Whole-Genome Sequencing Reveals a Novel GATA2 Mutation in Lower-Grade Glioma: Bioinformatics Analysis of Functional and Therapeutic Implications
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction
2.2. DNA Library Preparation and DNA Sequencing
2.3. Bioinformatics: NanoPlot Analysis
2.4. Mutagenesis of the Modeled Protein
2.5. Mutation Taster Analysis
2.6. HOPE Analysis
2.6.1. Amino Acid Analysis
2.6.2. Genomic Variants Analysis
2.6.3. Domain Identification
2.7. Drug Affinity Analysis
3. Results
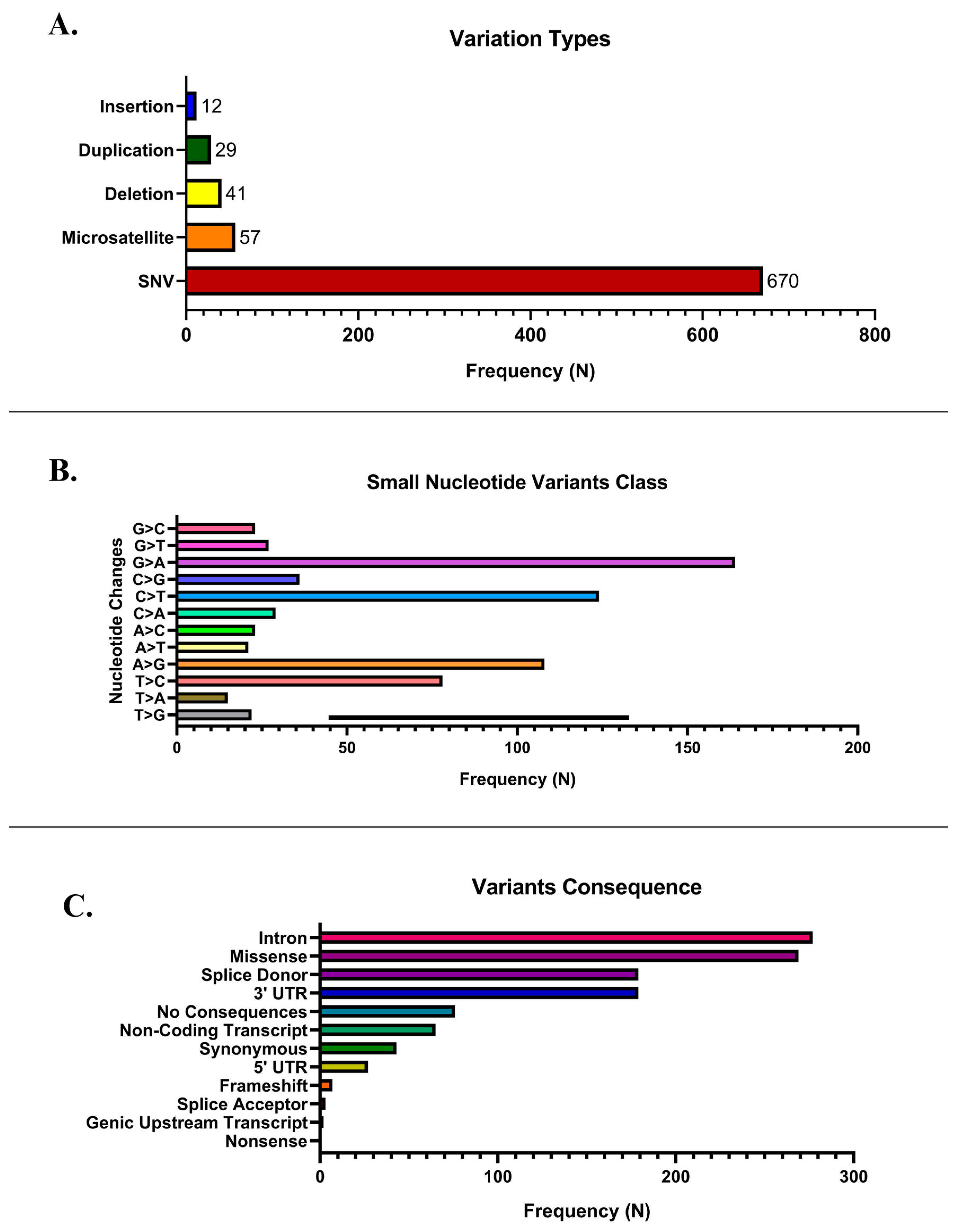
3.1. NanoPlot Analysis
3.1.1. Copy Number Variations
3.1.2. Single Nucleotide Variants
3.1.3. ClinVar Annotation
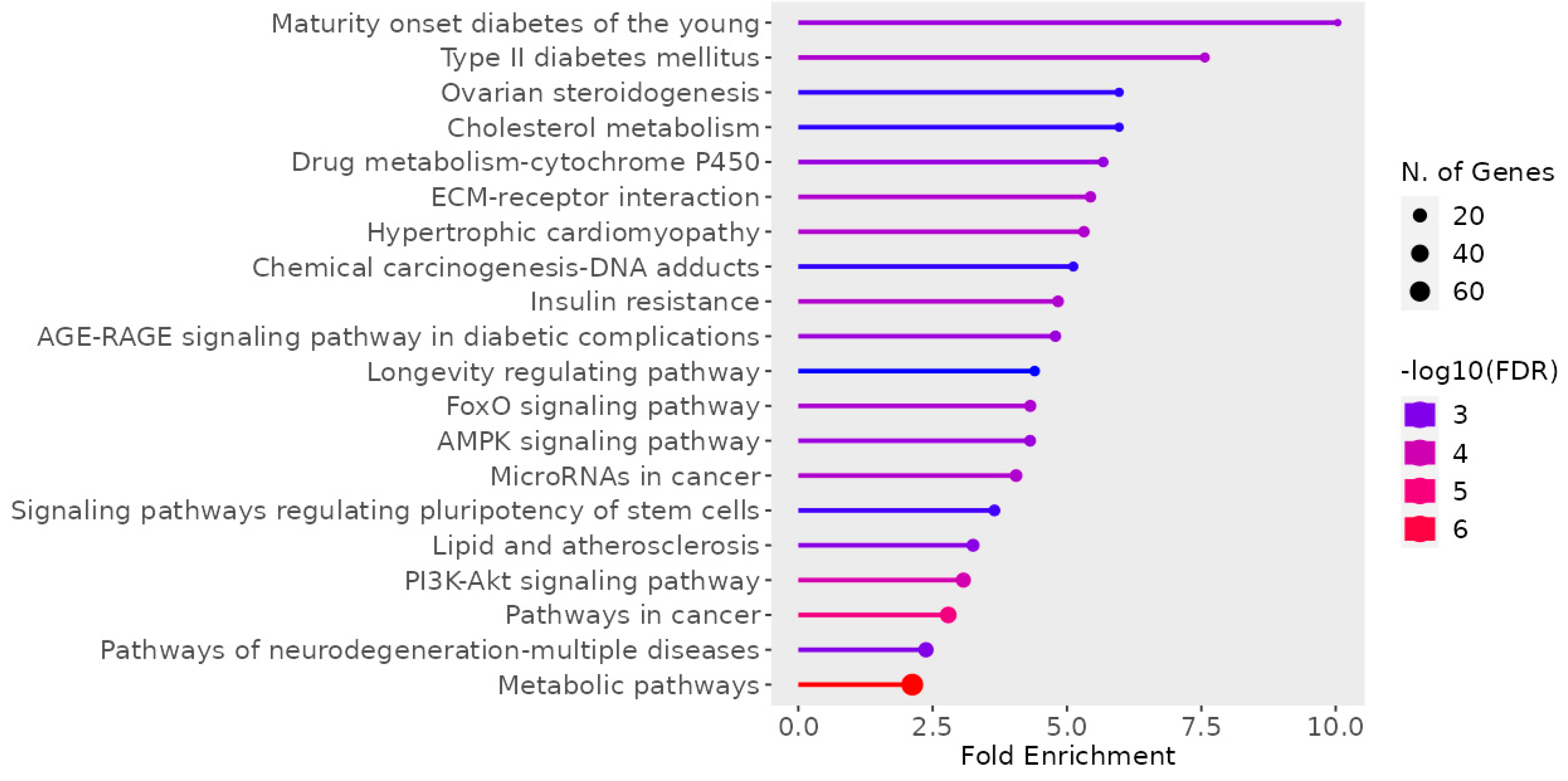
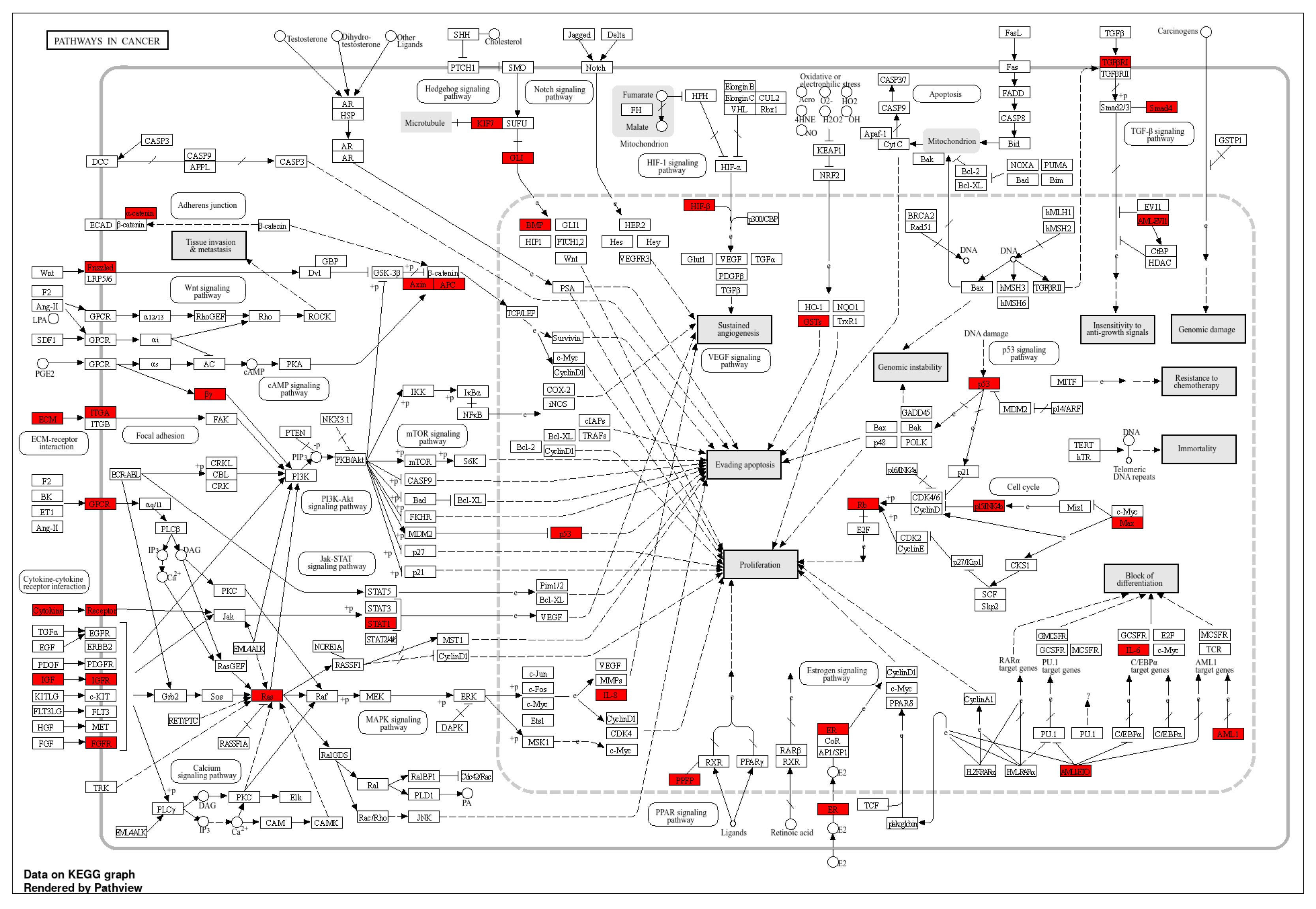
3.1.4. KEGG Pathway Enrichment Analysis
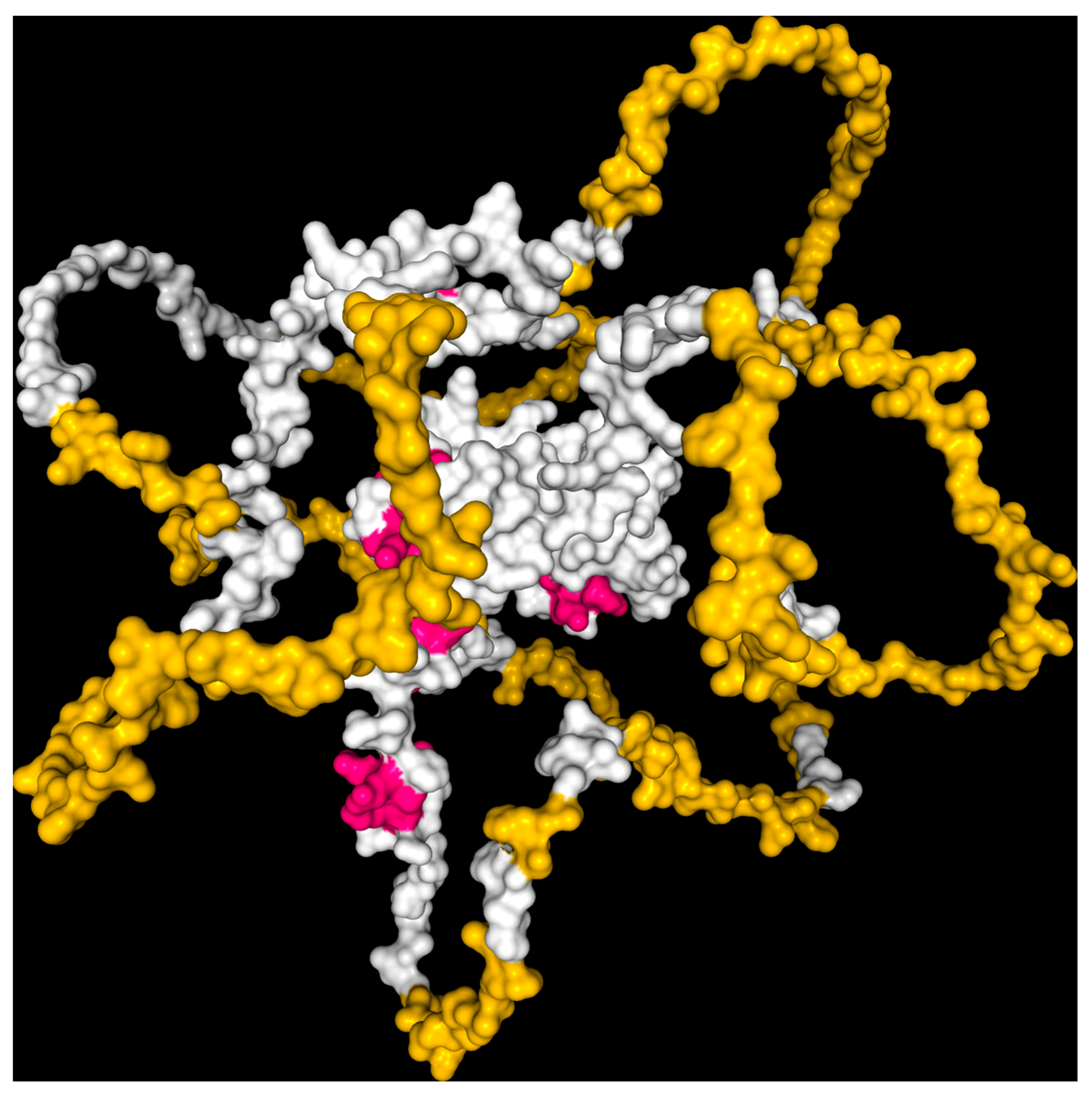
3.2. Mutagenesis of the Modeled Protein
3.3. Mutation Taster Analysis
3.4. HOPE Analysis
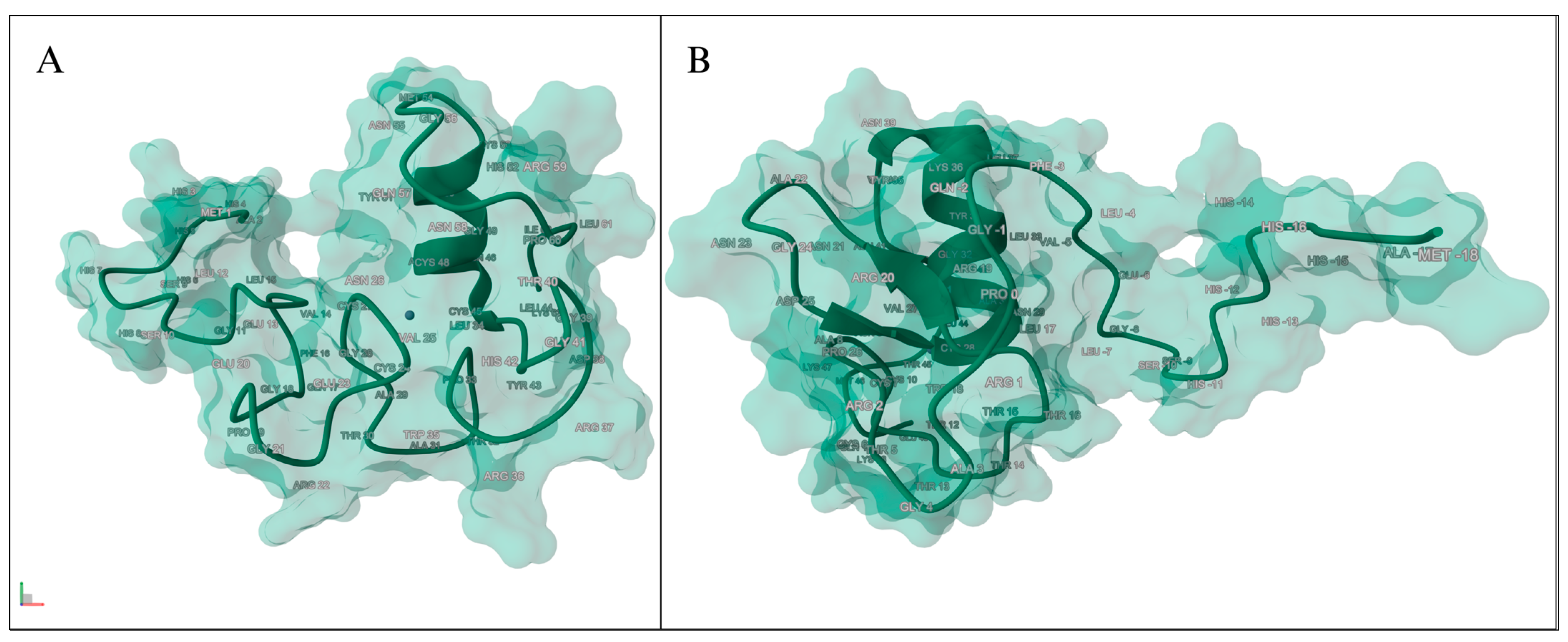
3.4.1. Amino Acid Analysis
3.4.2. Genomic Variants Analysis
3.4.3. Domain Identification
3.5. Drug Affinity Analysis
4. Discussion
4.1. Limitations
4.2. Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AML | Acute Myeloid |
| CNV | Copy Number Variation |
| IDH | Isocitrate Dehydrogenase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MDS | Myelodysplastic Syndrome |
| PBS | Phosphate-Buffer Saline |
| SNV | Single Nucleotide Variants |
| WGS | Whole-Genome Sequencing |
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States. Neuro-Oncol. 2014, 23, iii1–iii105. [Google Scholar] [CrossRef] [PubMed]
- Claus, E.B.; Walsh, K.M.; Wiencke, J.K.; Molinaro, A.M.; Wiemels, J.L.; Schildkraut, J.M.; Bondy, M.L.; Berger, M.; Jenkins, R.; Wrensch, M. Survival and Low-Grade Glioma: The Emergence of Genetic Information. Neurosurg. Focus 2015, 38, E6. [Google Scholar] [CrossRef] [PubMed]
- Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M. Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. Cancer Genome Atlas Res. Netw. 2015, 372, 2481–2498. [Google Scholar]
- Kinnersley, B.; Labussière, M.; Holroyd, A.; Di Stefano, A.L.; Broderick, P.; Vijayakrishnan, J.; Mokhtari, K.; Delattre, J.Y.; Gousias, K.; Schramm, J. Genomic Landscape of Diffuse Glioma Revealed by Whole Genome Sequencing. Nat. Commun. 2025, 16, 4233. [Google Scholar] [CrossRef]
- Kim, T.; Park, C.K.; Kim, S.H.; Choi, S.H.; Park, S.H.; Heo, J.; Kim, J.H.; Yoo, R.E.; Kang, K.M.; Yun, T.J. Comprehensive Molecular Genetic Analysis in Glioma Patients Using Multi-Gene Next-Generation Sequencing Panel. Brain Tumor Res. Treat. 2024, 12, 36–45. [Google Scholar] [CrossRef]
- Varachev, V.; Rybakov, A.; Gorbunova, A.; Ryzhova, M.; Zheludkova, O.; Kumirova, E.; Olyushin, V.; Bykova, A.; Shishkina, L.; Golanov, A. Genomic Profiling in Glioma Patients to Explore Clinically Actionable Alterations. Curr. Oncol. 2024, 31, 7789–7804. [Google Scholar]
- Sakthikumar, S.; Roy, A.; Haseeb, L.; Pettersson, M.E.; Sundström, E.; Marinescu, V.D.; Lindblad-Toh, K.; Forsberg-Nilsson, K. Whole-Genome Sequencing of Glioblastoma Reveals Enrichment of Non-Coding Constraint Mutations in Known and Novel Genes. Genome Biol. 2020, 21, 127. [Google Scholar] [CrossRef]
- Ocampo-Navia, M.I.; Cuellar-Giraldo, D.; García, E.; Rojas-Hernandez, J.P.; Hakim, F.; Pinilla-Monsalve, G.D.; García-Robledo, J.E.; Zorro, O.; Arango-Viana, J.C.; Jiménez-Hakim, E. Molecular Markers in Gliomas: A Practical Review and Guide for Pathologists and Clinicians. Interdiscip. Neurosurg. 2025, 41, 102062. [Google Scholar] [CrossRef]
- Papavassiliou, K.A.; Papavassiliou, A.G. Transcription Factors in Glioblastoma—Molecular Pathogenesis and Clinical Implications. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188667. [Google Scholar] [CrossRef]
- Lentjes, M.H.; Niessen, H.E.; Akiyama, Y.; De Bruïne, A.P.; Melotte, V.; Van Engeland, M. The Emerging Role of GATA Transcription Factors in Development and Disease. Expert Rev. Mol. Med. 2016, 18, e3. [Google Scholar] [CrossRef]
- Zheng, R.; Blobel, G.A. GATA Transcription Factors and Cancer. Genes Cancer 2010, 1, 1178–1188. [Google Scholar] [CrossRef]
- Vicente, C.; Conchillo, A.; García-Sánchez, M.A.; Odero, M.D. The Role of the GATA2 Transcription Factor in Normal and Malignant Hematopoiesis. Crit. Rev. Oncol. Hematol. 2012, 82, 1–17. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, C.J.; Kobayashi, D.K.; Johanns, T.M.; Bowman-Kirigin, J.A.; Schaettler, M.O.; Mao, D.D.; Bender, D.; Kelley, D.G.; Uppaluri, R.; et al. GATA2 Regulates Constitutive PD-L1 and PD-L2 Expression in Brain Tumors. Sci. Rep. 2020, 10, 9027. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yuan, H.; Sun, C.; Xu, L.; Chen, Y.; Zhu, Q.; Zhao, H.; Huang, Q.; Dong, J.; Lan, Q. GATA2 Promotes Glioma Progression through EGFR/ERK/Elk-1 Pathway. Med. Oncol. 2015, 32, 87. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation Prediction for the Deep-Sequencing Age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef]
- Venselaar, H.; te Beek, T.A.H.; Kuipers, R.K.P.; Hekkelman, M.L.; Vriend, G. Protein Structure Analysis of Mutations Causing Inheritable Diseases. An e-Science Approach with Life Scientist Friendly Interfaces. BMC Bioinform. 2010, 11, 548. [Google Scholar] [CrossRef]
- Li, C.; Zhi, D.; Wang, K.; Liu, X. MetaRNN: Differentiating Rare Pathogenic and Rare Benign Missense SNVs and InDels Using Deep Learning. bioRxiv 2021, 14, 115. [Google Scholar] [CrossRef]
- Liu, X.; Jian, X. DbNSFP: A Lightweight Database of Human Non-Synonymous SNPs and Their Functional Predictions. Hum. Mutat. 2011, 32, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. DbNSFP v4: A Comprehensive Database of Transcript-Specific Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.-H.; Liu, J.-X.; Liu, Y.; Chen, S.-W.; Dai, W.-T.; Xiao, Z.-X.; Cao, Y. DrugRep: An Automatic Virtual Screening Server for Drug Repurposing. Acta Pharmacol. Sin. 2023, 44, 888–896. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved Protein-Ligand Blind Docking by Integrating Cavity Detection, Docking and Homologous Template Fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, Y. Protein-Ligand Blind Docking Using CB-Dock2. Methods Mol. Biol. 2024, 2714, 113–125. [Google Scholar]
- Cho, Y.; Hazen, B.C.; Gandra, P.G.; Ward, S.R.; Schenk, S.; Russell, A.P.; Kralli, A. Perm1 Enhances Mitochondrial Biogenesis, Oxidative Capacity, and Fatigue Resistance in Adult Skeletal Muscle. FASEB J. 2016, 30, 674–687. [Google Scholar] [CrossRef]
- Bethke, L.; Webb, E.; Murray, A.; Feychting, M.; LonN, S.; Ahlbom, A.; Malmer, B.; Henriksson, R.; Auvinen, A.; Kiuru, A.; et al. Functional Polymorphisms in Folate Metabolism Genes Influence the Risk of Meningioma and Glioma. Cancer Epidemiol. Biomark. Prev. 2008, 17, 1195–1202. [Google Scholar] [CrossRef]
- Knott, T.J.; Pease, R.J.; Powell, L.M.; Wallis, S.C.; Rall, S.C., Jr.; Innerarity, T.L.; Blackhart, B.; Taylor, W.H.; Marcel, Y.; Milne, R.; et al. Complete Protein Sequence and Identification of Structural Domains of Human Apolipoprotein B. Nature 1986, 323, 734–738. [Google Scholar] [CrossRef]
- Irwin, D.M.; Tan, H. Molecular Evolution of the Vertebrate Hexokinase Gene Family: Identification of a Conserved Fifth Vertebrate Hexokinase Gene. Comp. Biochem. Physiol. Part D Genom. Proteom. 2008, 3, 96–107. [Google Scholar] [CrossRef]
- Kobilka, B.K.; Matsui, H.; Kobilka, T.S.; Yang-Feng, T.L.; Francke, U.; Caron, M.G.; Lefkowitz, R.J.; Regan, J.W. Cloning, Sequencing, and Expression of the Gene Coding for the Human Platelet Alpha 2-Adrenergic Receptor. Science 1987, 238, 650–656. [Google Scholar] [CrossRef]
- Schuchman, E.H.; Suchi, M.; Takahashi, T.; Sandhoff, K.; Desnick, R.J. Human Acid Sphingomyelinase. Isolation, Nucleotide Sequence and Expression of the Full-Length and Alternatively Spliced CDNAs. J. Biol. Chem. 1991, 266, 8531–8539. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.K.; Arioglu, E.; De Almeida, S.; Akkoc, N.; Taylor, S.I.; Bowcock, A.M.; Barnes, R.I.; Garg, A. BSCL2 Mutations in Congenital Generalized Lipodystrophy. Nat. Genet. 2002, 31, 365–369. [Google Scholar]
- Yuan, L.; Liu, J.G.; Zhao, J.; Brundell, E.; Daneholt, B.; Höög, C. The Murine SCP3 Gene Is Required for Synaptonemal Complex Assembly, Chromosome Synapsis, and Male Fertility. Mol. Cell 2000, 5, 73–83. [Google Scholar] [CrossRef]
- Kavitha, B.; Ranganathan, S.; Gopi, S.; Vetrivel, U.; Hemavathy, N.; Mohan, V.; Radha, V. Molecular Characterization and Re-Interpretation of HNF1A Variants Identified in Indian MODY Subjects towards Precision Medicine. Front. Endocrinol. 2023, 14, 1177268. [Google Scholar] [CrossRef]
- Kelsell, D.P.; Dunlop, J.; Stevens, H.P.; Lench, N.J.; Liang, J.N.; Parry, G.; Mueller, R.F.; Leigh, I.M. Connexin 26 Mutations in Hereditary Non-Syndromic Sensorineural Deafness. Nature 1997, 387, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.D.; Ulevitch, R.J.; Han, J.H. Primary Structure of BMK1: A New Mammalian MAP Kinase. Biochem. Biophys. Res. Commun. 1995, 213, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, W.; Seibold, A.; Antaramian, A.; Lonergan, M.; Arthus, M.F.; Hendy, G.N.; Birnbaumer, M.; Bichet, D.G. Molecular Identification of the Gene Responsible for Congenital Nephrogenic Diabetes Insipidus. Nature 1992, 359, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas (TCGA) Research Network. Cancer Genome Atlas Research Network Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Nurmohamed, S.S.; Broadhurst, R.W.; May, G.; Enver, T. Solution NMR Structure of Human GATA2 C-Terminal Zinc Finger Domain; RCSB Protein Data Bank: Piscataway, NJ, USA, 2019. [Google Scholar]
- Nurmohamed, S.S.; Broadhurst, R.W.; May, G.; Enver, T. Solution NMR Structure of Human GATA2 N-Terminal Zinc Finger Domain; RCSB Protein Data Bank: Piscataway, NJ, USA, 2021. [Google Scholar]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data Bank (Rcsb.org): Delivery of Experimentally-Determined PDB Structures alongside One Million Computed Structure Models of Proteins from Artificial Intelligence/Machine Learning. Nucleic Acids Res. 2023, 51, D488–D508. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing Structure Coverage for over 214 Million Protein Sequences. Nucleic Acids Res. 2024, 52, D368–D375. [Google Scholar] [CrossRef]
- Berger, T.R.; Wen, P.Y.; Lang-Orsini, M.; Chukwueke, U.N. World Health Organization 2021 Classification of Central Nervous System Tumors and Implications for Therapy for Adult-Type Gliomas. JAMA Oncol. 2022, 8, 1493. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.K.-Y.; Yao, Y.; Zhang, Z.; Chung, N.Y.-F.; Liu, J.S.-M.; Li, K.K.-W.; Shi, Z.; Chan, D.T.-M.; Poon, W.S.; Zhou, L.; et al. TERT Promoter Mutations Contribute to Subset Prognostication of Lower-Grade Gliomas. Mod. Pathol. 2015, 28, 177–186. [Google Scholar] [CrossRef]
- Johnson, K.D.; Jung, M.M.; Tran, V.L.; Bresnick, E.H. Interferon Regulatory Factor-8-Dependent Innate Immune Alarm Senses GATA2 Deficiency to Alter Hematopoietic Differentiation and Function. Curr. Opin. Hematol. 2023, 30, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Menendez-Gonzalez, J.B.; Vukovic, M.; Abdelfattah, A.; Saleh, L.; Almotiri, A.; Thomas, L.-A.; Agirre-Lizaso, A.; Azevedo, A.; Menezes, A.C.; Tornillo, G.; et al. Gata2 as a Crucial Regulator of Stem Cells in Adult Hematopoiesis and Acute Myeloid Leukemia. Stem Cell Rep. 2019, 13, 291–306. [Google Scholar] [CrossRef]
- Chong, C.-E.; Venugopal, P.; Stokes, P.H.; Lee, Y.K.; Brautigan, P.J.; Yeung, D.T.O.; Babic, M.; Engler, G.A.; Lane, S.W.; Klingler-Hoffmann, M.; et al. Differential Effects on Gene Transcription and Hematopoietic Differentiation Correlate with GATA2 Mutant Disease Phenotypes. Leukemia 2018, 32, 194–202. [Google Scholar] [CrossRef]
- Burak, N.; Jan, N.; Kessler, J.; Oei, E.; Patel, P.; Feldman, S. Diagnosis of GATA2 Deficiency in a Young Woman with Hemophagocytic Lymphohistiocytosis Triggered by Acute Systemic Cytomegalovirus Infection. Am. J. Case Rep. 2021, 22, e927087. [Google Scholar] [CrossRef]
- Vila, A.; Dapás, J.I.; Rivero, C.V.; Bocanegra, F.; Furnari, R.F.; Hsu, A.P.; Holland, S.M. Multiple Opportunistic Infections in a Woman with GATA2 Mutation. Int. J. Infect. Dis. 2017, 54, 89–91. [Google Scholar] [CrossRef][Green Version]
- Donadieu, J.; Lamant, M.; Fieschi, C.; de Fontbrune, F.S.; Caye, A.; Ouachee, M.; Beaupain, B.; Bustamante, J.; Poirel, H.A.; Isidor, B.; et al. Natural History of GATA2 Deficiency in a Survey of 79 French and Belgian Patients. Haematologica 2018, 103, 1278–1287. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, J.; Song, D.; You, Y.; Wang, Z. Haemophagocytic Lymphohistiocytosis Caused by GATA2 Deficiency: A Report on Three Patients. BMC Infect. Dis. 2024, 24, 482. [Google Scholar] [CrossRef]
- Hsu, A.P.; Sampaio, E.P.; Khan, J.; Calvo, K.R.; Lemieux, J.E.; Patel, S.Y.; Frucht, D.M.; Vinh, D.C.; Auth, R.D.; Freeman, A.F.; et al. Mutations in GATA2 Are Associated with the Autosomal Dominant and Sporadic Monocytopenia and Mycobacterial Infection (MonoMAC) Syndrome. Blood 2011, 118, 2653–2655. [Google Scholar] [CrossRef]
- Saida, S.; Zhen, T.; Kim, E.; Yu, K.; Lopez, G.; McReynolds, L.J.; Liu, P.P. Gata2 Deficiency Delays Leukemogenesis While Contributing to Aggressive Leukemia Phenotype in Cbfb-MYH11 Knockin Mice. Leukemia 2020, 34, 759–770. [Google Scholar] [CrossRef]
- Gioacchino, E.; Zhang, W.; Koyunlar, C.; Zink, J.; de Looper, H.; Gussinklo, K.J.; Hoogenboezem, R.; Bosch, D.; Bindels, E.; Touw, I.P.; et al. GATA2 Heterozygosity Causes an Epigenetic Feedback Mechanism Resulting in Myeloid and Erythroid Dysplasia. Br. J. Haematol. 2024, 205, 580–593. [Google Scholar] [CrossRef] [PubMed]
- Castaño, J.; Aranda, S.; Bueno, C.; Calero-Nieto, F.J.; Mejia-Ramirez, E.; Mosquera, J.L.; Blanco, E.; Wang, X.; Prieto, C.; Zabaleta, L.; et al. GATA2 Promotes Hematopoietic Development and Represses Cardiac Differentiation of Human Mesoderm. Stem Cell Rep. 2019, 13, 515–529. [Google Scholar] [CrossRef]
- Kanki, Y.; Kohro, T.; Jiang, S.; Tsutsumi, S.; Mimura, I.; Suehiro, J.-I.; Wada, Y.; Ohta, Y.; Ihara, S.; Iwanari, H.; et al. Epigenetically Coordinated GATA2 Binding Is Necessary for Endothelium-Specific Endomucin Expression. EMBO J. 2011, 30, 2582–2595. [Google Scholar] [CrossRef]
- Hou, A.; Zhao, L.; Zhao, F.; Wang, W.; Niu, J.; Li, B.; Zhou, Z.; Zhu, D. Expression of MECOM Is Associated with Unfavorable Prognosis in Glioblastoma Multiforme. Onco Targets Ther. 2016, 9, 315–320. [Google Scholar]
- Al Halabi, K.T.; Sievers, P.; Stichel, D.; Sommerkamp, A.C.; Sill, M.; Jäger, N.; Wittmann, A.; Kramm, C.; Snuderl, M.; Pfister, S.M.; et al. OS11.5.A PATZ1 Fusions Define a Novel Molecularly Distinct CNS Tumor Entity with a Broad Histological Spectrum. Neuro Oncol. 2021, 23, ii14–ii15. [Google Scholar] [CrossRef]
- Homan, C.C.; Venugopal, P.; Arts, P.; Shahrin, N.H.; Feurstein, S.; Rawlings, L.; Lawrence, D.M.; Andrews, J.; King-Smith, S.L.; Harvey, N.L.; et al. GATA2 Deficiency Syndrome: A Decade of Discovery. Hum. Mutat. 2021, 42, 1399–1421. [Google Scholar] [CrossRef] [PubMed]
- Kazenwadel, J.; Secker, G.A.; Liu, Y.J.; Rosenfeld, J.A.; Wildin, R.S.; Cuellar-Rodriguez, J.; Hsu, A.P.; Dyack, S.; Fernandez, C.V.; Chong, C.-E.; et al. Loss-of-Function Germline GATA2 Mutations in Patients with MDS/AML or MonoMAC Syndrome and Primary Lymphedema Reveal a Key Role for GATA2 in the Lymphatic Vasculature. Blood 2012, 119, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Katsumura, K.R.; Mehta, C.; Hewitt, K.J.; Soukup, A.A.; Fraga de Andrade, I.; Ranheim, E.A.; Johnson, K.D.; Bresnick, E.H. Human Leukemia Mutations Corrupt but Do Not Abrogate GATA-2 Function. Proc. Natl. Acad. Sci. USA 2018, 115, E10109–E10118. [Google Scholar] [CrossRef]
- Koyunlar, C.; de Pater, E. From Basic Biology to Patient Mutational Spectra of GATA2 Haploinsufficiencies: What Are the Mechanisms, Hurdles, and Prospects of Genome Editing for Treatment. Front. Genome Ed. 2020, 2, 602182. [Google Scholar] [CrossRef]
- Cortés-Lavaud, X.; Landecho, M.F.; Maicas, M.; Urquiza, L.; Merino, J.; Moreno-Miralles, I.; Odero, M.D. GATA2 Germline Mutations Impair GATA2 Transcription, Causing Haploinsufficiency: Functional Analysis of the p.Arg396Gln Mutation. J. Immunol. 2015, 194, 2190–2198. [Google Scholar] [CrossRef]
- Molenaar, R.J.; Wilmink, J.W. IDH1/2 Mutations in Cancer Stem Cells and Their Implications for Differentiation Therapy. J. Histochem. Cytochem. 2022, 70, 83–97. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef]
- Shin, H.; Sa, J.K.; Bae, J.S.; Koo, H.; Jin, S.; Cho, H.J.; Choi, S.W.; Kyoung, J.M.; Kim, J.Y.; Seo, Y.J.; et al. Clinical Targeted Next-Generation Sequencing Panels for Detection of Somatic Variants in Gliomas. Cancer Res. Treat. 2020, 52, 41–50. [Google Scholar] [CrossRef] [PubMed]
- GLASS Consortium. GLASS Consortium Glioma through the Looking GLASS: Molecular Evolution of Diffuse Gliomas and the Glioma Longitudinal Analysis Consortium. Neuro-Oncol. 2018, 20, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Wongsurawat, T.; Jenjaroenpun, P.; De Loose, A.; Alkam, D.; Ussery, D.W.; Nookaew, I.; Leung, Y.-K.; Ho, S.-M.; Day, J.D.; Rodriguez, A. A Novel Cas9-Targeted Long-Read Assay for Simultaneous Detection of IDH1/2 Mutations and Clinically Relevant MGMT Methylation in Fresh Biopsies of Diffuse Glioma. Acta Neuropathol. Commun. 2020, 8, 87. [Google Scholar] [CrossRef] [PubMed]



| Gene(s) | Significance | Type | Consequence | HGVSc | HGVSp | Previous Association | Reference |
|---|---|---|---|---|---|---|---|
| PERM1 | Pathogenic | SNV | Missense variant | NM_001291366.2:c.2330T>C | p.Val777Ala | Mitochondrial biogenesis | Cho et al. [25] |
| MTHFR | Pathogenic | SNV | Nonsense | NM_001330358.2:c.1531G>T | p.Glu511 * | Folate metabolism, glioma risk | Bethke et al. [26] |
| APOA2 | Pathogenic | SNV | No consequences found | NM_001643.2:c.-323C>T | - | Lipid metabolism | Knott et al. [27] |
| HKDC1, LOC101928994 | Pathogenic | SNV | Missense variant | NM_025130.4:c.173C>T | p.Thr58Met | Glucose metabolism | Irwin & Tan [28] |
| ADRA2A | Pathogenic | SNV | 3′ UTR variant | NM_000681.4:c. *427A>G | - | Adrenergic receptor | Kobilka et al. [29] |
| SMPD1 | Pathogenic | SNV | No consequences found | NM_000543.5:c.911T>C | p.Leu304Pro | Lysosomal enzyme | Schuchman et al. [30] |
| GNG3, BSCL2, HNRNPUL2-BSCL2 | Pathogenic | SNV | Intron variant | NM_012202.5:c.-431C>T | - | G-protein signaling | Agarwal et al. [31] |
| SYCP3 | Pathogenic | Deletion | Frameshift variant | NM_001177948.2:c.643delA | p.Ile215fs | DNA repair, meiosis | Yuan et al. [32] |
| HNF1A | Pathogenic | Deletion | Frameshift variant | NM_000545.8:c.872delC | p.Pro291fs | Transcription factor | Kavitha et al. [33] |
| GJB2 | Pathogenic | SNV | Missense variant | NM_004004.6:c.109G>A | p.Val37Ile | Gap junction protein | Kelsell et al. [34] |
| MAPK7 | Pathogenic | SNV | Missense variant | NM_139032.3:c.469G>A | p.Ala157Thr | MAPK signaling | Lee et al. [35] |
| AVPR2 | Pathogenic | Deletion | Frameshift variant, Non-coding transcript variant | NM_000054.7:c.738delG | p.Arg247fs | Hormone receptor | Rosenthal et al. [36] |
| IDH1 | Pathogenic/likely pathogenic | SNV | Missense variant | NM_001282386.1:c.395G>A | p.Arg132His | Well-established driver | Yan et al. [37] |
| GATA2 | Pathogenic/likely pathogenic | SNV | Missense variant | NM_001145661.2:c.1186C>T | p.Arg396Trp | Not previously reported | This study |
| TP53 | Pathogenic/likely pathogenic | SNV | Splice donor variant | NM_000546.6:c.559+1G>T | - | Tumor suppressor | TCGA [38] |
| Interpro Domain | Gene Ontology Term | Broad Gene Ontology Term |
|---|---|---|
| Zinc Finger, Gata-Type IPR000679 | Sequence-Specific DNA-Binding GO:0043565 | Binding GO:0005488 Molecular_Function GO:0003674 Nucleic Acid Binding GO:0003676 |
| Zinc Finger, Nhr/Gata-Type IPR013088 | Zinc Ion-Binding GO:0008270 | Molecular_Function GO:0003674 Binding GO:0005488 Ion Binding GO:0043167 |
| Transcription Factor, Gata-2/3 IPR016374 | DNA-Binding Transcription Factor Activity, Rna Polymerase Ii-Specific GO:0000981 | Molecular_Function GO:0003674 |
| Transcription Factor Gata IPR039355 | DNA-Binding Transcription Factor Activity GO:0003700 | Molecular_Function GO:0003674 |
| Drug/Compound | Wild-Type GATA2 | p.Arg396Trp-Mutated GATA2 | ||
|---|---|---|---|---|
| Vina | Dock Chain | Vina | Dock Chain | |
| Entrectinib C31H34F2N6O2 | −10.5 | Chain A: GLU294 CYS295 VAL296 ASN297 CYS298 ASP309 HIS313 LEU315 GLN328 ASN329 ARG330 PRO331 LEU332 ILE333 LYS334 PRO335 LYS336 ARG337 ARG338 SER340 ALA342 THR358 ILE393 GLN394 THR395 ARG396 ASN397 ARG398 LYS399 MET400 SER401 ASN402 | −9.8 | Chain A: VAL296 ASN297 CYS298 ASP309 THR311 HIS313 LEU315 GLN328 ASN329 ARG330 PRO331 LEU332 ILE333 LYS334 PRO335 LYS336 ARG337 ARG338 LEU339 SER340 ALA342 THR358 ASN371 ILE393 GLN394 THR395 TRP396 ASN397 ARG398 LYS399 MET400 SER401 ASN402 |
| Rucaparib C19H18FN3O | −8.9 | Chain A: GLU294 CYS295 VAL296 ASN297 CYS298 ASP309 THR311 HIS313 LEU315 HIS323 GLN328 ASN329 ARG330 PRO331 LEU332 ILE333 LYS334 PRO335 LYS336 ARG338 SER340 THR358 THR395 ARG396 ASN397 ARG398 LYS399 MET400 SER401 ASN402 | −8.3 | Chain A: VAL296 ASN297 CYS298 ASP309 THR311 HIS313 LEU315 HIS323 GLN328 ASN329 ARG330 PRO331 LEU332 LYS334 PRO335 LYS336 ARG337 ARG338 LEU339 SER340 THR358 ILE393 GLN394 THR395 TRP396 ASN397 ARG398 LYS399 MET400 SER401 |
| Vemurafenib C23H18ClF2N3O3S | −9.4 | Chain A: GLU294 VAL296 ASN297 CYS298 GLY299 ALA300 THR301 ALA302 ASN329 ARG330 LEU332 LYS334 PRO335 LYS336 ARG337 ARG338 LEU339 SER340 ALA342 ARG343 THR356 THR357 THR358 ILE393 GLN394 THR395 ARG396 ASN397 ARG398 LYS399 MET400 | −9.0 | Chain A: GLU294 VAL296 ASN297 CYS298 ASP309 HIS313 LEU315 GLN328 ASN329 ARG330 PRO331 LEU332 LYS334 PRO335 LYS336 ARG337 ARG338 SER340 THR358 GLN394 THR395 TRP396 ASN397 ARG398 LYS399 MET400 SER401 ASN402 |
| Brigatinib C29H39ClN7O2P | −9.1 | Chain A: VAL296 ASN297 CYS298 ASP309 THR311 HIS313 GLN328 ASN329 ARG330 PRO331 LEU332 LYS334 PRO335 LYS336 ARG337 ARG338 LEU339 SER340 ALA342 THR358 THR395 ARG396 ASN397 ARG398 LYS399 MET400 SER401 SER404 | −9.1 | Chain A: GLY292 GLU294 VAL296 ASN297 CYS298 ASP309 THR311 GLY312 HIS313 TYR314 GLN328 ASN329 ARG330 PRO331 LEU332 LYS334 PRO335 LYS336 ARG337 ARG338 LEU339 SER340 ALA342 THR357 THR358 ILE393 THR395 TRP396 ASN397 ARG398 LYS399 MET400 SER401 SER404 |
| Temozolamide C6H6N6O2 | −5.9 | Chain A: CYS352 THR354 THR356 THR357 THR358 LEU359 CYS370 ASN371 ALA372 CYS373 ARG384 MET388 LYS389 LYS390 GLU391 GLY392 ILE393 GLN394 | −6.1 | Chain A: CYS352 THR354 THR356 THR357 THR358 LEU359 CYS370 ASN371 ALA372 ARG384 MET388 LYS389 LYS390 GLU391 GLY392 ILE393 GLN394 TRP396 |
| Drug/Compound | Wild-Type GATA2 | p.Arg396Trp-Mutated GATA2 |
|---|---|---|
| Entrectinib C31H34F2N6O2 | 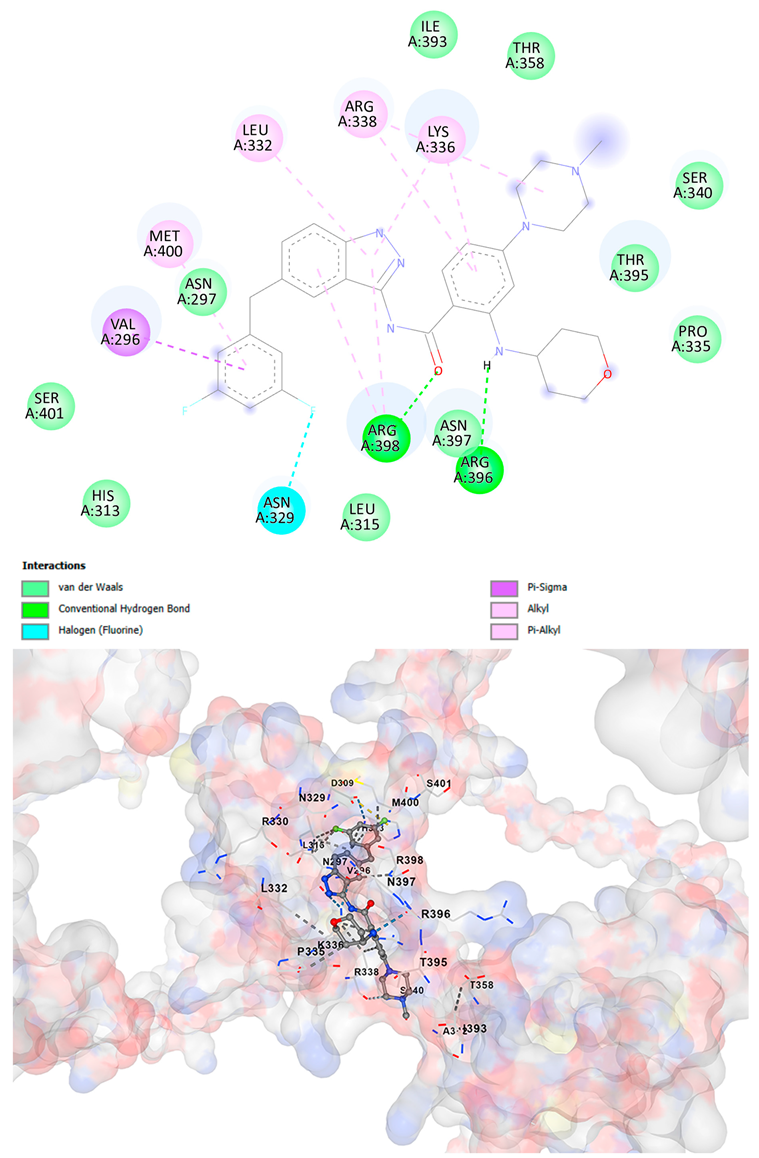 | 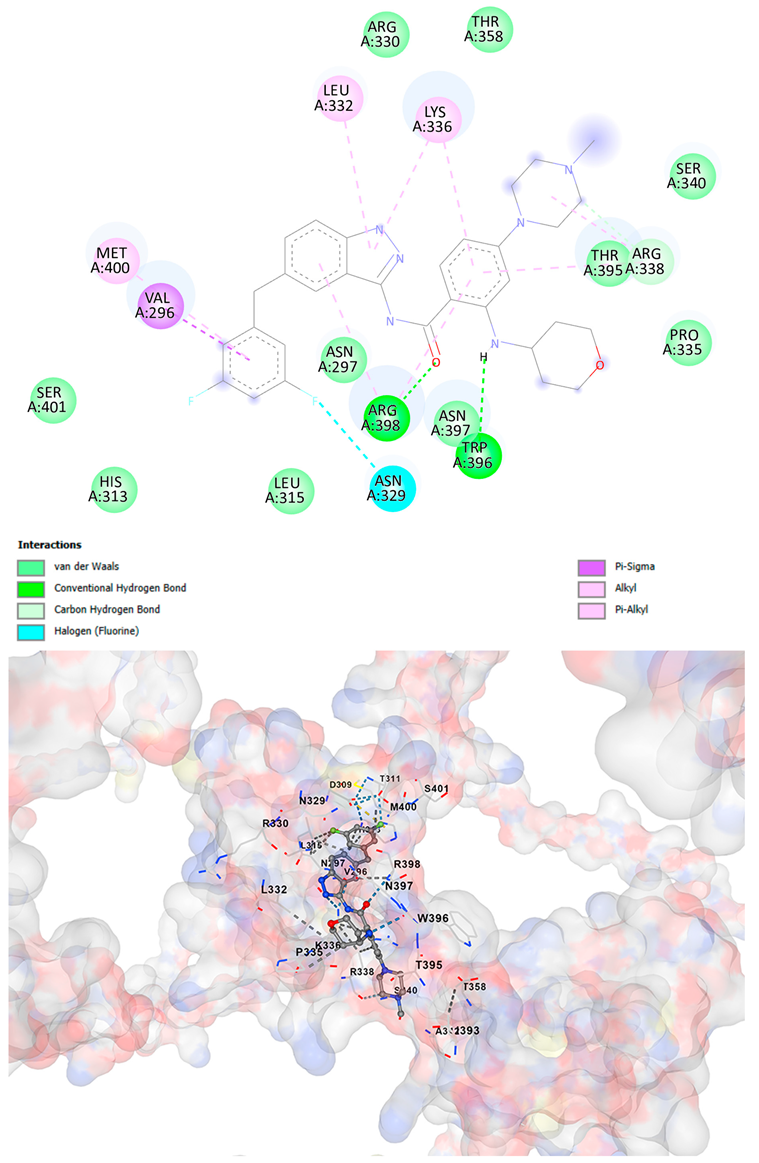 |
| Rucaparib C19H18FN3O | 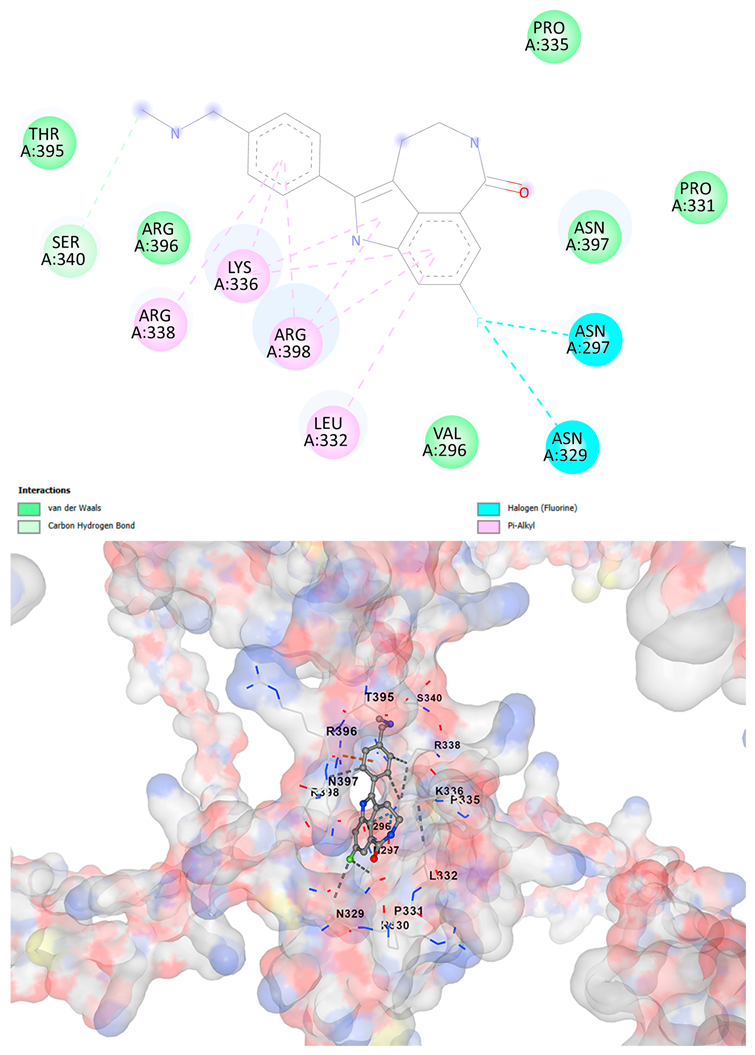 |  |
| Vemurafenib C23H18ClF2N3O3S | 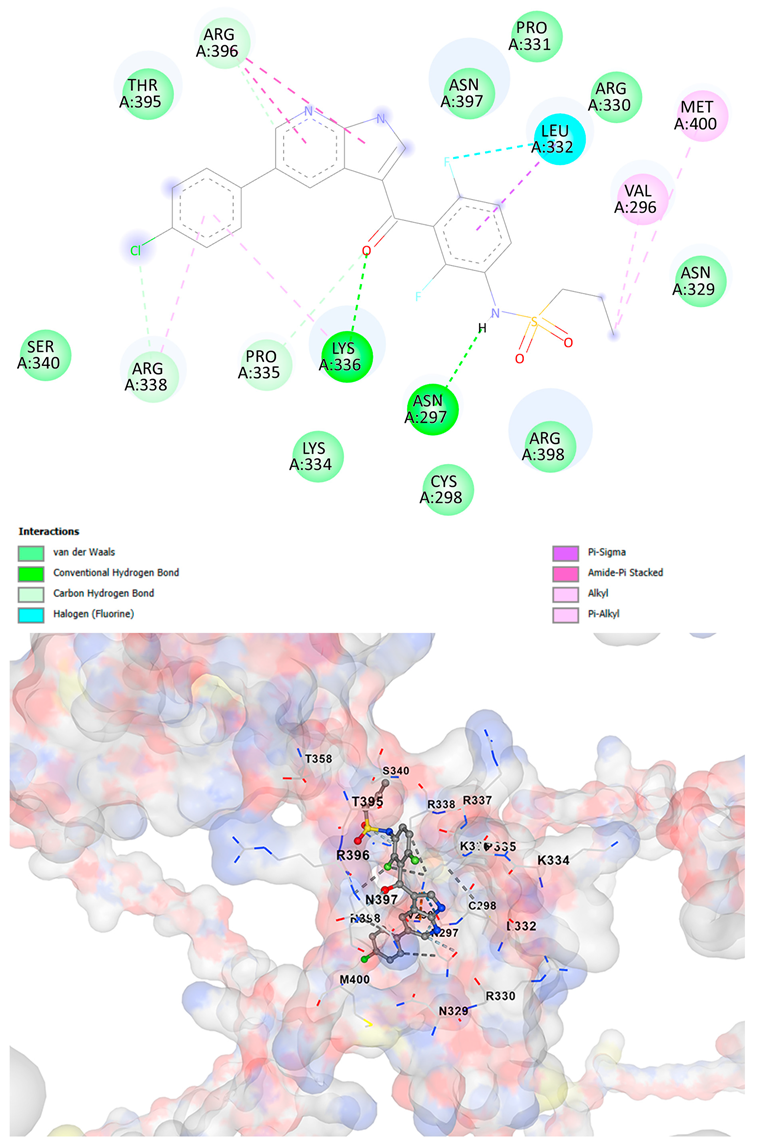 | 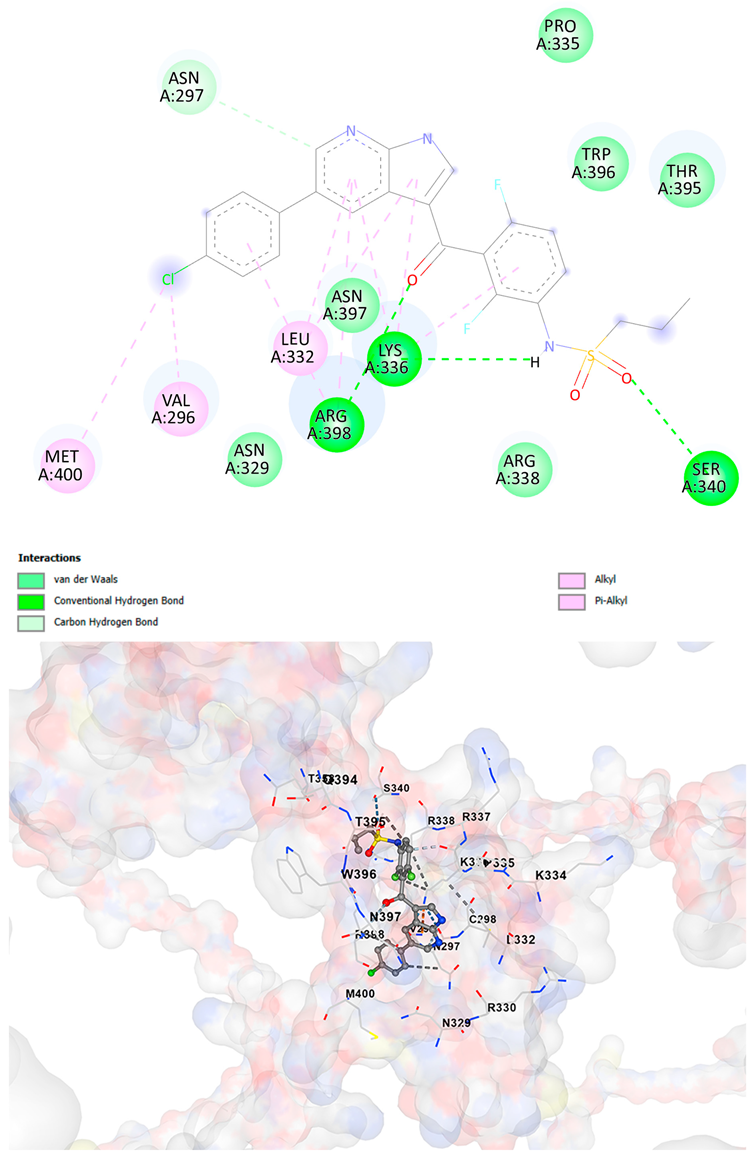 |
| Brigatinib C29H39ClN7O2P | 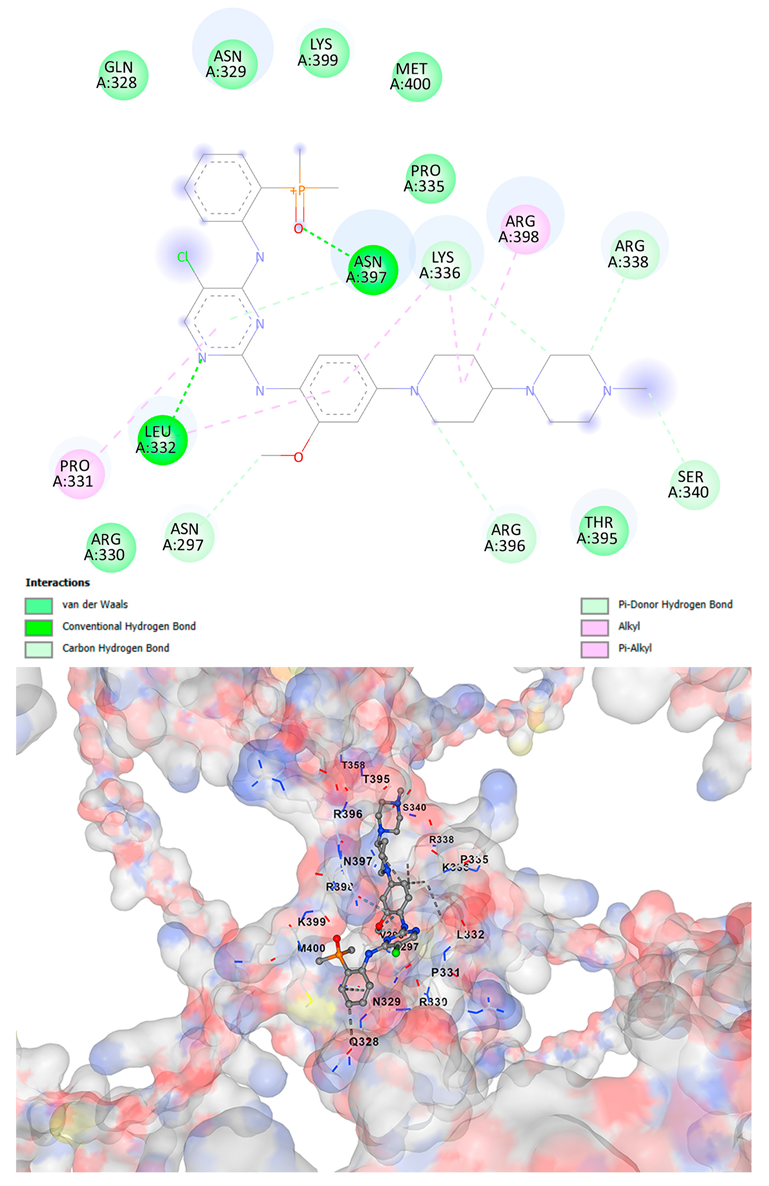 | 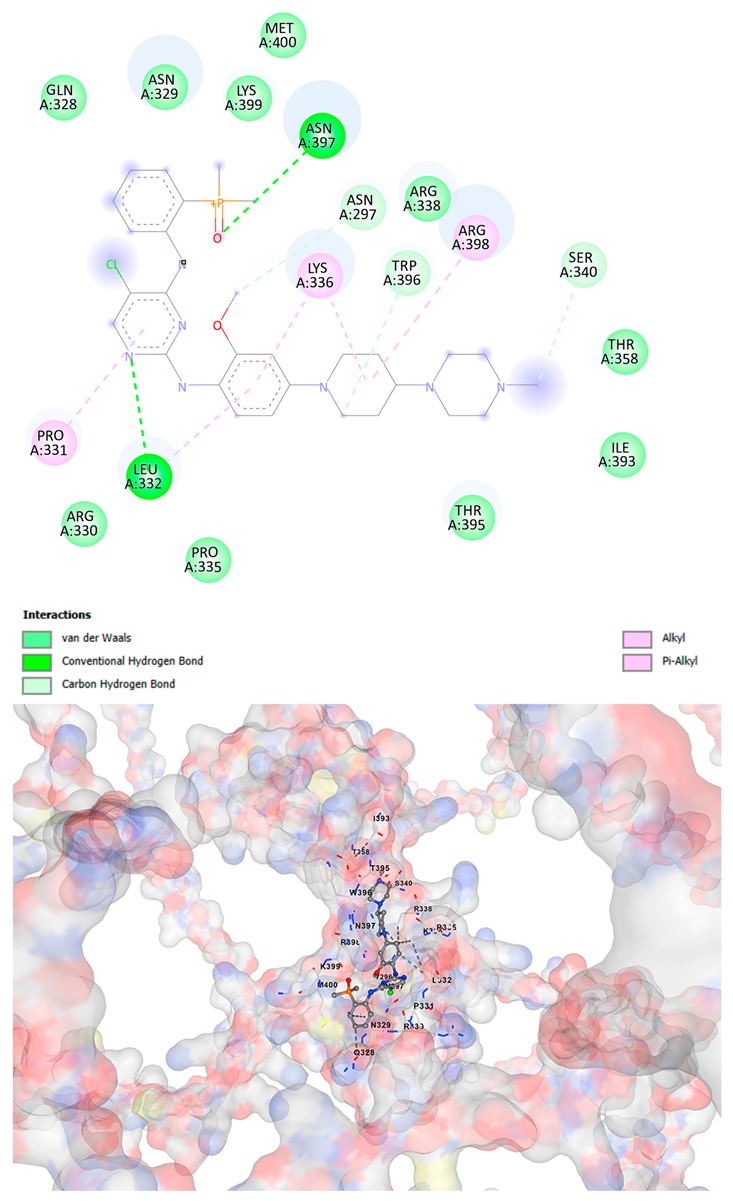 |
| Temozolamide C6H6N6O2 |  | 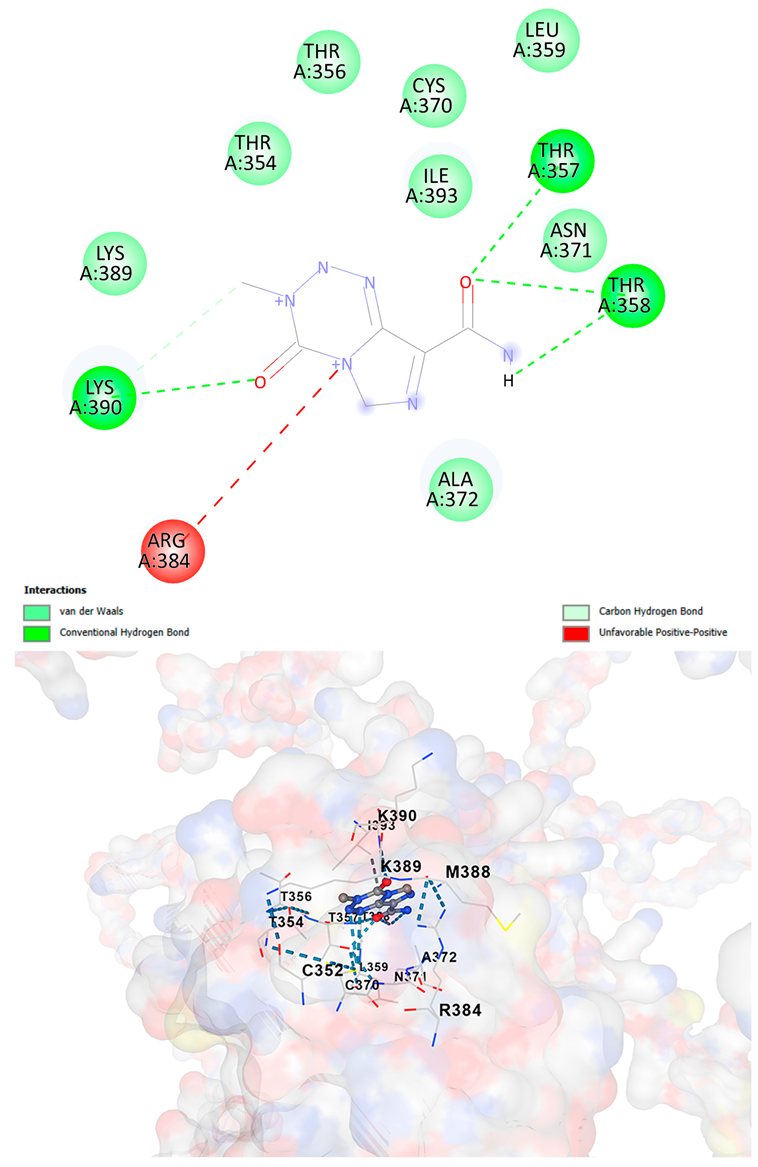 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Handoko; Lau, V.; Susanto, E.; Aman, R.A.; Heriyanto, D.S.; Gondhowiardjo, S.A. Whole-Genome Sequencing Reveals a Novel GATA2 Mutation in Lower-Grade Glioma: Bioinformatics Analysis of Functional and Therapeutic Implications. Cancers 2025, 17, 3338. https://doi.org/10.3390/cancers17203338
Handoko, Lau V, Susanto E, Aman RA, Heriyanto DS, Gondhowiardjo SA. Whole-Genome Sequencing Reveals a Novel GATA2 Mutation in Lower-Grade Glioma: Bioinformatics Analysis of Functional and Therapeutic Implications. Cancers. 2025; 17(20):3338. https://doi.org/10.3390/cancers17203338
Chicago/Turabian StyleHandoko, Vincent Lau, Eka Susanto, Renindra Ananda Aman, Didik Setyo Heriyanto, and Soehartati A. Gondhowiardjo. 2025. "Whole-Genome Sequencing Reveals a Novel GATA2 Mutation in Lower-Grade Glioma: Bioinformatics Analysis of Functional and Therapeutic Implications" Cancers 17, no. 20: 3338. https://doi.org/10.3390/cancers17203338
APA StyleHandoko, Lau, V., Susanto, E., Aman, R. A., Heriyanto, D. S., & Gondhowiardjo, S. A. (2025). Whole-Genome Sequencing Reveals a Novel GATA2 Mutation in Lower-Grade Glioma: Bioinformatics Analysis of Functional and Therapeutic Implications. Cancers, 17(20), 3338. https://doi.org/10.3390/cancers17203338






