Comparative Analysis of Targeted RNA-Seq and Optical Genome Mapping for Detecting Gene Rearrangements in Acute Leukemia
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. OGM Analysis
2.3. Targeted RNA-Seq
2.4. Variant Interpretation
2.5. Statistical Analysis
3. Results
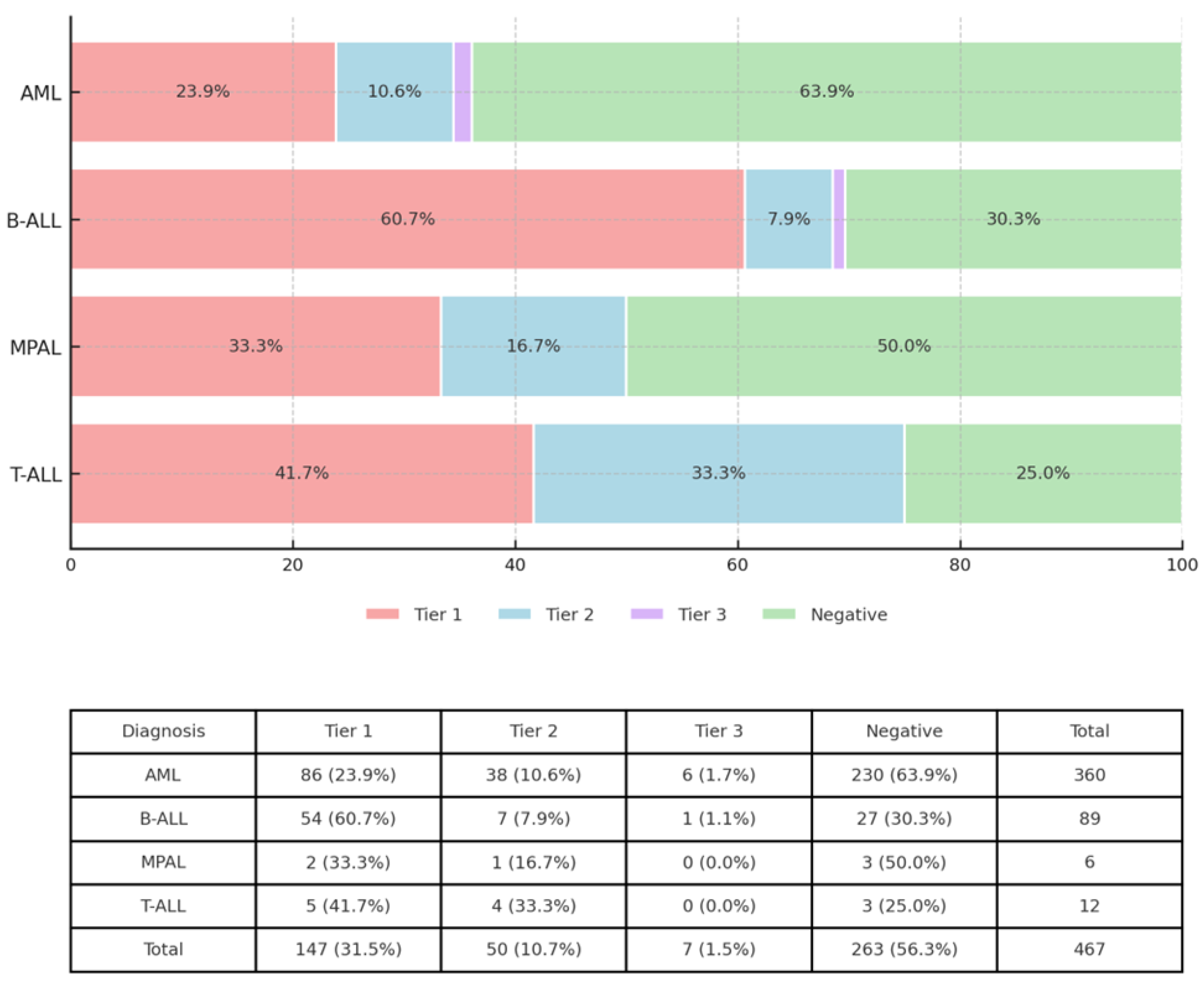
3.1. OGM and RNA Seq Analysis Revealed Clinically Significant Gene Rearrangements and Fusions
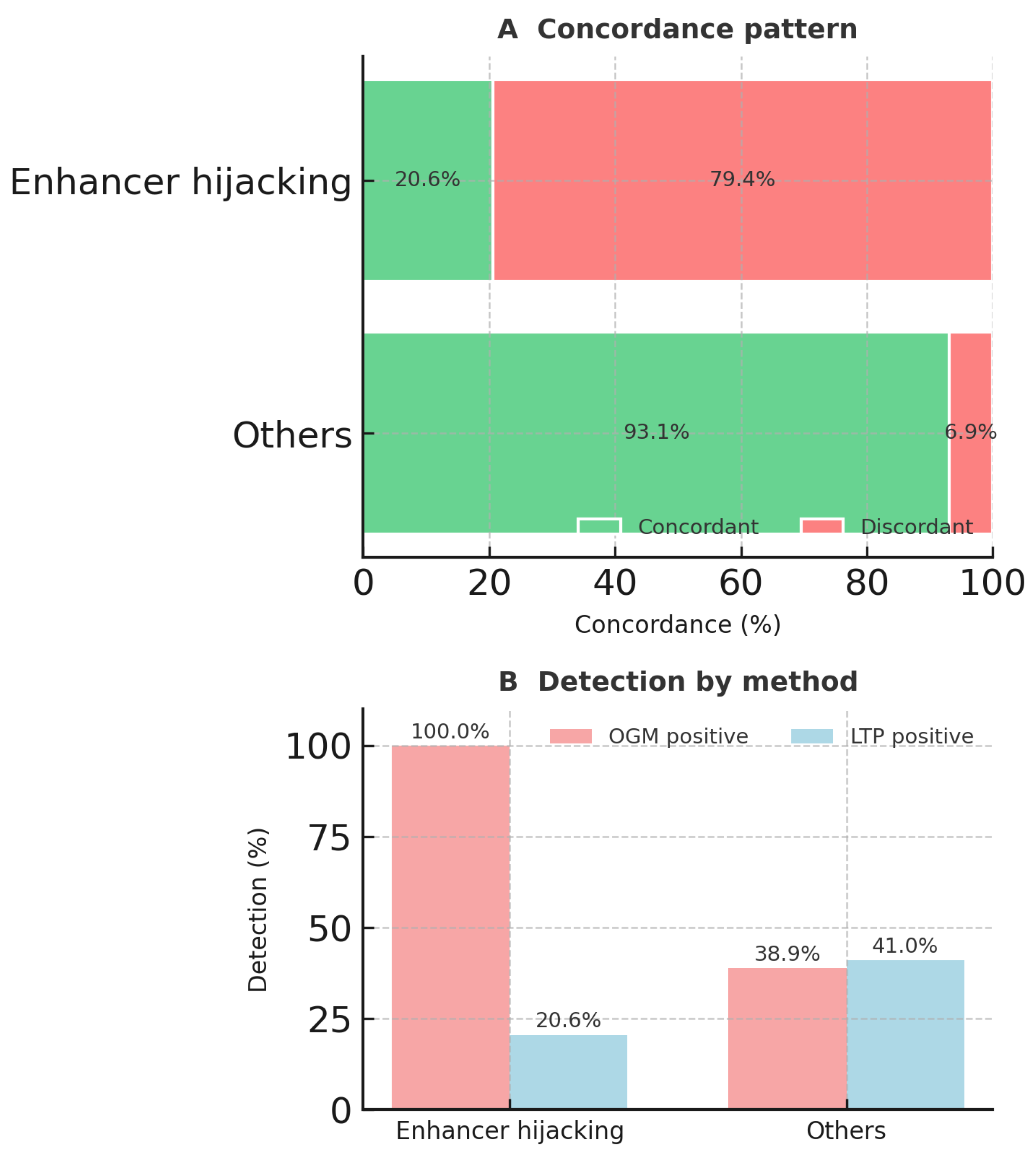
3.2. OGM and RNA-Seq Results Were Concordant in Most Cases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Voss, R.K.; Pastor Loyola, V.B.; Cardenas, M.F.; Kumar, P.; Maciaszek, J.L.; Namwanje, M.; Ma, J.; Neary, J.L.; Jin, M.; Umeda, M.; et al. Clinical experience of using integrated whole genome and transcriptome sequencing as a framework for pediatric and adolescent acute myeloid leukemia diagnosis and risk assessment. Leukemia 2025. [Google Scholar] [CrossRef]
- Tsai, M.M.; Kao, H.J.; Chen, H.H.; Yu, C.H.; Chien, Y.H.; Hwu, W.L.; Kwok, P.Y.; Lee, N.C.; Yang, Y.L. Optical genome mapping with whole genome sequencing identifies complex chromosomal structural variations in acute leukemia. Front. Genet. 2025, 16, 1496847. [Google Scholar] [CrossRef]
- Cao, X.; Huber, S.; Ahari, A.J.; Traube, F.R.; Seifert, M.; Oakes, C.C.; Secheyko, P.; Vilov, S.; Scheller, I.F.; Wagner, N.; et al. Analysis of 3760 hematologic malignancies reveals rare transcriptomic aberrations of driver genes. Genome Med. 2024, 16, 70. [Google Scholar] [CrossRef]
- Ryan, S.L.; Peden, J.F.; Kingsbury, Z.; Schwab, C.J.; James, T.; Polonen, P.; Mijuskovic, M.; Becq, J.; Yim, R.; Cranston, R.E.; et al. Whole genome sequencing provides comprehensive genetic testing in childhood B-cell acute lymphoblastic leukaemia. Leukemia 2023, 37, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Leongamornlert, D.; Gutiérrez-Abril, J.; Lee, S.; Barretta, E.; Creasey, T.; Gundem, G.; Levine, M.F.; Arango-Ossa, J.E.; Liosis, K.; Medina-Martinez, J.S.; et al. Diagnostic utility of whole genome sequencing in adults with B-other acute lymphoblastic leukemia. Blood Adv. 2023, 7, 3862–3873. [Google Scholar] [CrossRef]
- Llop, M.; Sargas, C.; Barragán, E. The role of next-generation sequencing in acute myeloid leukemia. Curr. Opin. Oncol. 2022, 34, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Thangavelu, T.; Wirta, V.; Orsmark-Pietras, C.; Cavelier, L.; Fioretos, T.; Barbany, G.; Olsson-Arvidsson, L.; Pandzic, T.; Staffas, A.; Rosenquist, R.; et al. Micro-costing of genetic diagnostics in acute leukemia in Sweden: From standard-of-care to whole-genome sequencing. J. Med. Econ. 2024, 27, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Salaverria, I.; Siebert, R.; Mrózek, K. Appraisal of current technologies for the study of genetic alterations in hematologic malignancies with a focus on chromosome analysis and structural variants. Med. Genet. 2024, 36, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Budurlean, L.; Tukaramrao, D.B.; Zhang, L.; Dovat, S.; Broach, J. Integrating Optical Genome Mapping and Whole Genome Sequencing in Somatic Structural Variant Detection. J. Pers. Med. 2024, 14, 291. [Google Scholar] [CrossRef] [PubMed]
- Rezayee, F.; Eisfeldt, J.; Skaftason, A.; Öfverholm, I.; Sayyab, S.; Syvänen, A.C.; Maqbool, K.; Lilljebjörn, H.; Johansson, B.; Olsson-Arvidsson, L.; et al. Feasibility to use whole-genome sequencing as a sole diagnostic method to detect genomic aberrations in pediatric B-cell acute lymphoblastic leukemia. Front. Oncol. 2023, 13, 1217712. [Google Scholar] [CrossRef] [PubMed]
- Kogure, Y.; Kameda, T.; Koya, J.; Yoshimitsu, M.; Nosaka, K.; Yasunaga, J.I.; Imaizumi, Y.; Watanabe, M.; Saito, Y.; Ito, Y.; et al. Whole-genome landscape of adult T-cell leukemia/lymphoma. Blood 2022, 139, 967–982. [Google Scholar] [CrossRef]
- Jobanputra, V.; Wrzeszczynski, K.O.; Buttner, R.; Caldas, C.; Cuppen, E.; Grimmond, S.; Haferlach, T.; Mullighan, C.; Schuh, A.; Elemento, O. Clinical interpretation of whole-genome and whole-transcriptome sequencing for precision oncology. Semin. Cancer Biol. 2022, 84, 23–31. [Google Scholar] [CrossRef]
- Duncavage, E.J.; Bagg, A.; Hasserjian, R.P.; DiNardo, C.D.; Godley, L.A.; Iacobucci, I.; Jaiswal, S.; Malcovati, L.; Vannucchi, A.M.; Patel, K.P.; et al. Genomic profiling for clinical decision making in myeloid neoplasms and acute leukemia. Blood 2022, 140, 2228–2247. [Google Scholar] [CrossRef] [PubMed]
- Berglund, E.; Barbany, G.; Orsmark-Pietras, C.; Fogelstrand, L.; Abrahamsson, J.; Golovleva, I.; Hallböök, H.; Höglund, M.; Lazarevic, V.; Levin, L.; et al. A Study Protocol for Validation and Implementation of Whole-Genome and -Transcriptome Sequencing as a Comprehensive Precision Diagnostic Test in Acute Leukemias. Front. Med. 2022, 9, 842507. [Google Scholar] [CrossRef]
- Duncavage, E.J.; Schroeder, M.C.; O’Laughlin, M.; Wilson, R.; MacMillan, S.; Bohannon, A.; Kruchowski, S.; Garza, J.; Du, F.; Hughes, A.E.O.; et al. Genome Sequencing as an Alternative to Cytogenetic Analysis in Myeloid Cancers. N. Engl. J. Med. 2021, 384, 924–935. [Google Scholar] [CrossRef]
- Toruner, G.A.; Hu, S.; Loghavi, S.; Ok, C.Y.; Tang, Z.; Wei, Q.; Kanagal-Shamanna, R.; Medeiros, L.J.; Tang, G. Clinical Utility of Optical Genome Mapping as an Additional Tool in a Standard Cytogenetic Workup in Hematological Malignancies. Cancers 2025, 17, 1436. [Google Scholar] [CrossRef] [PubMed]
- Kanagal-Shamanna, R.; Puiggros, A.; Granada, I.; Raca, G.; Rack, K.; Mallo, M.; Dewaele, B.; Smith, A.C.; Akkari, Y.; Levy, B.; et al. Integration of Optical Genome Mapping in the Cytogenomic and Molecular Work-Up of Hematological Malignancies: Expert Recommendations From the International Consortium for Optical Genome Mapping. Am. J. Hematol. 2025, 100, 1029–1048. [Google Scholar] [CrossRef] [PubMed]
- Loghavi, S.; Wei, Q.; Ravandi, F.; Quesada, A.E.; Routbort, M.J.; Hu, S.; Toruner, G.A.; Wang, S.A.; Wang, W.; Miranda, R.N.; et al. Optical genome mapping improves the accuracy of classification, risk stratification, and personalized treatment strategies for patients with acute myeloid leukemia. Am. J. Hematol. 2024, 99, 1959–1968. [Google Scholar] [CrossRef] [PubMed]
- Vieler, L.M.; Nilius-Eliliwi, V.; Schroers, R.; Vangala, D.B.; Nguyen, H.P.; Gerding, W.M. Optical Genome Mapping Reveals and Characterizes Recurrent Aberrations and New Fusion Genes in Adult ALL. Genes 2023, 14, 686. [Google Scholar] [CrossRef] [PubMed]
- Soler, G.; Ouedraogo, Z.G.; Goumy, C.; Lebecque, B.; Aspas Requena, G.; Ravinet, A.; Kanold, J.; Véronèse, L.; Tchirkov, A. Optical Genome Mapping in Routine Cytogenetic Diagnosis of Acute Leukemia. Cancers 2023, 15, 2131. [Google Scholar] [CrossRef]
- Nilius-Eliliwi, V.; Gerding, W.M.; Schroers, R.; Nguyen, H.P.; Vangala, D.B. Optical Genome Mapping for Cytogenetic Diagnostics in AML. Cancers 2023, 15, 1684. [Google Scholar] [CrossRef]
- Levy, B.; Baughn, L.B.; Akkari, Y.; Chartrand, S.; LaBarge, B.; Claxton, D.; Lennon, P.A.; Cujar, C.; Kolhe, R.; Kroeger, K.; et al. Optical genome mapping in acute myeloid leukemia: A multicenter evaluation. Blood Adv. 2023, 7, 1297–1307. [Google Scholar] [CrossRef]
- Coccaro, N.; Anelli, L.; Zagaria, A.; Tarantini, F.; Cumbo, C.; Tota, G.; Minervini, C.F.; Minervini, A.; Conserva, M.R.; Redavid, I.; et al. Feasibility of Optical Genome Mapping in Cytogenetic Diagnostics of Hematological Neoplasms: A New Way to Look at DNA. Diagnostics 2023, 13, 1841. [Google Scholar] [CrossRef] [PubMed]
- Suttorp, J.; Lühmann, J.L.; Behrens, Y.L.; Göhring, G.; Steinemann, D.; Reinhardt, D.; Neuhoff, N.V.; Schneider, M. Optical Genome Mapping as a Diagnostic Tool in Pediatric Acute Myeloid Leukemia. Cancers 2022, 14, 2058. [Google Scholar] [CrossRef] [PubMed]
- Neveling, K.; Mantere, T.; Vermeulen, S.; Oorsprong, M.; van Beek, R.; Kater-Baats, E.; Pauper, M.; van der Zande, G.; Smeets, D.; Weghuis, D.O.; et al. Next-generation cytogenetics: Comprehensive assessment of 52 hematological malignancy genomes by optical genome mapping. Am. J. Hum. Genet. 2021, 108, 1423–1435. [Google Scholar] [CrossRef]
- Lühmann, J.L.; Stelter, M.; Wolter, M.; Kater, J.; Lentes, J.; Bergmann, A.K.; Schieck, M.; Göhring, G.; Möricke, A.; Cario, G.; et al. The Clinical Utility of Optical Genome Mapping for the Assessment of Genomic Aberrations in Acute Lymphoblastic Leukemia. Cancers 2021, 13, 4388. [Google Scholar] [CrossRef] [PubMed]
- Lestringant, V.; Duployez, N.; Penther, D.; Luquet, I.; Derrieux, C.; Lutun, A.; Preudhomme, C.; West, M.; Ouled-Haddou, H.; Devoldere, C.; et al. Optical genome mapping, a promising alternative to gold standard cytogenetic approaches in a series of acute lymphoblastic leukemias. Genes Chromosomes Cancer 2021, 60, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Sreedharanunni, S.; Thakur, V.; Balakrishnan, A.; Sachdeva, M.U.S.; Kaur, P.; Raina, S.; Jamwal, M.; Singh, C.; Sharma, P.; Mallik, N.; et al. Effective Utilization of a Customized Targeted Hybrid Capture RNA Sequencing in the Routine Molecular Categorization of Adolescent and Adult B-Lineage Acute Lymphoblastic Leukemia: A Real-World Experience. Mol. Diagn. Ther. 2025, 29, 407–418. [Google Scholar] [CrossRef]
- Vu, M.; Degeling, K.; Ryland, G.L.; Hofmann, O.; Ng, A.P.; Westerman, D.; Ijzerman, M.J. Economic Impact of Whole Genome Sequencing and Whole Transcriptome Sequencing Versus Routine Diagnostic Molecular Testing to Stratify Patients with B-Cell Acute Lymphoblastic Leukemia. J. Mol. Diagn. 2024, 26, 673–684. [Google Scholar] [CrossRef]
- Singh, M.; Sharma, P.; Bhatia, P.; Trehan, A.; Thakur, R.; Sreedharanunni, S. Integrated analysis of transcriptome and genome variations in pediatric T cell acute lymphoblastic leukemia: Data from north Indian tertiary care center. BMC Cancer 2024, 24, 325. [Google Scholar] [CrossRef]
- Cuppen, E.; Elemento, O.; Rosenquist, R.; Nikic, S.; Ijzerman, M.; Zaleski, I.D.; Frederix, G.; Levin, L.; Mullighan, C.G.; Buettner, R.; et al. Implementation of Whole-Genome and Transcriptome Sequencing Into Clinical Cancer Care. JCO Precis. Oncol. 2022, 6, e2200245. [Google Scholar] [CrossRef] [PubMed]
- Meggendorfer, M.; Walter, W.; Haferlach, T. WGS and WTS in leukaemia: A tool for diagnostics? Best Pract. Res. Clin. Haematol. 2020, 33, 101190. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.C.; Herborg, L.L.; Hansen, M.; Roug, A.S.; Hokland, P. Combination of RNA- and exome sequencing: Increasing specificity for identification of somatic point mutations and indels in acute leukaemia. Leuk. Res. 2016, 51, 27–31. [Google Scholar] [CrossRef]
- Ngo, T.Q.; Goh, A.F.N.; Dorwal, P.; Leong, E.; Shortt, J.; Fedele, P.L.; Gilbertson, M.; Fong, C.Y.; Shanmuganathan, N.; Kumar, B.; et al. Next-generation sequencing RNA fusion panel for the diagnosis of haematological malignancies. Pathology 2025, 57, 340–347. [Google Scholar] [CrossRef]
- Koduru, P.; Chen, W.; Fuda, F.; Kaur, G.; Awan, F.; John, S.; Garcia, R.; Gagan, J. RNASeq Analysis for Accurate Identification of Fusion Partners in Tumor Specific Translocations Detected by Standard FISH Probes in Hematologic Malignancies. Clin. Pathol. 2024, 17, 1–9. [Google Scholar] [CrossRef]
- Yeung, C.; Qu, X.; Sala-Torra, O.; Woolston, D.; Radich, J.; Fang, M. Mutational profiling in acute lymphoblastic leukemia by RNA sequencing and chromosomal genomic array testing. Cancer Med. 2021, 10, 5629–5642. [Google Scholar] [CrossRef] [PubMed]
- Hayette, S.; Grange, B.; Vallee, M.; Bardel, C.; Huet, S.; Mosnier, I.; Chabane, K.; Simonet, T.; Balsat, M.; Heiblig, M.; et al. Performances of Targeted RNA Sequencing for the Analysis of Fusion Transcripts, Gene Mutation, and Expression in Hematological Malignancies. Hemasphere 2021, 5, e522. [Google Scholar] [CrossRef] [PubMed]
- Archer™ Analysis, v7.2; Integrated DNA Technologies, Inc.: Boulder, CO, USA, 2024. Available online: https://sfvideo.blob.core.windows.net/sitefinity/docs/default-source/user-guide-manual/archer-analysis-user-guide.pdf?sfvrsn=b36f207_3 (accessed on 21 October 2025).
- Mikhail, F.M.; Biegel, J.A.; Cooley, L.D.; Dubuc, A.M.; Hirsch, B.; Horner, V.L.; Newman, S.; Shao, L.; Wolff, D.J.; Raca, G. Technical laboratory standards for interpretation and reporting of acquired copy-number abnormalities and copy-neutral loss of heterozygosity in neoplastic disorders: A joint consensus recommendation from the American College of Medical Genetics and Genomics (ACMG) and the Cancer Genomics Consortium (CGC). Genet. Med. 2019, 21, 1903–1916. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Mulet-Lazaro, R.; Delwel, R. Oncogenic Enhancers in Leukemia. Blood Cancer Discov. 2024, 5, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Fears, S.; Mathieu, C.; Zeleznik-Le, N.; Huang, S.; Rowley, J.D.; Nucifora, G. Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc. Natl. Acad. Sci. USA 1996, 93, 1642–1647. [Google Scholar] [CrossRef]
- Bordereaux, D.; Fichelson, S.; Tambourin, P.; Gisselbrecht, S. Alternative splicing of the Evi-1 zinc finger gene generates mRNAs which differ by the number of zinc finger motifs. Oncogene 1990, 5, 925–927. [Google Scholar] [PubMed]
- Bard-Chapeau, E.A.; Jeyakani, J.; Kok, C.H.; Muller, J.; Chua, B.Q.; Gunaratne, J.; Batagov, A.; Jenjaroenpun, P.; Kuznetsov, V.A.; Wei, C.L.; et al. Ecotopic viral integration site 1 (EVI1) regulates multiple cellular processes important for cancer and is a synergistic partner for FOS protein in invasive tumors. Proc. Natl. Acad. Sci. USA 2012, 109, 2168–2173. [Google Scholar] [CrossRef]
- Bard-Chapeau, E.A.; Gunaratne, J.; Kumar, P.; Chua, B.Q.; Muller, J.; Bard, F.A.; Blackstock, W.; Copeland, N.G.; Jenkins, N.A. EVI1 oncoprotein interacts with a large and complex network of proteins and integrates signals through protein phosphorylation. Proc. Natl. Acad. Sci. USA 2013, 110, E2885–E2894. [Google Scholar] [CrossRef]
- Glass, C.; Wuertzer, C.; Cui, X.; Bi, Y.; Davuluri, R.; Xiao, Y.Y.; Wilson, M.; Owens, K.; Zhang, Y.; Perkins, A. Global Identification of EVI1 Target Genes in Acute Myeloid Leukemia. PLoS ONE 2013, 8, e67134. [Google Scholar] [CrossRef]
- Di Giacomo, D.; La Starza, R.; Gorello, P.; Pellanera, F.; Kalender Atak, Z.; De Keersmaecker, K.; Pierini, V.; Harrison, C.J.; Arniani, S.; Moretti, M.; et al. 14q32 rearrangements deregulating BCL11B mark a distinct subgroup of T-lymphoid and myeloid immature acute leukemia. Blood 2021, 138, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, L.E.; Bendig, S.; Gu, Z.; Chen, X.; Polonen, P.; Ma, X.; Murison, A.; Zeng, A.; Garcia-Prat, L.; Dickerson, K.; et al. Enhancer Hijacking Drives Oncogenic BCL11B Expression in Lineage-Ambiguous Stem Cell Leukemia. Cancer Discov. 2021, 11, 2846–2867. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Hu, S.; Xu, J.; Loghavi, S.; Daver, N.; Toruner, G.A.; Wang, W.; Medeiros, L.J.; Tang, G. Detection of KMT2A Partial Tandem Duplication by Optical Genome Mapping in Myeloid Neoplasms: Associated Cytogenetics, Gene Mutations, Treatment Responses, and Patient Outcomes. Cancers 2024, 16, 4193. [Google Scholar] [CrossRef] [PubMed]
- Cools, J.; DeAngelo, D.J.; Gotlib, J.; Stover, E.H.; Legare, R.D.; Cortes, J.; Kutok, J.; Clark, J.; Galinsky, I.; Griffin, J.D.; et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N. Engl. J. Med. 2003, 348, 1201–1214. [Google Scholar] [CrossRef] [PubMed]

| AML | B-ALL (Continued) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aberration | Tier | N | Concordant | Discordant | Concordance | Aberration | Tier | N | Concordant | Discordant | Concordance |
| KMT2A-R | I | 22 | 21 | 1 | 95.5 | ABL2-R | I | 2 | 2 | 0 | 100 |
| MECOM-R | I | 22 | 6 | 16 | 27.3 | JAK2-R | I | 2 | 2 | 0 | 100 |
| CBFB::MYH11 | I | 16 | 16 | 0 | 100 | MEF2D-R | I | 2 | 2 | 0 | 100 |
| NUP98-R | I | 5 | 5 | 0 | 100 | IGH::P2RY8 | I | 1 | 1 | 0 | 100 |
| PML::RARA | I | 5 | 5 | 0 | 100 | PAX5alt | I | 1 | 1 | 0 | 100 |
| RUNX1::RUNX1T1 | I | 5 | 5 | 0 | 100 | PICALM::MLLT10 | I | 1 | 1 | 0 | 100 |
| DEK::NUP214 | I | 4 | 4 | 0 | 100 | IGH::BCL2 | I | 1 | 0 | 1 | 0 |
| CBFA2T3::GLIS2 | I | 2 | 2 | 0 | 100 | P2RY8::IGH AS | I | 1 | 0 | 1 | 0 |
| BCR::ABL1 | I | 1 | 1 | 0 | 100 | IKZF1 del | II | 23 | 23 | 0 | 100 |
| KAT6A::CREBBP | I | 1 | 1 | 0 | 100 | IGH::CEBPA | II | 2 | 1 | 1 | 50 |
| PICALM::MLLT10 | I | 1 | 1 | 0 | 100 | KMT2A-PTD | II | 1 | 1 | 0 | 100 |
| BCL11B-R | I | 1 | 0 | 1 | 0 | IGH::CEBPB | II | 1 | 0 | 1 | 0 |
| CDK6::MNX1 | I | 1 | 0 | 1 | 0 | SET::NUP214 | II | 1 | 0 | 1 | 0 |
| KMT2A-PTD | II | 30 | 26 | 4 | 86.7 | IKZF2-R | III | 1 | 0 | 1 | 0 |
| Variant RUNX1 | II | 5 | 3 | 2 | 60 | NF1::RHOT1 | III | 1 | 0 | 1 | 0 |
| NPM1::MLF1 | II | 1 | 1 | 0 | 100 | Positive | 86 | 69 | 17 | 80.2 | |
| ETV6-R | II | 1 | 0 | 1 | 0 | Negative | 27 | 27 | 0 | 100 | |
| IKZF1 del | II | 1 | 0 | 1 | 0 | Total (B-ALL) | 113 | 96 | 17 | 85 | |
| SET::NUP214 | II | 1 | 0 | 1 | 0 | MPAL | |||||
| CSTF3::WT1 | III | 1 | 1 | 0 | 100 | BCR::ABL1 | I | 1 | 1 | 0 | 100 |
| IKZF1::LRBA | III | 1 | 1 | 0 | 100 | MECOM-R | I | 1 | 0 | 1 | 0 |
| KPNA1::TP63 | III | 1 | 1 | 0 | 100 | ETV6-R | II | 1 | 0 | 1 | 0 |
| CDKN2A::SLC24A2 | III | 1 | 0 | 1 | 0 | Positive | 3 | 1 | 2 | 33.3 | |
| EPOR-R | III | 1 | 0 | 1 | 0 | Negative | 3 | 3 | 0 | 100 | |
| EPS15L1::KLF2 | III | 1 | 0 | 1 | 0 | Total (MPAL) | 6 | 4 | 2 | 66.7 | |
| FAR2::CCND2 | III | 1 | 0 | 1 | 0 | T-ALL | |||||
| LMO1::RIC3 | III | 1 | 0 | 1 | 0 | BCL11B-R | I | 5 | 0 | 5 | 0 |
| Positive | 133 | 100 | 33 | 75.2 | ETV6-R | II | 4 | 3 | 1 | 75 | |
| Negative | 230 | 230 | 0 | 100 | NUP214::ABL1 | II | 2 | 1 | 1 | 50 | |
| Total (AML) | 363 | 330 | 33 | 90.9 | SET::NUP214 | II | 1 | 1 | 0 | 100 | |
| Positive | 12 | 5 | 7 | 41.7 | |||||||
| B-ALL | Negative | 3 | 3 | 0 | 100 | ||||||
| BCR::ABL1 | I | 22 | 22 | 0 | 100 | Total (T-ALL) | 15 | 8 | 7 | 53.3 | |
| IGH::CRLF2 | I | 6 | 2 | 4 | 33.3 | ||||||
| KMT2A-R | I | 4 | 4 | 0 | 100 | TOTAL | |||||
| TCF3-R | I | 4 | 4 | 0 | 100 | Positive | 234 | 175 | 59 | 74.7 | |
| ETV6::RUNX1 | I | 3 | 3 | 0 | 100 | Negative | 263 | 263 | 0 | 100 | |
| IGH::EPOR | I | 2 | 2 | 0 | 100 | Grand Total | 497 | 438 | 59 | 88.1 | |
| P2YR8::CRLF2 | I | 3 | 0 | 3 | 0 | ||||||
| Aberration | Tier | OGM Only | LTP Only | Both | Total | Aberration | Tier | OGM Only | LTP Only | Both | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KMT2A-R | I | 0 (0.0%) | 1 (3.8%) | 25 (96.2%) | 26 | KMT2A-PTD | II | 2 (6.5%) | 2 (6.5%) | 27 (87.1%) | 31 |
| BCR::ABL1 | I | 0 (0.0%) | 0 (0.0%) | 24 (100.0%) | 24 | Z | II | 0 (0.0%) | 1 (4.3%) | 22 (95.7%) | 23 |
| MECOM-R | I | 17 (73.9%) | 0 (0.0%) | 6 (26.1%) | 23 | ETV6-R | II | 1 (16.7%) | 2 (33.3%) | 3 (50.0%) | 6 |
| CBFB::MYH11 | I | 0 (0.0%) | 0 (0.0%) | 16 (100.0%) | 16 | Variant RUNX1 | II | 1 (20.0%) | 1 (20.0%) | 3 (60.0%) | 5 |
| BCL11B-R | I | 6 (100.0%) | 0 (0.0%) | 0 (0.0%) | 6 | SET::NUP214 | II | 0 (0.0%) | 2 (66.7%) | 1 (33.3%) | 3 |
| IGH::CRLF2 | I | 4 (66.7%) | 0 (0.0%) | 2 (33.3%) | 6 | IGH::CEBPA | II | 1 (50.0%) | 0 (0.0%) | 1 (50.0%) | 2 |
| NUP98-R | I | 0 (0.0%) | 0 (0.0%) | 5 (100.0%) | 5 | NUP214::ABL1 | II | 0 (0.0%) | 1 (50.0%) | 1 (50.0%) | 2 |
| PML::RARA | I | 0 (0.0%) | 0 (0.0%) | 5 (100.0%) | 5 | IGH::CEBPB | II | 1 (100.0%) | 0 (0.0%) | 0 (0.0%) | 1 |
| RUNX1::RUNX1T1 | I | 0 (0.0%) | 0 (0.0%) | 5 (100.0%) | 5 | NPM1::MLF1 | II | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) | 1 |
| DEK::NUP214 | I | 0 (0.0%) | 0 (0.0%) | 4 (100.0%) | 4 | CDKN2A::SLC24A2 | III | 0 (0.0%) | 1 (100.0%) | 0 (0.0%) | 1 |
| TCF3-R | I | 0 (0.0%) | 0 (0.0%) | 4 (100.0%) | 4 | CSTF3::WT1 | III | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) | 1 |
| ETV6::RUNX1 | I | 0 (0.0%) | 0 (0.0%) | 3 (100.0%) | 3 | EPOR-R | III | 0 (0.0%) | 1 (100.0%) | 0 (0.0%) | 1 |
| IGH::EPOR | I | 2 (66.7%) | 1 (33.3%) | 0 (0.0%) | 3 | EPS15L1::KLF2 | III | 0 (0.0%) | 1 (100.0%) | 0 (0.0%) | 1 |
| P2YR8::CRLF2 | I | 0 (0.0%) | 3 (100.0%) | 0 (0.0%) | 3 | FAR2::CCND2 | III | 0 (0.0%) | 1 (100.0%) | 0 (0.0%) | 1 |
| ABL2-R | I | 0 (0.0%) | 0 (0.0%) | 2 (100.0%) | 2 | IKZF1::LRBA | III | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) | 1 |
| CBFA2T3::GLIS2 | I | 0 (0.0%) | 0 (0.0%) | 2 (100.0%) | 2 | IKZF2 = R | III | 0 (0.0%) | 1 (100.0%) | 0 (0.0%) | 1 |
| JAK2-R | I | 0 (0.0%) | 0 (0.0%) | 2 (100.0%) | 2 | KPNA1::TP63 | III | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) | 1 |
| MEF2D-R | I | 0 (0.0%) | 0 (0.0%) | 2 (100.0%) | 2 | LMO1::RIC3 | III | 0 (0.0%) | 1 (100.0%) | 0 (0.0%) | 1 |
| PICALM::MLLT10 | I | 0 (0.0%) | 0 (0.0%) | 2 (100.0%) | 2 | NF1::RHOT1 | III | 0 (0.0%) | 1 (100.0%) | 0 (0.0%) | 1 |
| CDK6::MNX1 | I | 1 (100.0%) | 0 (0.0%) | 0 (0.0%) | 1 | ||||||
| IGH::BCL2 | I | 1 (100.0%) | 0 (0.0%) | 0 (0.0%) | 1 | Total | 37 (15.8%) | 22 (9.4%) | 175 (74.7%) | 234 | |
| IGH::P2RY8 | I | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) | 1 | ||||||
| KAT6A::CREBBP | I | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) | 1 | ||||||
| P2RY8::IGH AS | I | 0 (0.0%) | 1 (100.0%) | 0 (0.0%) | 1 | ||||||
| PAX5alt | I | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ok, C.Y.; Tang, G.; Loghavi, S.; Hu, S.; Wei, Q.; Quesada, A.E.; Routbort, M.J.; Kanagal-Shamanna, R.; Yin, C.C.; Sarami, I.; et al. Comparative Analysis of Targeted RNA-Seq and Optical Genome Mapping for Detecting Gene Rearrangements in Acute Leukemia. Cancers 2025, 17, 3458. https://doi.org/10.3390/cancers17213458
Ok CY, Tang G, Loghavi S, Hu S, Wei Q, Quesada AE, Routbort MJ, Kanagal-Shamanna R, Yin CC, Sarami I, et al. Comparative Analysis of Targeted RNA-Seq and Optical Genome Mapping for Detecting Gene Rearrangements in Acute Leukemia. Cancers. 2025; 17(21):3458. https://doi.org/10.3390/cancers17213458
Chicago/Turabian StyleOk, Chi Young, Guilin Tang, Sanam Loghavi, Shimin Hu, Qing Wei, Andres E. Quesada, Mark J. Routbort, Rashmi Kanagal-Shamanna, C. Cameron Yin, Iman Sarami, and et al. 2025. "Comparative Analysis of Targeted RNA-Seq and Optical Genome Mapping for Detecting Gene Rearrangements in Acute Leukemia" Cancers 17, no. 21: 3458. https://doi.org/10.3390/cancers17213458
APA StyleOk, C. Y., Tang, G., Loghavi, S., Hu, S., Wei, Q., Quesada, A. E., Routbort, M. J., Kanagal-Shamanna, R., Yin, C. C., Sarami, I., Garces, S., Agarwal, N. K., Luthra, R., Fang, H., Jelloul, F. Z., Bryan, J., Medeiros, L. J., Patel, K. P., & Toruner, G. A. (2025). Comparative Analysis of Targeted RNA-Seq and Optical Genome Mapping for Detecting Gene Rearrangements in Acute Leukemia. Cancers, 17(21), 3458. https://doi.org/10.3390/cancers17213458









