Circulating Tumor DNA as a Prognostic and Predictive Biomarker in Lung Cancer
Simple Summary
Abstract
1. Introduction
2. Applications
2.1. Detection/Screening
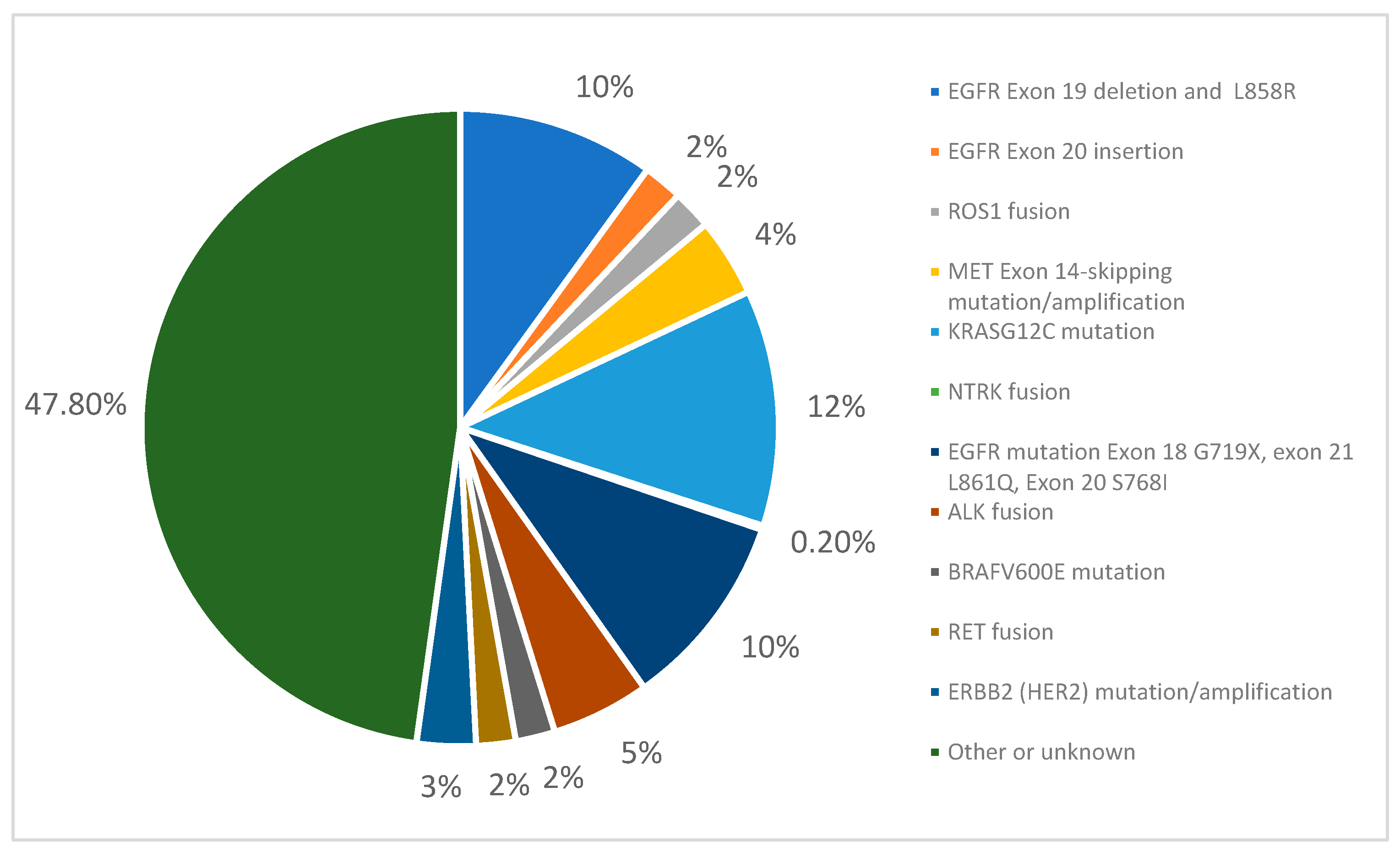
2.2. Genotyping and Detecting Actionable Genomic Alterations (AGAs)
2.3. Utilizing ctDNA in Resectable Disease
2.3.1. Chemoimmunotherapy in Resectable Disease
2.3.2. Targeted Therapies in Resectable Disease
2.4. Locally Advanced, Metastatic, and Recurrent NSCLC
2.5. Resistance
3. Future Directions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGA | Actionable genomic alterations. |
| BL | Baseline. |
| CFDNA | Cell-free DNA. |
| CICI | Consolidation immune checkpoint inhibition. |
| CNS | Central nervous system. |
| CPI | Checkpoint inhibitor. |
| CRT | Chemoradiation therapy. |
| CTDNA | Circulating tumor DNA. |
| DFS | Disease-free survival. |
| DNA | Deoxyribonucleic acid. |
| EGFR | Epidermal growth factor receptor. |
| ICI | Immune checkpoint inhibition. |
| MPR | Major pathologic response. |
| MR | Molecular response. |
| MRD | Minimal residual disease. |
| NSCLC | Non-small cell lung cancer. |
| OS | Overall survival. |
| ORR | Overall response rate. |
| pCR | Pathologic complete response. |
| PD | Progression of disease. |
| PFS | Progression-free survival. |
| PT/PTs | Patient/Patients. |
| RECIST | Response Evaluation Criteria in Solid Tumors. |
| RFS | Recurrence-free survival. |
| RT | Radiotherapy. |
| TKI | Tyrosine kinase inhibitors. |
| USPSTF | United States Preventive Services Taskforce. |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. U.S. Cancer Statistics Lung Cancer Stat Bite; U.S. Department of Health and Human Services: Washington, DC, USA, 2025. Available online: https://www.cdc.gov/lung-cancer/statistics/index.html (accessed on 25 July 2025).
- Zhang, Y.-L.; Yuan, J.-Q.; Wang, K.-F.; Fu, X.-H.; Han, X.-R.; Threapleton, D.; Yang, Z.-Y.; Mao, C.; Tang, J.-L. The prevalence of EGFR mutation in patients with non-small cell lung cancer: A systematic review and meta-analysis. Oncotarget 2016, 7, 78985–78993. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Hiatt, J.B.; Doebley, A.-L.; Arnold, H.U.; Adil, M.; Sandborg, H.; Persse, T.W.; Ko, M.; Wu, F.; Villalonga, A.Q.; Santana-Davila, R.; et al. Molecular phenotyping of small cell lung cancer using targeted cfDNA profiling of transcriptional regulatory regions. Sci. Adv. 2024, 10, eadk2082. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J. Circulating tumor DNA (ctDNA) as a biomarker for lung cancer: Early detection, monitoring and therapy prediction. Tumor Biol. 2023, 46, S283–S295. [Google Scholar] [CrossRef]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic Characteristics of the DNA Found in the Plasma of Cancer Patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Gray, J.E.; Markovets, A.; Reungwetwattana, T.; Majem, M.; Nogami, N.; Peled, N.; Lee, J.-S.; Cho, B.C.; Chewaskulyong, B.; John, T.; et al. Longitudinal Analyses of Circulating Tumor DNA for the Detection of EGFR Mutation-Positive Advanced NSCLC Progression During Treatment: Data from FLAURA and AURA3. J. Thorac. Oncol. 2024, 19, 1525–1538. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement From the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Pellini, B.; Chaudhuri, A.A. Circulating Tumor DNA Minimal Residual Disease Detection of Non–Small-Cell Lung Cancer Treated with Curative Intent. J. Clin. Oncol. 2022, 40, 567–575. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef]
- Bestvina, C.M.; Garassino, M.C.; Neal, J.W.; Wakelee, H.A.; Diehn, M.; Vokes, E.E. Early-Stage Lung Cancer: Using Circulating Tumor DNA to Get Personal. J. Clin. Oncol. 2023, 41, 4093–4096. [Google Scholar] [CrossRef]
- Diaz, L.A., Jr.; Bardelli, A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Li, S.; Cheng, H. Circulating DNA in EGFR-mutated lung cancer. Ann. Transl. Med. 2017, 5, 379. [Google Scholar] [CrossRef]
- Singh, A.P.; Cheng, H.; Guo, X.; Levy, B.; Halmos, B. Circulating Tumor DNA in Non–Small-Cell Lung Cancer: A Primer for the Clinician. JCO Precis. Oncol. 2017, 1, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Verzè, M.; Pluchino, M.; Leonetti, A.; Corianò, M.; Bonatti, F.; Armillotta, M.P.; Perrone, F.; Casali, M.; Minari, R.; Tiseo, M. Role of ctDNA for the detection of minimal residual disease in resected non-small cell lung cancer: A systematic review. Transl. Lung Cancer Res. 2022, 11, 2588–2600. [Google Scholar] [CrossRef] [PubMed]
- Marinello, A.; Tagliamento, M.; Pagliaro, A.; Conci, N.; Cella, E.; Vasseur, D.; Remon, J.; Levy, A.; Dall’oLio, F.G.; Besse, B. Circulating tumor DNA to guide diagnosis and treatment of localized and locally advanced non-small cell lung cancer. Cancer Treat. Rev. 2024, 129, 102791. [Google Scholar] [CrossRef]
- Mathios, D.; Johansen, J.S.; Cristiano, S.; Medina, J.E.; Phallen, J.; Larsen, K.R.; Bruhm, D.C.; Niknafs, N.; Ferreira, L.; Adleff, V.; et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021, 12, 5060. [Google Scholar] [CrossRef]
- Mazzone, P.J.; Bach, P.B.; Carey, J.; Schonewolf, C.A.; Bognar, K.; Ahluwalia, M.S.; Cruz-Correa, M.; Gierada, D.; Kotagiri, S.; Lloyd, K.; et al. Clinical Validation of a Cell-Free DNA Fragmentome Assay for Augmentation of Lung Cancer Early Detection. Cancer Discov. 2024, 14, 2224–2242. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Li, Y.; Fan, Z.; Meng, Y.; Liu, S.; Zhan, H. Blood-based DNA methylation signatures in cancer: A systematic review. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2022, 1869, 166583. [Google Scholar] [CrossRef] [PubMed]
- Barlesi, F.; Gervais, R.; Lena, H.; Hureaux, J.; Berard, H.; Paillotin, D.; Bota, S.; Monnet, I.; Chajara, A.; Robinet, G. Pemetrexed and cisplatin as first-line chemotherapy for advanced non-small-cell lung cancer (NSCLC) with asymptomatic inoperable brain metastases: A multicenter phase II trial (GFPC 07-01). Ann. Oncol. 2011, 22, 2466–2470. [Google Scholar] [CrossRef] [PubMed]
- De Mattos-Arruda, L.; Mayor, R.; Ng, C.K.Y.; Weigelt, B.; Martínez-Ricarte, F.; Torrejon, D.; Oliveira, M.; Arias, A.; Raventos, C.; Tang, J.; et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 2015, 6, 8839. [Google Scholar] [CrossRef]
- Sasaki, S.; Yoshioka, Y.; Ko, R.; Katsura, Y.; Namba, Y.; Shukuya, T.; Kido, K.; Iwakami, S.; Tominaga, S.; Takahashi, K. Diagnostic significance of cerebrospinal fluid EGFR mutation analysis for leptomeningeal metastasis in non-small-cell lung cancer patients harboring an active EGFR mutation following gefitinib therapy failure. Respir. Investig. 2015, 54, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Nanjo, S.; Okuda, C.; Kaji, R.; Masago, K.; Fujita, S.; Irie, K.; Okada, H.; Fukushima, S.; Katakami, N. 451PD Osimertinib at 80 mg for refractory leptomeningeal metastases in T790M-positive EGFR-mutant non-small cell lung cancer. Ann. Oncol. 2016, 27, mdw594.015. [Google Scholar] [CrossRef][Green Version]
- Reckamp, K.L.; Melnikova, V.O.; Karlovich, C.; Sequist, L.V.; Camidge, D.R.; Wakelee, H.; Perol, M.; Oxnard, G.R.; Kosco, K.; Croucher, P.; et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J. Thorac. Oncol. 2016, 11, 1690–1700. [Google Scholar] [CrossRef]
- Zhao, J.; Ye, X.; Xu, Y.; Chen, M.; Zhong, W.; Sun, Y.; Yang, Z.; Zhu, G.; Gu, Y.; Wang, M. EGFR mutation status of paired cerebrospinal fluid and plasma samples in EGFR mutant non-small cell lung cancer with leptomeningeal metastases. Cancer Chemother. Pharmacol. 2016, 78, 1305–1310. [Google Scholar] [CrossRef]
- Wei, F.; Lin, C.-C.; Joon, A.; Feng, Z.; Troche, G.; Lira, M.E.; Chia, D.; Mao, M.; Ho, C.-L.; Su, W.-C.; et al. Noninvasive Saliva-based EGFR Gene Mutation Detection in Patients with Lung Cancer. Am. J. Respir. Crit. Care Med. 2014, 190, 1117–1126. [Google Scholar] [CrossRef]
- Park, S.; Hur, J.Y.; Lee, K.Y.; Lee, J.C.; Rho, J.K.; Shin, S.H.; Choi, C.-M. Assessment of EGFR mutation status using cell-free DNA from bronchoalveolar lavage fluid. CCLM 2017, 55, 1489–1495. [Google Scholar] [CrossRef]
- García-Pardo, M.; Czarnecka-Kujawa, K.; Law, J.H.; Salvarrey, A.M.; Fernandes, R.; Fan, Z.J.; Waddell, T.K.; Yasufuku, K.; Liu, G.; Donahoe, L.L.; et al. Association of Circulating Tumor DNA Testing Before Tissue Diagnosis with Time to Treatment Among Patients with Suspected Advanced Lung Cancer. JAMA Netw. Open 2023, 6, e2325332. [Google Scholar] [CrossRef] [PubMed]
- Moding, E.J.; Nabet, B.Y.; Alizadeh, A.A.; Diehn, M. Detecting Liquid Remnants of Solid Tumors: Circulating Tumor DNA Minimal Residual Disease. Cancer Discov. 2021, 11, 2968–2986. [Google Scholar] [CrossRef]
- Bartolomucci, A.; Nobrega, M.; Ferrier, T.; Dickinson, K.; Kaorey, N.; Nadeau, A.; Castillo, A.; Burnier, J.V. Circulating tumor DNA to monitor treatment response in solid tumors and advance precision oncology. npj Precis. Oncol. 2025, 9, 84. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Xue, R.; Li, X.; Yang, L.; Yang, M.; Zhang, B.; Zhang, X.; Li, L.; Duan, X.; Yan, R.; He, X.; et al. Evaluation and integration of cell-free DNA signatures for detection of lung cancer. Cancer Lett. 2024, 604, 217216. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.C.; Aggarwal, C.; Wong, J.; Nimgaonkar, V.; Hwang, W.-T.; Andronov, M.; Dibardino, D.M.; Hutchinson, C.T.; Ma, K.C.; Lanfranco, A.; et al. Plasma Genotyping at the Time of Diagnostic Tissue Biopsy Decreases Time-to-Treatment in Patients with Advanced NSCLC—Results From a Prospective Pilot Study. JTO Clin. Res. Rep. 2022, 3, 100301. [Google Scholar] [CrossRef]
- Raez, L.E.; Brice, K.; Dumais, K.; Lopez-Cohen, A.; Wietecha, D.; Izquierdo, P.A.; Santos, E.S.; Powery, H.W. Liquid Biopsy Versus Tissue Biopsy to Determine Front Line Therapy in Metastatic Non-Small Cell Lung Cancer (NSCLC). Clin. Lung Cancer 2022, 24, 120–129. [Google Scholar] [CrossRef]
- Chen, K.; Zhao, H.; Shi, Y.; Yang, F.; Wang, L.T.; Kang, G.; Nie, Y.; Wang, J. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin. Cancer Res. 2019, 25, 7058–7067. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, N.; Daaboul, N. Lung Cancer: Targeted Therapy in 2025. Curr. Oncol. 2025, 32, 146. [Google Scholar] [CrossRef]
- Li, R.-Y.; Liang, Z.-Y. Circulating tumor DNA in lung cancer: Real-time monitoring of disease evolution and treatment response. Chin. Med. J. 2020, 133, 2476–2485. [Google Scholar] [CrossRef]
- Zviran, A.; Schulman, R.C.; Shah, M.; Hill, S.T.K.; Deochand, S.; Khamnei, C.C.; Maloney, D.; Patel, K.; Liao, W.; Widman, A.J.; et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 2020, 26, 1114–1124. [Google Scholar] [CrossRef]
- Kato, R.; Hayashi, H.; Sakai, K.; Suzuki, S.; Haratani, K.; Takahama, T.; Tanizaki, J.; Nonagase, Y.; Tanaka, K.; Yoshida, T.; et al. CAPP-seq analysis of circulating tumor DNA from patients with EGFR T790M–positive lung cancer after osimertinib. Int. J. Clin. Oncol. 2021, 26, 1628–1639. [Google Scholar] [CrossRef]
- Peng, M.; Huang, Q.; Yin, W.; Tan, S.; Chen, C.; Liu, W.; Tang, J.; Wang, X.; Zhang, B.; Zou, M.; et al. Circulating Tumor DNA as a Prognostic Biomarker in Localized Non-small Cell Lung Cancer. Front. Oncol. 2020, 10, 561598. [Google Scholar] [CrossRef]
- Kan, C.F.K.; Unis, G.D.; Li, L.Z.; Gunn, S.; Li, L.; Soyer, H.P.; Stark, M.S. Circulating Biomarkers for Early Stage Non-Small Cell Lung Carcinoma Detection: Supplementation to Low-Dose Computed Tomography. Front. Oncol. 2021, 11, 555331. [Google Scholar] [CrossRef]
- Kuang, P.P.; Li, N.; Liu, Z.; Sun, T.Y.; Wang, S.Q.; Hu, J.; Ou, W.; Wang, S.Y. Circulating Tumor DNA Analyses as a Potential Marker of Recurrence and Effectiveness of Adjuvant Chemotherapy for Resected Non-Small-Cell Lung Cancer. Front. Oncol. 2021, 10, 595650. [Google Scholar] [CrossRef]
- Provencio, M.; Serna-Blasco, R.; Nadal, E.; Insa, A.; García-Campelo, M.R.; Casal Rubio, J.; Dómine, M.; Majem, M.; Rodríguez-Abreu, D.; Martínez-Martí, A.; et al. Overall survival and biomarker analysis of neoadjuvant nivolumab plus chemotherapy in operable stage IIIA non-small-cell lung cancer (NADIM phase II trial). J. Clin. Oncol. 2022, 40, 2924–2933. [Google Scholar] [CrossRef]
- Reck, M.; Gale, D.; Harpole, D.; Taube, J.; Mitsudomi, T.; Hochmair, M.; Winder, T.; Zhu, Z.; Lai, Z.; Stewart, R.; et al. LBA49 Associations of ctDNA clearance (CL) during neoadjuvant Tx with pathological response and event-free survival (EFS) in pts with resectable NSCLC (R-NSCLC): Expanded analyses from AEGEAN. Ann. Oncol. 2024, 35, S1239. [Google Scholar] [CrossRef]
- Deutsch, J.S.; Cimino-Mathews, A.; Thompson, E.; Provencio, M.; Forde, P.M.; Spicer, J.; Girard, N.; Wang, D.; Anders, R.A.; Gabrielson, E.; et al. Association between pathologic response and survival after neoadjuvant therapy in lung cancer. Nat. Med. 2024, 30, 218–228. [Google Scholar] [CrossRef]
- Forde, P.M.; Spicer, J.; Lu, S.; Provencio, M.; Mitsudomi, T.; Awad, M.M.; Felip, E.; Broderick, S.R.; Brahmer, J.R.; Swanson, S.J.; et al. CheckMate 816 Investigators. Neoadjuvant nivolumab plus chemotherapy in resectable lung cancer. N. Engl. J. Med. 2022, 386, 1973–1985. [Google Scholar] [CrossRef]
- Herbst, R.S.; Wang, M.; Chen, L. When immunotherapy meets surgery in non-small cell lung cancer. Cancer Cell 2022, 40, 603–605. [Google Scholar] [CrossRef]
- Zhou, C.; Das Thakur, M.; Srivastava, M.; Zou, W.; Xu, H.; Ballinger, M.; Felip, E.; Wakelee, H.; Altorki, N.; Reck, M.; et al. 2O IMpower010: Biomarkers of disease-free survival (DFS) in a phase III study of atezolizumab (atezo) vs. best supportive care (BSC) after adjuvant chemotherapy in stage IB-IIIA NSCLC. Ann. Oncol. 2021, 32, S134. [Google Scholar] [CrossRef]
- Abbosh, C.; Frankell, A.M.; Harrison, T.; Kisistok, J.; Garnett, A.; Johnson, L.; Veeriah, S.; Moreau, M.; Chesh, A.; Chaunzwa, T.L.; et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature 2023, 616, 553–562. [Google Scholar] [CrossRef]
- Jung, H.-A.; Ku, B.M.; Kim, Y.J.; Park, S.; Sun, J.-M.; Lee, S.-H.; Ahn, J.S.; Cho, J.H.; Kim, H.K.; Choi, Y.S.; et al. Longitudinal Monitoring of Circulating Tumor DNA From Plasma in Patients with Curative Resected Stages I to IIIA EGFR-Mutant Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2023, 18, 1199–1208. [Google Scholar] [CrossRef]
- Guo, N.; Lou, F.; Ma, Y.; Li, J.; Yang, B.; Chen, W.; Ye, H.; Zhang, J.-B.; Zhao, M.-Y.; Wu, W.-J.; et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci. Rep. 2016, 6, 33519. [Google Scholar] [CrossRef]
- Waldeck, S.; Mitschke, J.; Wiesemann, S.; Rassner, M.; Andrieux, G.; Deuter, M.; Mutter, J.; Lüchtenborg, A.M.; Kottmann, D.; Titze, L.; et al. Early assessment of circulating tumor DNA after curative-intent resection predicts tumor recurrence in early-stage and locally advanced non-small-cell lung cancer. Mol. Oncol. 2022, 16, 527–537. [Google Scholar] [CrossRef]
- Gale, D.; Heider, K.; Ruiz-Valdepenas, A.; Hackinger, S.; Perry, M.; Marsico, G.; Rundell, V.; Wulff, J.; Sharma, G.; Knock, H.; et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann. Oncol. 2022, 33, 500–510. [Google Scholar] [CrossRef]
- Oh, Y.; Yoon, S.M.; Lee, J.; Park, J.H.; Lee, S.; Hong, T.; Chung, L.I.; Sudhaman, S.; Riddell, T.; Palsuledesai, C.C.; et al. Personalized, tumor-informed, circulating tumor DNA assay for detecting minimal residual disease in non-small cell lung cancer patients receiving curative treatments. Thorac. Cancer 2024, 15, 1095–1102. [Google Scholar] [CrossRef]
- Ohara, S.; Suda, K.; Sakai, K.; Nishino, M.; Chiba, M.; Shimoji, M.; Takemoto, T.; Fujino, T.; Koga, T.; Hamada, A.; et al. Prognostic implications of preoperative versus postoperative circulating tumor DNA in surgically resected lung cancer patients: A pilot study. Transl. Lung Cancer Res. 2020, 9, 1915–1923. [Google Scholar] [CrossRef]
- Tan, A.C.; Lai, G.G.Y.; Saw, S.P.L.; Chua, K.L.M.; Takano, A.; Ong, B.H.; Koh, T.P.T.; Jain, A.; Tan, W.L.; Ng, Q.S.; et al. Detection of circulating tumor DNA with ultradeep sequencing of plasma cell-free DNA for monitoring minimal residual disease and early detection of recurrence in early-stage lung cancer. Cancer 2024, 130, 1758–1765. [Google Scholar] [CrossRef]
- Rosenlund, L.; Guldbrandsen, K.; Ahlborn, L.B.; Bloch, M.; Skougaard, K.; Albrecht-Beste, E.; Nellemann, H.M.; Krakauer, M.; Gørtz, P.M.; Fledelius, J.; et al. ctDNA can detect minimal residual disease in curative treated non-small cell lung cancer patients using a tumor agnostic approach. Lung Cancer 2025, 203, 108528. [Google Scholar] [CrossRef]
- Schuurbiers, M.M.F.; Smith, C.G.; Hartemink, K.J.; Rintoul, R.C.; Gale, D.; Monkhorst, K.; Mandos, B.L.R.; Paterson, A.L.; Broek, D.v.D.; Rosenfeld, N.; et al. Recurrence prediction using circulating tumor DNA in patients with early-stage non-small cell lung cancer after treatment with curative intent: A retrospective validation study. PLoS Med. 2025, 22, e1004574. [Google Scholar] [CrossRef]
- Tan, A.; Lai, G.; Saw, S.; Chua, K.; Takano, A.; Ong, B.; Koh, T.; Jain, A.; Tan, W.; Ng, Q.; et al. MA07.06 Circulating Tumor DNA for Monitoring Minimal Residual Disease and Early Detection of Recurrence in Early Stage Lung Cancer. J. Thorac. Oncol. 2021, 16, S907. [Google Scholar] [CrossRef]
- Zhang, J.-T.; Dong, S.; Gu, W.Q.; Zhao, N.; Liang, Y.; Tang, W.F.; Liu, S.Y.; Wang, F.; Wang, G.S.; Peng, B.; et al. Adjuvant Therapy-Free Strategy for Stage IB to IIIA Non–Small-Cell Lung Cancer Patients After Radical Resection Based on Longitudinal Undetectable Molecular Residual Disease: Prospective, Multicenter, Single-Arm Study (CTONG 2201). Clin. Lung Cancer. 2024, 25, e1–e4. [Google Scholar] [CrossRef]
- Zhao, X.; Dai, F.; Mei, L.; Huang, D.; Shen, X.; Zhang, H.; She, X.; Ma, Z. The Potential Use of Dynamics Changes of ctDNA and cfDNA in the Perioperative Period to Predict the Recurrence Risk in Early NSCLC. Front. Oncol. 2021, 11, 671963. [Google Scholar] [CrossRef]
- Li, N.; Wang, B.X.; Li, J.; Shao, Y.; Li, M.T.; Li, J.J.; Kuang, P.P.; Liu, Z.; Sun, T.Y.; Wu, H.Q.; et al. Perioperative circulating tumor DNA as a potential prognostic marker for operable stage I to IIIA non-small cell lung cancer. Cancer 2022, 128, 708–718. [Google Scholar] [CrossRef]
- Xia, L.; Mei, J.; Kang, R.; Deng, S.; Chen, Y.; Yang, Y.; Feng, G.; Deng, Y.; Gan, F.; Lin, Y.; et al. Perioperative ctDNA-Based Molecular Residual Disease Detection for Non-Small Cell Lung Cancer: A Prospective Multicenter Cohort Study (LUNGCA-1). Clin. Cancer Res. 2022, 28, 3308–3317. [Google Scholar] [CrossRef]
- Qiu, B.; Guo, W.; Zhang, F.; Lv, F.; Ji, Y.; Peng, Y.; Chen, X.; Bao, H.; Xu, Y.; Shao, Y.; et al. Dynamic recurrence risk and adjuvant chemotherapy benefit prediction by ctDNA in resected NSCLC. Nat. Commun. 2021, 12, 6770. [Google Scholar] [CrossRef]
- Moding, E.J.; Liu, Y.; Nabet, B.Y.; Chabon, J.J.; Chaudhuri, A.A.; Hui, A.B.; Bonilla, R.F.; Ko, R.B.; Yoo, C.H.; Gojenola, L.; et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat. Cancer 2020, 1, 176–183. [Google Scholar] [CrossRef]
- Desai, A.; Vázquez, T.A.; Arce, K.M.; Corassa, M.; Mack, P.C.; Gray, J.E.; Pellini, B. ctDNA for the Evaluation and Management of EGFR-Mutant Non-Small Cell Lung Cancer. Cancers 2024, 16, 940. [Google Scholar] [CrossRef]
- O’Kane, G.M.; Bradbury, P.A.; Feld, R.; Leighl, N.B.; Liu, G.; Pisters, K.-M.; Kamel-Reid, S.; Tsao, M.S.; Shepherd, F.A. Uncommon EGFR mutations in advanced non-small cell lung cancer. Lung Cancer 2017, 109, 137–144. [Google Scholar] [CrossRef]
- Arcila, M.E.; Nafa, K.; Chaft, J.E.; Rekhtman, N.; Lau, C.; Reva, B.A.; Zakowski, M.F.; Kris, M.G.; Ladanyi, M. EGFR Exon 20 Insertion Mutations in Lung Adenocarcinomas: Prevalence, Molecular Heterogeneity, and Clinicopathologic Characteristics. Mol. Cancer Ther. 2013, 12, 220–229. [Google Scholar] [CrossRef]
- Felip, E.; Cho, B.C.; Gutiérrez, V.; Alip, A.; Besse, B.; Lu, S.; Spira, A.I.; Girard, N.; Califano, R.; Gadgeel, A.M.; et al. Amivantamab plus lazertinib versus osimertinib in first-line EGFR-mutant advanced non-small-cell lung cancer with biomarkers of high-risk disease: A secondary analysis from MARIPOSA. Ann. Oncol. 2024, 35, 805–816. [Google Scholar] [CrossRef]
- Liu, S.-Y.; Erazo, T.; Jee, J.; Arfe, A.; Gupta, A.; Pike, L.R.; Santini, F.C.; Daly, B.; Schoenfeld, A.; Eichholz, J.; et al. Optimal systemic treatment and real-world clinical application of ctDNA in patients with metastatic HER2-mutant lung cancer. Eur. J. Cancer 2024, 210, 114257. [Google Scholar] [CrossRef]
- Anagnostou, V.; Ho, C.; Nicholas, G.; Juergens, R.A.; Sacher, A.; Fung, A.S.; Wheatley-Price, P.; Laurie, S.A.; Levy, B.; Brahmer, J.R.; et al. ctDNA response after pembrolizumab in non-small cell lung cancer: Phase 2 adaptive trial results. Nat. Med. 2023, 29, 2559–2569. [Google Scholar] [CrossRef]
- Lovly, C.M.; Salama, A.K.; Salgia, R. Tumor heterogeneity and therapeutic resistance. Am. Soc. Clin. Oncol. Educ. Book 2016, 36, e585–e593. [Google Scholar] [CrossRef]
- Song, Y.; Hu, C.; Xie, Z.; Wu, L.; Zhu, Z.; Rao, C.; Liu, L.; Chen, Y.; Liang, N.; Chen, J.; et al. Written on behalf of AME Lung Cancer Collaborative Group. Circulating tumor DNA clearance predicts prognosis across treatment regimen in a large real-world longitudinally monitored advanced non-small cell lung cancer cohort. Transl. Lung Cancer Res. 2020, 9, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in Non–Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef]
- Jun, S.; Shukla, N.A.; Durm, G.; Hui, A.B.; Cao, S.; Ganti, A.K.; Jabbour, S.K.; Kunder, C.; Alizadeh, A.A.; Hanna, N.H.; et al. Analysis of Circulating Tumor DNA Predicts Outcomes of Short-Course Consolidation Immunotherapy in Unresectable Stage III NSCLC. J. Thorac. Oncol. 2024, 19, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhang, J.-T.; Gao, X.; Chen, Z.-Y.; Yan, B.; Tan, P.-X.; Yang, X.-R.; Gao, W.; Gong, Y.; Tian, Z.; et al. Dynamic circulating tumor DNA during chemoradiotherapy predicts clinical outcomes for locally advanced non-small cell lung cancer patients. Cancer Cell 2023, 41, 1763–1773.e4. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, T.; Wang, J.; Wang, J.; Xu, Y.; Zhao, X.; Ou, Q.; Shao, Y.; Wang, X.; Wu, Y.; et al. The clinical utility of dynamic ctDNA monitoring in inoperable localized NSCLC patients. Mol. Cancer 2022, 21, 117. [Google Scholar] [CrossRef] [PubMed]
- Horndalsveen, H.; Haakensen, V.D.; Madebo, T.; Gronberg, B.H.; Halvorsen, T.O.; Koivunen, J.; Oselin, K.; Cicenas, S.; Helbekkmo, N.; Aanerud, M.; et al. ctDNA-based MRD detection in unresectable NSCLC undergoing curatively intended chemoradiotherapy and durvalumab. J. Clin. Oncol. 2025, 43, 8011. [Google Scholar] [CrossRef]
- Ernst, S.M.; van Marion, R.; Atmodimedjo, P.N.; de Jonge, E.; Mathijssen, R.H.J.; Paats, M.S.; de Bruijn, P.; Koolen, S.L.; von der Thüsen, J.H.; Aerts, J.G.J.V.; et al. Clinical utility of circulating tumor DNA in patients with advanced KRASG12C-mutated NSCLC treated with sotorasib. J. Thorac. Oncol. 2024, 19, 995–1006. [Google Scholar] [CrossRef]
- Yamaguchi, O.; Kasahara, N.; Soda, H.; Imai, H.; Naruse, I.; Yamaguchi, H.; Itai, M.; Taguchi, K.; Uchida, M.; Sunaga, N.; et al. Predictive significance of circulating tumor DNA against patients with T790M-positive EGFR-mutant NSCLC receiving osimertinib. Sci. Rep. 2023, 13, 20848. [Google Scholar] [CrossRef]
- Shaw, A.T.; Solomon, B.J.; Besse, B.; Bauer, T.M.; Lin, C.C.; Soo, R.A.; Riely, G.J.; Ou, S.H.I.; Clancy, J.S.; Li, S.; et al. ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2019, 37, 1370–1379. [Google Scholar] [CrossRef]
- Vendrell, J.A.; Quantin, X.; Aussel, A.; Solassol, I.; Serre, I.; Solassol, J. EGFR-dependent mechanisms of resistance to osimertinib determined by ctDNA NGS analysis identify patients with better outcome. Transl. Lung Cancer Res. 2021, 10, 4084–4094. [Google Scholar] [CrossRef]
- Remon, J.; Besse, B.; Aix, S.P.; Callejo, A.; Al-Rabi, K.; Bernabe, R.; Greillier, L.; Majem, M.; Reguart, N.; Monnet, I.; et al. Osimertinib treatment based on plasma T790M monitoring in patients with EGFR-mutant non-small-cell lung cancer (NSCLC): EORTC Lung Cancer Group 1613 APPLE phase II randomized clinical trial. Ann. Oncol. 2023, 34, 468–476. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S.-J.; Tsui, D.W.Y.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.-F.; Kingsbury, Z.; Wong, A.S.C.; et al. Non-Invasive Analysis of Acquired Resistance to Cancer Therapy by Sequencing of Plasma DNA. Nature 2013, 497, 108–112. [Google Scholar] [CrossRef]
- Gridelli, C.; Nuccio, E.; Casaluce, F. The Role of Osimertinib in Stage I–II Non-Small-Cell Lung Cancer with Activating EGFR Mutation. Targets 2025, 3, 20. [Google Scholar] [CrossRef]
- Zhang, E.W.; Dagogo-Jack, I.; Kuo, A.; Rooney, M.M.; Shaw, A.T.; Digumarthy, S.R. Association between circulating tumor DNA burden and disease burden in patients with ALK-positive lung cancer. Cancer 2020, 126, 4473–4484. [Google Scholar] [CrossRef]
- Dziadziuszko, R.; Mok, T.; Peters, S.; Han, J.Y.; Alatorre-Alexander, J.; Leighl, N.; Sriuranpong, V.; Pérol, M.; de Castro Junior, G.; Nadal, E.; et al. Blood First Assay Screening Trial (BFAST) in treatment-naive advanced or metastatic NSCLC: Initial Results of the Phase 2 ALK-Positive Cohort. J. Thorac. Oncol. 2021, 16, 2040–2050. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Brannon, A.R.; Ferris, L.A.; Campbell, C.D.; Lin, J.J.; Schultz, K.R.; Ackil, J.; Stevens, S.; Dardaei, L.; Yoda, S.; et al. Tracking the evolution of resistance to ALK Tyrosine Kinase inhibitors through longitudinal analysis of circulating tumor DNA. JCO Precis. Oncol. 2018, 2, 1–14. [Google Scholar] [CrossRef]
- Heeke, S.; Gandhi, S.; Tran, H.T.; Lam, V.K.; Byers, L.A.; Gibbons, D.L.; Gay, C.M.; Altan, M.; Antonoff, M.B.; Le, X.; et al. Longitudinal Tracking of ALK-Rearranged NSCLC From Plasma Using Circulating Tumor RNA and Circulating Tumor DNA. JTO Clin. Res. Rep. 2025, 6, 100795. [Google Scholar] [CrossRef]
- Chmielecki, J.; Gray, J.E.; Cheng, Y.; Ohe, Y.; Imamura, F.; Cho, B.C.; Lin, M.-C.; Majem, M.; Shah, R.; Rukazenkov, Y. Can-didate mechanisms of acquired resistance to first-line osimertinib in EGFR-mutated advanced non-small cell lung cancer. Nat. Commun. 2023, 14, 1070. [Google Scholar] [CrossRef]
- Sivapalan, L.; Murray, J.C.; Canzoniero, J.V.; Landon, B.; Jackson, J.; Scott, S.; Lam, V.; Levy, B.P.; Sausen, M.; Anagnostou, V. Liquid biopsy approaches to capture tumor evolution and clinical outcomes during cancer immunotherapy. J. Immunother. Cancer 2023, 11, e005924. [Google Scholar] [CrossRef]
- Planchard, D.; Loriot, Y.; Andre, F.; Gobert, A.; Auger, N.; Lacroix, L.; Soria, J. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Ann. Oncol. 2015, 26, 2073–2078. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]
| Study | Study Design, Stage, and/or Population | Outcome |
|---|---|---|
| Guo et al., 2016 [55] | Prospective; all stages | ctDNA from stage IA and IB patients had more dramatic decrease post-op compared with mutation frequency in more advanced cancer pts |
| Abbosh et al., 2017 [13] | Prospective; IIA–IIIB | Tumor volume correlated with the mean plasma VAF of clonal SNVs in ctDNA-positive NSCLCs |
| Chaudhuri et al., 2017 [11] | Retrospective; IB–IIIB, | Posttreatment ctDNA detection preceded radiographic progression in 72% of patients by a median of 5.2 months |
| Chen et al., 2019 [39] | Prospective; I–IIIA | Rapid decrease in ctDNA occurred after radical tumor resection. Median ctDNA half-life was 35 min. RFS in pts with detectable ctDNA was shorter than in those without |
| Waldeck et al., 2022 [56] | Prospective; IA–IIIB | Of ctDNA-negative pts, 33% experienced relapse post-op. Positive ctDNA in early post-op plasma samples was associated with shorter PFS and OS |
| Gale et al., 2022 [57] | Prospective; IA–IIIB | ctDNA detection had clinical specificity >98.5% and preceded clinical recurrence by a median of 212.5 days |
| Abbosh et al., 2023 [53] | Prospective; II–III | Results showed that 3- to 6-monthly ctDNA surveillance identified impending disease relapse in 20% of landmark-negative patients |
| Jung et al., 2023 [54] | Prospective; I-IIIA EGFR mutant | Of patients with baseline +ctDNA, 76% had clearance at 4 weeks after surgery. MRD was detected before radiological recurrence in 69% of patients with exon 19 deletion and in 20% with L858R mutation |
| Provencio et al., 2022 [47] | Phase II trial; IIIA or IIIB | ctDNA levels were associated with differences in PFS/OS; 67% of the experimental group and 44% of the control group were ctDNA-negative after neoadjuvant treatment. |
| Oh et al., 2024 [58] | Retrospective; I–IV | Pts with ctDNA+ were more likely to experience recurrence compared to ctDNA- patients. ctDNA+ was associated with poorer RFS than persistently ctDNA- patients |
| Ohara et al., 2020 [59] | Prospective; II–III | ctDNA+ pts had disease recurrence within median of 9.1 months and shorter RFS/OS than those without detectable ctDNA. Three pts who had CNS-only metastases did not have detectable ctDNA |
| Tan et al., 2024 [60] | Retrospective; I–III | ctDNA detection preop was associated with shorter RFS. ctDNA positivity preceded radiological findings by a median lead time of 2.8 months |
| Herbst et al., 2022 [51] | Phase III post hoc; IB–IIIA; EGFR mutant | MRD+ had clinical sensitivity of 65% and specificity of 95% and preceded a DFS event by a median of 4.7 months |
| Rosenlund et al., 2025 [61] | Prospective; I–III | Detectable ctDNA post-treatment was significantly associated with increased risk of tumor recurrence and shorter RFS. Post RT/CRT, ctDNA detection was significantly linked to shorter RFS in MRD analysis |
| Schuurbiers et al., 2025 [62] | Retrospective; I–III | Before treatment, ctDNA was detected in 48% LEMA and 51% LUCID pts. ctDNA detection after treatment was associated with shorter recurrence-free survival and overall survival |
| Study | Study Design, Stage, and/or Population | Outcome |
|---|---|---|
| Liu et al., 2024 [74] | Retrospective; Metastatic HER2-mutant | HER2 mutations tracked longitudinally correlated with treatment response. Pts with detectable BL ctDNA had shorter OS |
| Song et al., 2020 [77] | Prospective; IIIB to IV | Higher ctDNA at BL was associated with shorter OS. ctDNA- was associated with longer PFS/OS regardless of type of treatment |
| Paik et al., 2020 [78] | Phase II; advanced/metastatic MET exon 14 skipping mutation | A total of 67% of patients had a molecular cfDNA response; of those, 71% had a radiographic response. ctDNA and imaging PD were highly concordant |
| Anagnostou et al., 2023 [75] | Phase II; advanced/metastatic PD-L1 expression level of ≥1% | Median time to ctDNA response was 2.1 months; pts with MR had longer PFS and OS |
| Jun et al., 2024 [79] | Phase II post hoc; III | ctDNA+ predicted inferior PFS after completion of CRT and at the end of CPI |
| Moding et al., 2020 [69] | Stage IIB-IIIB NSCLC | Pts who were ctDNA- after CRT had better outcomes regardless of CICI. Patients with MRD+ after CRT who received CICI had significantly better outcomes than pts who did not receive CICI |
| Pan et al., 2023 [80] | Stage IIB- IIIC | Longitudinal undetectable MRD was found in 20.1% of patients. The 2-year PFS of these pts was 88.4% |
| Yang et al., 2022 [81] | Stage III NSCLC | ctDNA collected 1 month post-CRT/RT was the optimal choice to predict pts’ PFS and OS, and the dynamic change in ctDNA was closely associated with clinical outcomes |
| Horndalsveen et al., 2025 [82] (abstract) | Phase II; III | Preliminary OS data shows that detectable ctDNA during the first four months post-CRT significantly increased the odds of death within 24 months |
| Ernst et al., 2024 [83] | Prospective; advanced KRASG12C-Mutated | Pretreatment, KRASG12C ctDNA was detected in 76% of pts. The disease control rate was significantly higher in those with a molecular response |
| Yamaguchi et al., 2023 [84] | Prospective; III-IV | Detection of T790M at PD after osimertinib initiation was a significant independent prognostic factor for predicting shorter prognosis |
| Shaw et al., 2019 [85] | Phase II; ALK mutation | PFS was similar in pts with and without ALK mutations on plasma genotyping but longer in patients with ALK mutations on tissue genotyping |
| Vendrell et al., 2021 [86] | Prospective; IV | Pts who developed an EGFR-dependent mechanism of resistance responded longer to osimertinib than EGFR-independent resistant pts |
| Gray et al., 2024 [9] | Phase III post hoc; advanced stage | ctDNA PD preceded or co-occurred with RECIST-defined PD: 64% in FLAURA and 56% in AURA3 |
| Remon et al., 2023 [87] | Prospective; EGFR mutant | Molecular PD before RECIST PD led to an earlier switch to osimertinib in 17% of patients with satisfactory PFS/OS outcomes |
| Trial/Study (with NCT if Available) | Study Type | Objective/Goal |
|---|---|---|
| Resectable | ||
| NCT06358222 | Observational | Develop tumor-naïve MRD panel to predict nodal disease |
| NCT06426511 | Phase II | Personalize consolidation toripalimab based on MRD |
| NCT06979661 | Early Phase I | Use HaystackTM MRD to guide post-op RT/systemic therapy |
| NCT05536505 | Phase II | De-escalate in MRD−; treat MRD+ with icotinib/osimertinib |
| NCT04317534 | Phase II | Observation vs. pembrolizumab in stage I with ctDNA correlatives |
| NCT04712877 | Observational | Validate biomarker-integrated peri-operative care pathways including ctDNA |
| *NCT05079022 | Phase I/II | Test benefits of adjuvant furmonertinib specifically in MRD+ stage I |
| *NCT04385368 | Phase III | Determine if adding durvalumab to adjuvant chemotherapy improves DFS in MRD+ post-resection |
| *NCT04642469 | Phase III | Test durvalumab vs. placebo in patients who become MRD+ during surveillance after curative therapy |
| NCT06053099 | Prospective cohort | Define prognostic and mechanistic correlates of relapse during/after adjuvant osimertinib |
| *NCT04037150 | Observational | Evaluate peri-operative ctDNA as a surveillance tool |
| NCT06323148 | Phase III | Test adjuvant osimertinib specifically in MRD+ |
| NCT05457049 | Observational | Evaluate therapy-free surveillance if MRD remains undetectable |
| NCT05059444 | Observational | Validate a novel commercial ctDNA assay for surveillance |
| *NCT04351555 | Phase III | Evaluate benefits of MRD in neoadjuvant osimertinib ± chemo vs. chemo |
| NCT04966663 | Phase II | Test adjuvant therapy in ctDNA+ vs. observation |
| NCT05167604 | Observational | Explore MRD status and correlation with outcomes after adjuvant chemo in early NSCLC |
| NCT04367311 | Phase II | Assess intensified adjuvant chemo + atezolizumab for post-op ctDNA+ pts |
| *NCT04625699 | Phase II | Evaluate peri-operative IO doublet for patients who develop detectable ctDNA after standard adjuvant therapy |
| NCT05236114 | Observational | Map optimal peri-op timepoints for ctDNA MRD |
| NCT04638582 | Phase II | Define benefits of preop pembrolizumab with ctDNA correlatives in early NSCLC |
| NCT05254782 | Observational | Quantify detection rate and prognostic value of peri-operative ctDNA |
| NCT05460195 | Phase II | Peri-op sintilimab and anlotinib based on MRD |
| NCT04585477 | Phase II | ctDNA-adapted adjuvant immunotherapy strategies |
| Advanced/Metastatic | ||
| NCT04841811 | Phase III | Evaluate almonertinib in unresectable stage III EGFRm NSCLC; evaluate dynamic MRD-guided maintenance therapy with almonertinib |
| *NCT06020989 | Phase II | Determine if early chemo add-on for ctDNA-positive patients improves outcomes vs. lazertinib alone |
| NCT04912687 | Observational | Implement and evaluate baseline ctDNA testing at initial diagnosis |
| NCT05334277 | Phase II | Test ctDNA-guided escalation after furmonertinib induction |
| *NCT03865511 | Phase II | Characterize on-treatment resistance using paired biopsies and ctDNA with osimertinib |
| NCT04737382 | Observational | Define concordance and clinical utility of tumor NGS vs. plasma ctDNA |
| NCT05281406 | Phase II | Assess benefit of adding platinum + pemetrexed for early ctDNA persistence on 1 L osimertinib |
| NCT05598528 | Observational | Identify predictors of primary resistance to 3rd-gen EGFR TKIs |
| NCT05257967 | Observational | Quantify incremental value of CSF ctDNA in leptomeningeal disease |
| NCT05401110 | Phase I | Determine safety/activity of carotuximab + osimertinib; follow ctDNA+ |
| NCT05534113 | Phase II | Evaluate sequential envafolimab after ctDNA clearance on almonertinib |
| NCT05813522 | Phase II | Assess high-dose furmonertinib efficacy in LM and validate CSF ctDNA monitoring |
| NCT04585490 | Phase III | Personalize consolidation durvalumab after CRT using ctDNA |
| NCT05198154 | Observational | Anticipate progression using ctDNA in long-responders |
| Screening/Risk Stratification | ||
| *NCT05117840 | Observational | Evaluate blood-based GuardantLUNAR-2 screening assay |
| NCT06163846 | Observational | Integrate imaging and liquid biopsy for stratification |
| *NCT05020275 | Observational | Map resistance to inform subsequent therapy |
| NCT03774758 | Observational | Validate ctDNA signature for risk stratification |
| *NCT02194738 | Screening (ALCHEMIST) | Test resected NSCLC for genetic mutations to facilitate accrual to randomized adjuvant studies of adjuvant treatment trials for resected NSCLC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhillon, P.; Du, S.; Cheng, H. Circulating Tumor DNA as a Prognostic and Predictive Biomarker in Lung Cancer. Cancers 2025, 17, 3327. https://doi.org/10.3390/cancers17203327
Dhillon P, Du S, Cheng H. Circulating Tumor DNA as a Prognostic and Predictive Biomarker in Lung Cancer. Cancers. 2025; 17(20):3327. https://doi.org/10.3390/cancers17203327
Chicago/Turabian StyleDhillon, Puneet, Simo Du, and Haiying Cheng. 2025. "Circulating Tumor DNA as a Prognostic and Predictive Biomarker in Lung Cancer" Cancers 17, no. 20: 3327. https://doi.org/10.3390/cancers17203327
APA StyleDhillon, P., Du, S., & Cheng, H. (2025). Circulating Tumor DNA as a Prognostic and Predictive Biomarker in Lung Cancer. Cancers, 17(20), 3327. https://doi.org/10.3390/cancers17203327






