CRISPR with a Double Mismatch Guide RNA Enhances Detection Sensitivity for Low-Frequency Single-Base EGFR Mutation in Circulating Cell-Free DNA of Lung Cancer Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Purification of LbCas12a Recombinant Protein
2.2. In Vitro Transcription of crRNA
2.3. Preparations of PCR Amplicon and In Vitro Cleavage
2.4. Cell-Free DNA Purification and Quantification
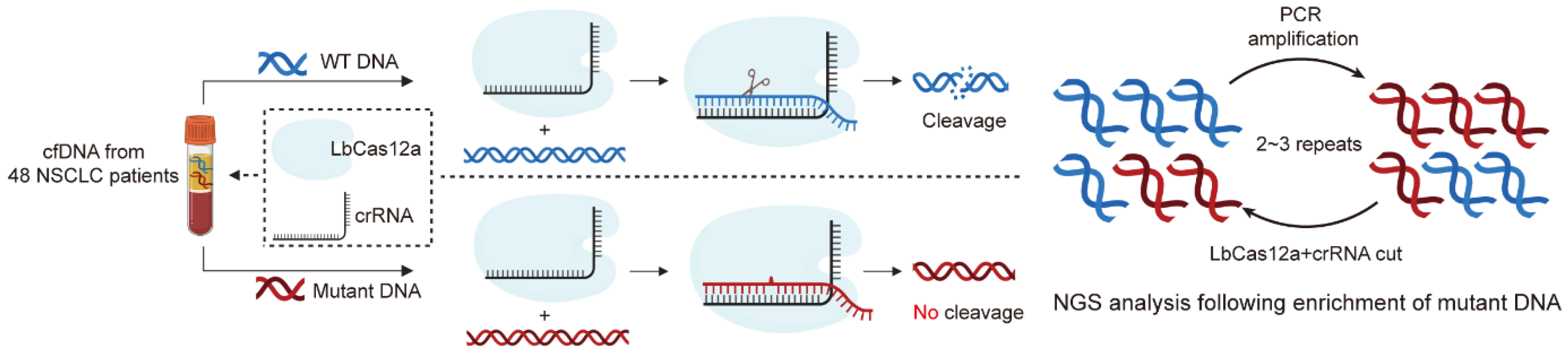
2.5. Enrichment of Mutated DNA Through the CRISPR/Cas12a Amplification
3. Result
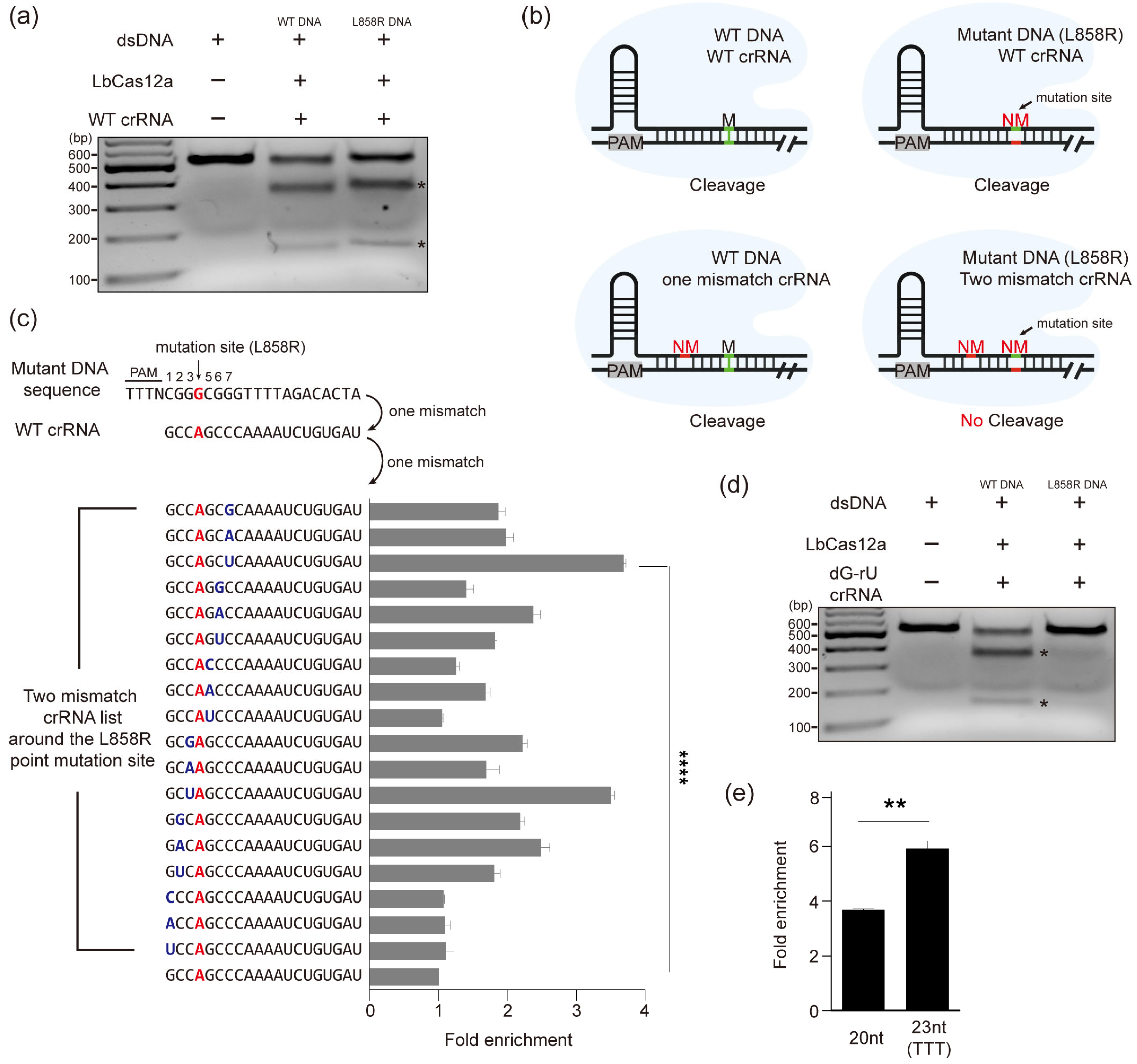
3.1. Establishment of a CRISPR/Cas12a-Based Diagnostic System for Enrichment of Mutated DNA
3.2. Selective Enrichment of Mutant DNA Through Mismatch crRNA Targeting
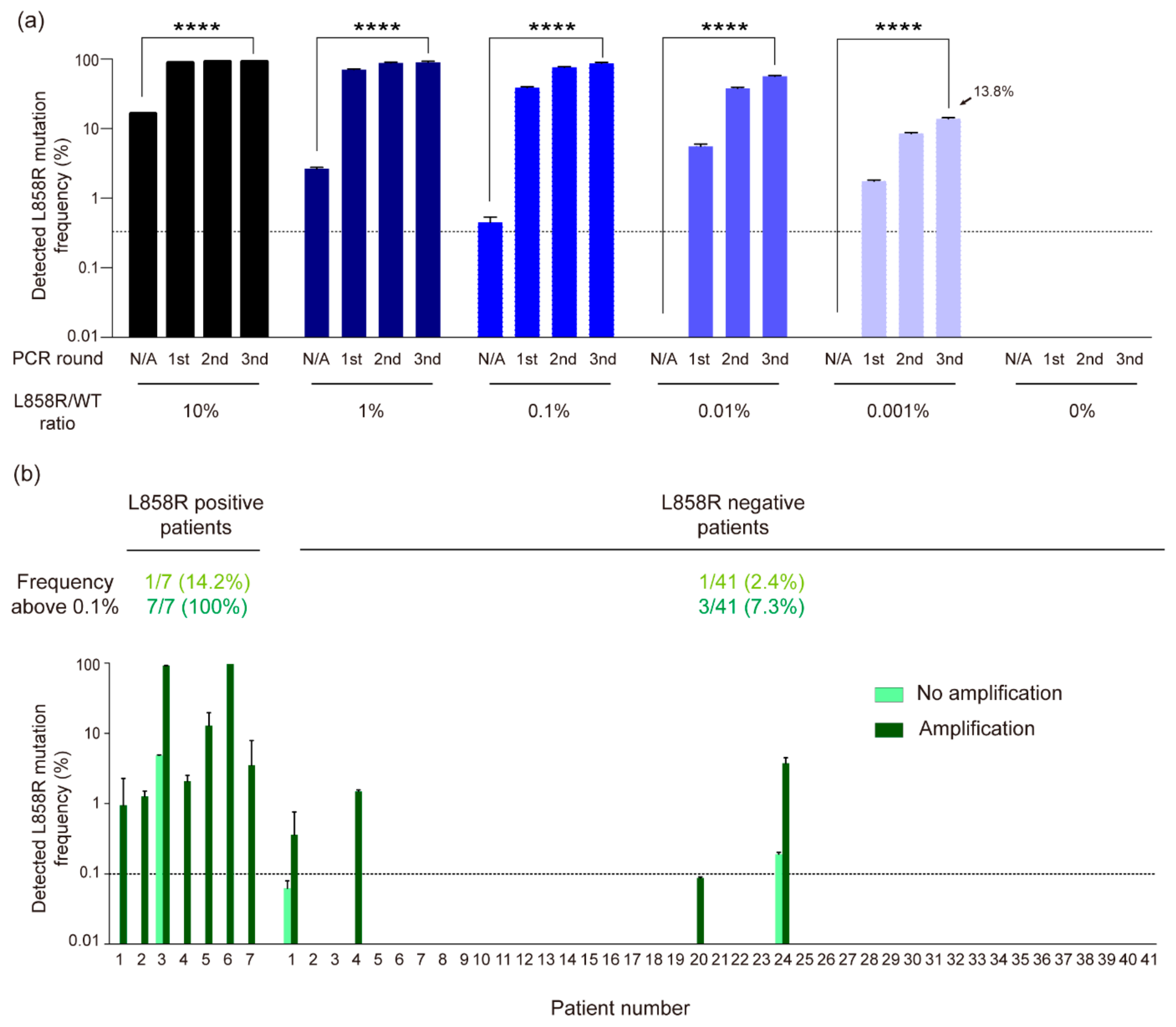
3.3. High-Sensitivity Detection of EGFR L858R Mutation in NSCLC Liquid Biopsy Samples Using CRISPR/Cas12a-Based Diagnostic System
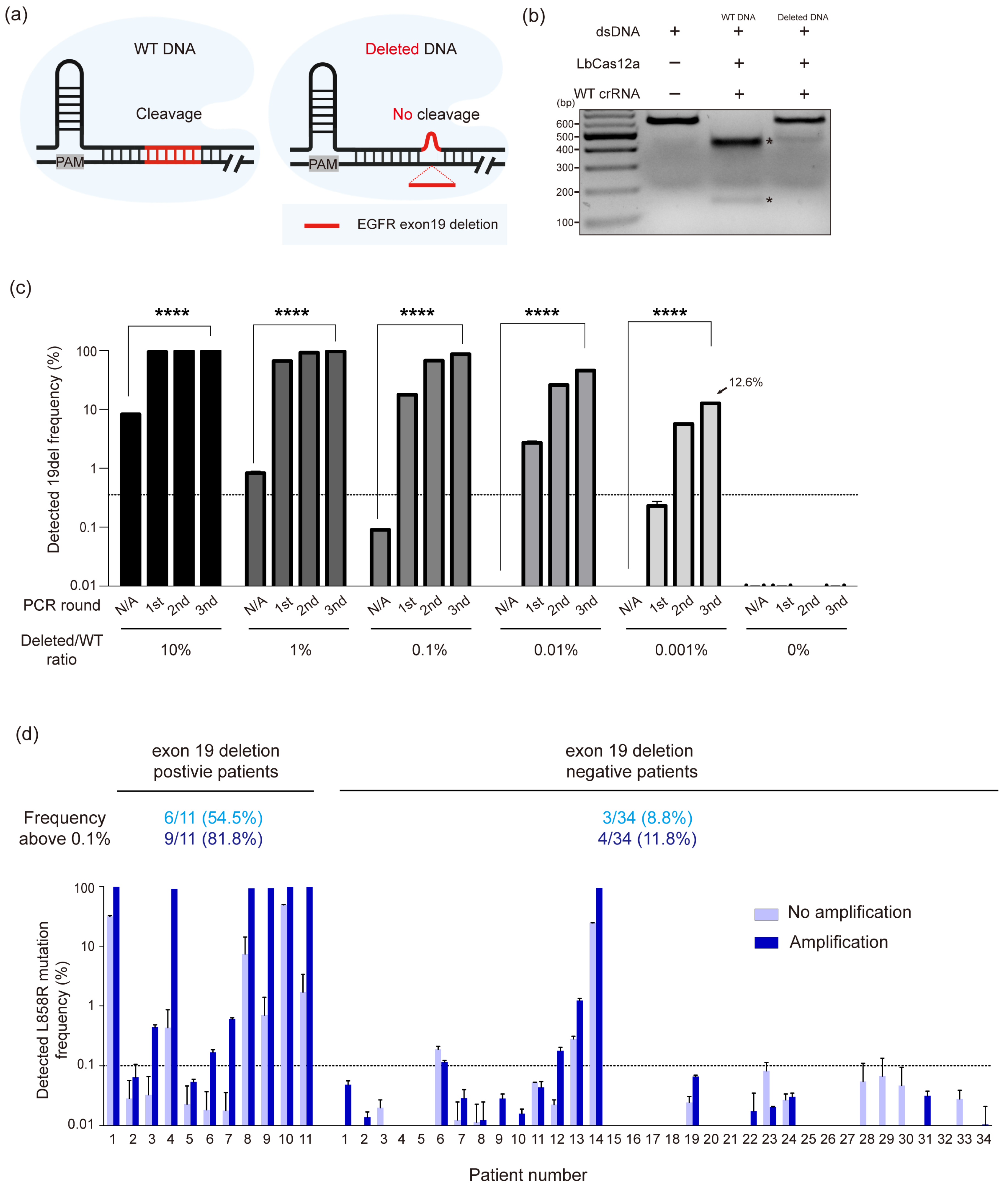
3.4. Extending CRISPR/Cas12a-Based Diagnostic System to EGFR Exon 19 Deletions in NSCLC
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kruglyak, K.M.; Lin, E.; Ong, F.S. Next-generation sequencing in precision oncology: Challenges and opportunities. Expert Rev. Mol. Diagn. 2014, 14, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Kricka, L.J.; Fortina, P. Next-generation sequencing in the clinic. Nat. Biotechnol. 2013, 31, 990–992. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, J.; Yamazaki, K.; Kinoshita, I.; Yokouchi, H.; Dosaka-Akita, H.; Nishimura, M. Non-responsiveness to gefitinib in a patient with lung adenocarcinoma having rare EGFR mutations S768I and V769L. Lung Cancer 2006, 54, 419–422. [Google Scholar]
- Naidoo, J.; Sima, C.S.; Rodriguez, K.; Busby, N.; Nafa, K.; Ladanyi, M.; Riely, G.J.; Kris, M.G.; Arcila, M.E.; Yu, H.A. Epidermal growth factor receptor exon 20 insertions in advanced lung adenocarcinomas: Clinical outcomes and response to erlotinib. Cancer 2015, 121, 3212–3220. [Google Scholar] [CrossRef]
- Yang, J.C.; Sequist, L.V.; Geater, S.L.; Tsai, C.M.; Mok, T.S.; Schuler, M.; Yamamoto, N.; Yu, C.J.; Ou, S.H.; Zhou, C.; et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: A combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015, 16, 830–838. [Google Scholar] [CrossRef]
- Sorber, L.; Zwaenepoel, K.; Deschoolmeester, V.; Van Schil, P.E.; Van Meerbeeck, J.; Lardon, F.; Rolfo, C.; Pauwels, P. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer 2017, 107, 100–107. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Caldas, C. Cell-free circulating tumour DNA as a liquid biopsy in breast cancer. Mol. Oncol. 2016, 10, 464–474. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef]
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Schwarzenbach, H.; Pantel, K. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 2012, 63, 199–215. [Google Scholar] [CrossRef]
- Freidin, M.B.; Freydina, D.V.; Leung, M.; Montero Fernandez, A.; Nicholson, A.G.; Lim, E. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin. Chem. 2015, 61, 1299–1304. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.C.; Weiss, J.; Hudson, C.; Christophi, C.; Cebon, J.; Behren, A.; Dobrovic, A. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci. Rep. 2015, 5, 11198. [Google Scholar] [CrossRef] [PubMed]
- Chabon, J.J.; Simmons, A.D.; Lovejoy, A.F.; Esfahani, M.S.; Newman, A.M.; Haringsma, H.J.; Kurtz, D.M.; Stehr, H.; Scherer, F.; Karlovich, C.A.; et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat. Commun. 2016, 7, 11815. [Google Scholar] [CrossRef]
- Lanman, R.B.; Mortimer, S.A.; Zill, O.A.; Sebisanovic, D.; Lopez, R.; Blau, S.; Collisson, E.A.; Divers, S.G.; Hoon, D.S.; Kopetz, E.S.; et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS ONE 2015, 10, e0140712. [Google Scholar]
- Joshi, J.; Raval, A.; Desai, U.; Upadhyay, V.; Bhavsar, M.; Shah, K.; Rawal, R.; Panchal, H.; Shah, F. EGFR Mutation Analysis in Non-small Cell Lung Carcinoma Patients: A Liquid Biopsy Approach. Indian J. Clin. Biochem. 2021, 36, 51–58. [Google Scholar]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement From the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef]
- Ulivi, P.; Petracci, E.; Canale, M.; Priano, I.; Capelli, L.; Calistri, D.; Chiadini, E.; Cravero, P.; Rossi, A.; Delmonte, A.; et al. Liquid Biopsy for EGFR Mutation Analysis in Advanced Non-Small-Cell Lung Cancer Patients: Thoughts Drawn from a Real-Life Experience. Biomedicines 2021, 9, 1299. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Paweletz, C.P.; Kuang, Y.; Mach, S.L.; O’Connell, A.; Messineo, M.M.; Luke, J.J.; Butaney, M.; Kirschmeier, P.; Jackman, D.M.; et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. 2014, 20, 1698–1705. [Google Scholar] [CrossRef]
- Narayan, A.; Carriero, N.J.; Gettinger, S.N.; Kluytenaar, J.; Kozak, K.R.; Yock, T.I.; Muscato, N.E.; Ugarelli, P.; Decker, R.H.; Patel, A.A. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012, 72, 3492–3498. [Google Scholar] [CrossRef]
- Lee, S.H.; Yu, J.; Hwang, G.H.; Kim, S.; Kim, H.S.; Ye, S.; Kim, K.; Park, J.; Park, D.Y.; Cho, Y.K.; et al. CUT-PCR: CRISPR-mediated, ultrasensitive detection of target DNA using PCR. Oncogene 2017, 36, 6823–6829. [Google Scholar] [CrossRef]
- Gu, W.; Crawford, E.D.; O’Donovan, B.D.; Wilson, M.R.; Chow, E.D.; Retallack, H.; DeRisi, J.L. Depletion of Abundant Sequences by Hybridization (DASH): Using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016, 17, 41. [Google Scholar] [CrossRef]
- Fan, H.C.; Blumenfeld, Y.J.; Chitkara, U.; Hudgins, L.; Quake, S.R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA 2008, 105, 16266–16271. [Google Scholar] [CrossRef]
- Park, J.; Lim, K.; Kim, J.S.; Bae, S. Cas-Analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics 2017, 33, 286–288. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.X.; St Onge, R.P.; Fire, A.Z.; Smith, J.D. Distinct patterns of Cas9 mismatch tolerance in vitro and in vivo. Nucleic Acids Res. 2016, 44, 5365–5377. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Cradick, T.J.; Brown, M.T.; Deshmukh, H.; Ranjan, P.; Sarode, N.; Wile, B.M.; Vertino, P.M.; Stewart, F.J.; Bao, G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014, 42, 7473–7485. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Fortin, J.P.; Tan, J.; Gascoigne, K.E.; Haverty, P.M.; Forrest, W.F.; Costa, M.R.; Martin, S.E. Multiple-gene targeting and mismatch tolerance can confound analysis of genome-wide pooled CRISPR screens. Genome Biol. 2019, 20, 21. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Kim, H.J.; Lee, S.J. CRISPR-Cas9-mediated pinpoint microbial genome editing aided by target-mismatched sgRNAs. Genome Res. 2020, 30, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015, 33, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, V.; Lin, S.; Guilinger, J.P.; Ma, E.; Doudna, J.A.; Liu, D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013, 31, 839–843. [Google Scholar] [CrossRef]
- Zheng, T.; Hou, Y.; Zhang, P.; Zhang, Z.; Xu, Y.; Zhang, L.; Niu, L.; Yang, Y.; Liang, D.; Yi, F.; et al. Profiling single-guide RNA specificity reveals a mismatch sensitive core sequence. Sci. Rep. 2017, 7, 40638. [Google Scholar] [CrossRef]
- Lazzarotto, C.R.; Malinin, N.L.; Li, Y.; Zhang, R.; Yang, Y.; Lee, G.; Cowley, E.; He, Y.; Lan, X.; Jividen, K.; et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR-Cas9 genome-wide activity. Na.t Biotechnol. 2020, 38, 1317–1327. [Google Scholar] [CrossRef]
- Kim, H.J.; Oh, S.Y.; Lee, S.J. Single-Base Genome Editing in Corynebacterium glutamicum with the Help of Negative Selection by Target-Mismatched CRISPR/Cpf1. J. Microbiol. Biotechnol. 2020, 30, 1583–1591. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Tsai, S.Q.; Prew, M.S.; Nguyen, N.T.; Welch, M.M.; Lopez, J.M.; McCaw, Z.R.; Aryee, M.J.; Joung, J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016, 34, 869–874. [Google Scholar] [CrossRef]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.H.; Kim, J.S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–868. [Google Scholar] [CrossRef]
- Kim, J.M.; Kim, D.; Kim, S.; Kim, J.S. Genotyping with CRISPR-Cas-derived RNA-guided endonucleases. Nat. Commun. 2014, 5, 3157. [Google Scholar] [CrossRef]
- Moon, S.B.; Lee, J.M.; Kang, J.G.; Lee, N.E.; Ha, D.I.; Kim, D.Y.; Kim, S.H.; Yoo, K.; Kim, D.; Ko, J.H.; et al. Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3 ‘-overhang. Nat. Commun. 2018, 9, 3651. [Google Scholar] [CrossRef]
- Marchetti, A.; Del Grammastro, M.; Filice, G.; Felicioni, L.; Rossi, G.; Graziano, P.; Sartori, G.; Leone, A.; Malatesta, S.; Iacono, M.; et al. Complex mutations & subpopulations of deletions at exon 19 of EGFR in NSCLC revealed by next generation sequencing: Potential clinical implications. PLoS ONE 2012, 7, e42164. [Google Scholar]
- Tian, Y.; Zhao, J.; Ren, P.; Wang, B.; Zhao, C.; Shi, C.; Wei, B.; Ma, J.; Guo, Y. Different subtypes of EGFR exon19 mutation can affect prognosis of patients with non-small cell lung adenocarcinoma. PLoS ONE 2018, 13, e0201682. [Google Scholar] [CrossRef] [PubMed]
- Tokudome, N.; Koh, Y.; Akamatsu, H.; Fujimoto, D.; Okamoto, I.; Nakagawa, K.; Hida, T.; Imamura, F.; Morita, S.; Yamamoto, N. Differential significance of molecular subtypes which were classified into EGFR exon 19 deletion on the first line afatinib monotherapy. BMC Cancer 2020, 20, 103. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Toschi, L.; Finocchiaro, G.; Di Noia, V.; Bonomi, M.; Cerchiaro, E.; Ceresoli, G.L.; Beretta, G.D.; D’Argento, E.; Santoro, A. Impact of Exon 19 Deletion Subtypes in EGFR-Mutant Metastatic Non-Small-Cell Lung Cancer Treated With First-Line Tyrosine Kinase Inhibitors. Clin. Lung Cancer 2019, 20, 82–87. [Google Scholar] [CrossRef]
- Zhao, C.; Jiang, T.; Li, J.; Wang, Y.; Su, C.; Chen, X.; Ren, S.; Li, X.; Zhou, C. The impact of EGFR exon 19 deletion subtypes on clinical outcomes in non-small cell lung cancer. Transl. Lung Cancer Res. 2020, 9, 1149–1158. [Google Scholar] [CrossRef]
- Lee Yu, H.; Cao, Y.; Lu, X.; Hsing, I.M. Detection of rare variant alleles using the AsCas12a double-stranded DNA trans-cleavage activity. Biosens. Bioelectron. 2021, 189, 113382. [Google Scholar] [CrossRef]
- Feng, C.; Liang, W.; Liu, F.; Xiong, Y.; Chen, M.; Feng, P.; Guo, M.; Wang, Y.; Li, Z.; Zhang, L. A Simple and Highly Sensitive Naked-Eye Analysis of EGFR 19del via CRISPR/Cas12a Triggered No-Nonspecific Nucleic Acid Amplification. ACS Synth. Biol. 2022, 11, 867–876. [Google Scholar] [CrossRef]
- Kim, H.; Lee, W.J.; Oh, Y.; Kang, S.H.; Hur, J.K.; Lee, H.; Song, W.; Lim, K.S.; Park, Y.H.; Song, B.S.; et al. Enhancement of target specificity of CRISPR-Cas12a by using a chimeric DNA-RNA guide. Nucleic Acids Res. 2020, 48, 8601–8616. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef]
- Meshalkina, D.A.; Glushchenko, A.S.; Kysil, E.V.; Mizgirev, I.V.; Frolov, A. SpCas9- and LbCas12a-mediated DNA editing produce distinct mutational outcomes in zebrafish embryos. Genes 2020, 11, 740. [Google Scholar] [CrossRef]
- Su, H.; Wang, Y.; Xu, J.; Omar, A.A.; Grosser, J.W.; Wang, N. Cas12a RNP-mediated co-transformation enables efficient CRISPR editing in plants. Front. Plant Sci. 2024, 15, 1448807. [Google Scholar]
- Wang, L.; Wang, Y.; Chen, J.; Zhu, Y.; Qin, H.; Liu, J.; Ai, Y.; Lai, J.; Lian, Z.; Han, H. An engineered CRISPR–Cas12i tool for efficient multiplexed genome editing in embryos. Nucleic Acids Res. 2025, 53, gkaf806. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Been, K.W.; Kang, S.; Bae, T.; Hong, S.; Kim, G.; Hur, J.K.; Hwang, W.; Chang, B. CRISPR with a Double Mismatch Guide RNA Enhances Detection Sensitivity for Low-Frequency Single-Base EGFR Mutation in Circulating Cell-Free DNA of Lung Cancer Patients. Cancers 2025, 17, 3343. https://doi.org/10.3390/cancers17203343
Been KW, Kang S, Bae T, Hong S, Kim G, Hur JK, Hwang W, Chang B. CRISPR with a Double Mismatch Guide RNA Enhances Detection Sensitivity for Low-Frequency Single-Base EGFR Mutation in Circulating Cell-Free DNA of Lung Cancer Patients. Cancers. 2025; 17(20):3343. https://doi.org/10.3390/cancers17203343
Chicago/Turabian StyleBeen, Kyung Wook, Seunghun Kang, Taegeun Bae, Sumin Hong, Garyeong Kim, Junho K. Hur, Woochang Hwang, and Boksoon Chang. 2025. "CRISPR with a Double Mismatch Guide RNA Enhances Detection Sensitivity for Low-Frequency Single-Base EGFR Mutation in Circulating Cell-Free DNA of Lung Cancer Patients" Cancers 17, no. 20: 3343. https://doi.org/10.3390/cancers17203343
APA StyleBeen, K. W., Kang, S., Bae, T., Hong, S., Kim, G., Hur, J. K., Hwang, W., & Chang, B. (2025). CRISPR with a Double Mismatch Guide RNA Enhances Detection Sensitivity for Low-Frequency Single-Base EGFR Mutation in Circulating Cell-Free DNA of Lung Cancer Patients. Cancers, 17(20), 3343. https://doi.org/10.3390/cancers17203343







