual Disease Using ctDNA in Lung Cancer: Current Evidence and Future Directions. J. Thorac. Oncol. 2019, 14, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Al Bakir, M.; Huebner, A.; Martínez-Ruiz, C.; Grigoriadis, K.; Watkins, T.B.K.; Pich, O.; Moore, D.A.; Veeriah, S.; Ward, S.; Laycock, J.; et al. The Evolution of Non-Small Cell Lung Cancer Metastases in TRACERx. Nature 2023, 616, 534–542. [Google Scholar] [CrossRef]
- Fusco, N.; Venetis, K.; Pepe, F.; Shetty, O.; Fariñas, S.C.; Heeke, S.; Burnier, J.V.; Patton, S.J.; Heitzer, E.; Nigita, G.; et al. International Society of Liquid Biopsy (ISLB) Perspective on Minimal Requirements for ctDNA Testing in Solid Tumors. J. Liq. Biopsy 2025, 8, 100301. [Google Scholar] [CrossRef]
- Fernández-Murga, L.; Garde-Noguera, J.; Llors, P.; Vidal, J.; Pellicer, A.; Iranzo, J.; García-Sánchez, J.; Cabrera, E.; Albert, A.; Llombart-Cussac, A.; et al. Impact on the Therapeutic Decision of Plasma NGS in Advanced Cancer. ISLB Abstracts 2024, 5, PP01.25. [Google Scholar]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merrit, R.E.; et al. An Ultrasensitive Method for Quantitating Circulating Tumor DNA with Broad Patient Coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Sabari, J.K.; Offin, M.; Stephens, D.; Ni, A.; Lee, A.; Pavlakis, N.; Clarke, S.; Diakos, C.I.; Datta, S.; Tandon, N.; et al. A Prospective Study of Circulating Tumor DNA to Guide Matched Targeted Therapy in Lung Cancers. J. Natl. Cancer Inst. 2019, 111, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Zill, O.A.; Banks, K.C.; Fairclough, S.R.; Mortimer, S.A.; Vowles, J.V.; Mokhtari, R.; Gandara, D.R.; Mack, P.C.; Odegaard, J.I.; Nagy, R.J.; et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin. Cancer Res. 2018, 24, 3528–3538. [Google Scholar] [CrossRef]
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.; Fan, R.; Yee, S.S.; Chien, A.L.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical Implications of Plasma-Based Genotyping with the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 173–180. [Google Scholar] [CrossRef]
- Andrews, H.S.; Zariffa, N.; Nishimura, K.K.; Choi, S.H.; Deng, S.; Eisele, M.; Espenschied, C.R.; Goren, E.M.; Guha, M.; Hong, S.; et al. ctDNA Clearance as an Early Indicator of Improved Clinical Outcomes in Advanced NSCLC Treated with TKI: Findings from an Aggregate Analysis of Eight Clinical Trials. Clin. Cancer Res. 2025, 31, OF1–OF11. [Google Scholar] [CrossRef]
- Jee, J.; Lebow, E.S.; Yeh, R.; Das, J.P.; Namakydoust, A.; Paik, P.K.; Chaft, J.E.; Jayakumaran, G.; Rose Brannon, A.; Benayed, R.; et al. Overall Survival with Circulating Tumor DNA-Guided Therapy in Advanced Non-Small-Cell Lung Cancer. Nat. Med. 2022, 28, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Iwama, E.; Sakai, K.; Hidaka, N.; Inoue, K.; Fujii, A.; Nakagaki, N.; Ota, K.; Toyozawa, R.; Azuma, K.; Nakatomi, K.; et al. Longitudinal Monitoring of Somatic Genetic Alterations in Circulating Cell-Free DNA during Treatment with Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors. Cancer 2020, 126, 219–227. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | N (%) |
|---|---|
| Total Patients | 78 |
| Male | 52 (66.7) |
| Female | 26 (33.3) |
| Age (in years) | |
| Median | 63.5 |
| Range | 29–90 |
| Histology | |
| Adenocarcinoma | 67 (85.9) |
| Squamous carcinoma | 5 (6.4) |
| Other | 6 (7.8) |
| Smoking Status | |
| Non-smoker | 12 (15.4) |
| Former smoker | 38 (48.7) |
| Active smoker | 26 (33.3) |
| ECOG PS | |
| 0–1 | 63 (80.8) |
| 2 | 13 (16.7) |
| 3 | 2 (2.6) |
| Previous Treatment | |
| 0 | 58 (65.4) |
| 1 | 15 (19.2) |
| ≥2 | 5 (6.4) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llor-Rodriguez, P.; Blasco-Cordellat, A.; Macia-Escalante, S.; Fernández-Murga, L.; Vidal-Martinez, J.; Garde-Noguera, J.; García-Sánchez, J.; Llombart-Cussac, A. The Impact on the Therapeutic Decision of Massive Gene Sequencing (NGS) in Plasma from Patients with Advanced Non-Small Cell Lung Cancer (NSCLC). Cancers 2025, 17, 3469. https://doi.org/10.3390/cancers17213469
Llor-Rodriguez P, Blasco-Cordellat A, Macia-Escalante S, Fernández-Murga L, Vidal-Martinez J, Garde-Noguera J, García-Sánchez J, Llombart-Cussac A. The Impact on the Therapeutic Decision of Massive Gene Sequencing (NGS) in Plasma from Patients with Advanced Non-Small Cell Lung Cancer (NSCLC). Cancers. 2025; 17(21):3469. https://doi.org/10.3390/cancers17213469
Chicago/Turabian StyleLlor-Rodriguez, Paula, Ana Blasco-Cordellat, Sonia Macia-Escalante, Leonor Fernández-Murga, José Vidal-Martinez, Javier Garde-Noguera, José García-Sánchez, and Antonio Llombart-Cussac. 2025. "The Impact on the Therapeutic Decision of Massive Gene Sequencing (NGS) in Plasma from Patients with Advanced Non-Small Cell Lung Cancer (NSCLC)" Cancers 17, no. 21: 3469. https://doi.org/10.3390/cancers17213469
APA StyleLlor-Rodriguez, P., Blasco-Cordellat, A., Macia-Escalante, S., Fernández-Murga, L., Vidal-Martinez, J., Garde-Noguera, J., García-Sánchez, J., & Llombart-Cussac, A. (2025). The Impact on the Therapeutic Decision of Massive Gene Sequencing (NGS) in Plasma from Patients with Advanced Non-Small Cell Lung Cancer (NSCLC). Cancers, 17(21), 3469. https://doi.org/10.3390/cancers17213469








