Application of Single-Cell Sequencing and Machine Learning in Prognosis and Immune Profiling of Lung Adenocarcinoma: Exploring Disease Mechanisms and Treatment Strategies Based on Circadian Rhythm Gene Signatures
Simple Summary
Abstract
1. Introduction
2. Method
2.1. Strategy for Transcriptomic Data Acquisition and Correction of Batch Effects
2.2. Identification of a Circadian Rhythm-Driven Gene Signature for Prognostic Stratification
2.3. External Validation Datasets
2.4. Clinical Correlation and Prognostic Evaluation
2.5. Analysis of Immune Cell Infiltration and the Tumor Microenvironment
2.6. Single-Cell Atlas Construction and CRGs Activity Scoring
2.7. Intercellular Communication Profiling
2.8. Integration of Copy Number Variation and Mutation Burden for Prognostic Stratification
2.9. Multidimensional Analysis of Predictive Features for Immunotherapy Response
2.10. Assessment of Drug Response Variability Across Risk Subgroups
2.11. Cultivation and Transfection of LUAD Cell Lines
2.12. Quantitative Real-Time PCR
2.13. Transwell Migration and Invasion Assays
2.14. Colony Formation Assay
2.15. Statistical Methods and Data Interpretation
3. Results
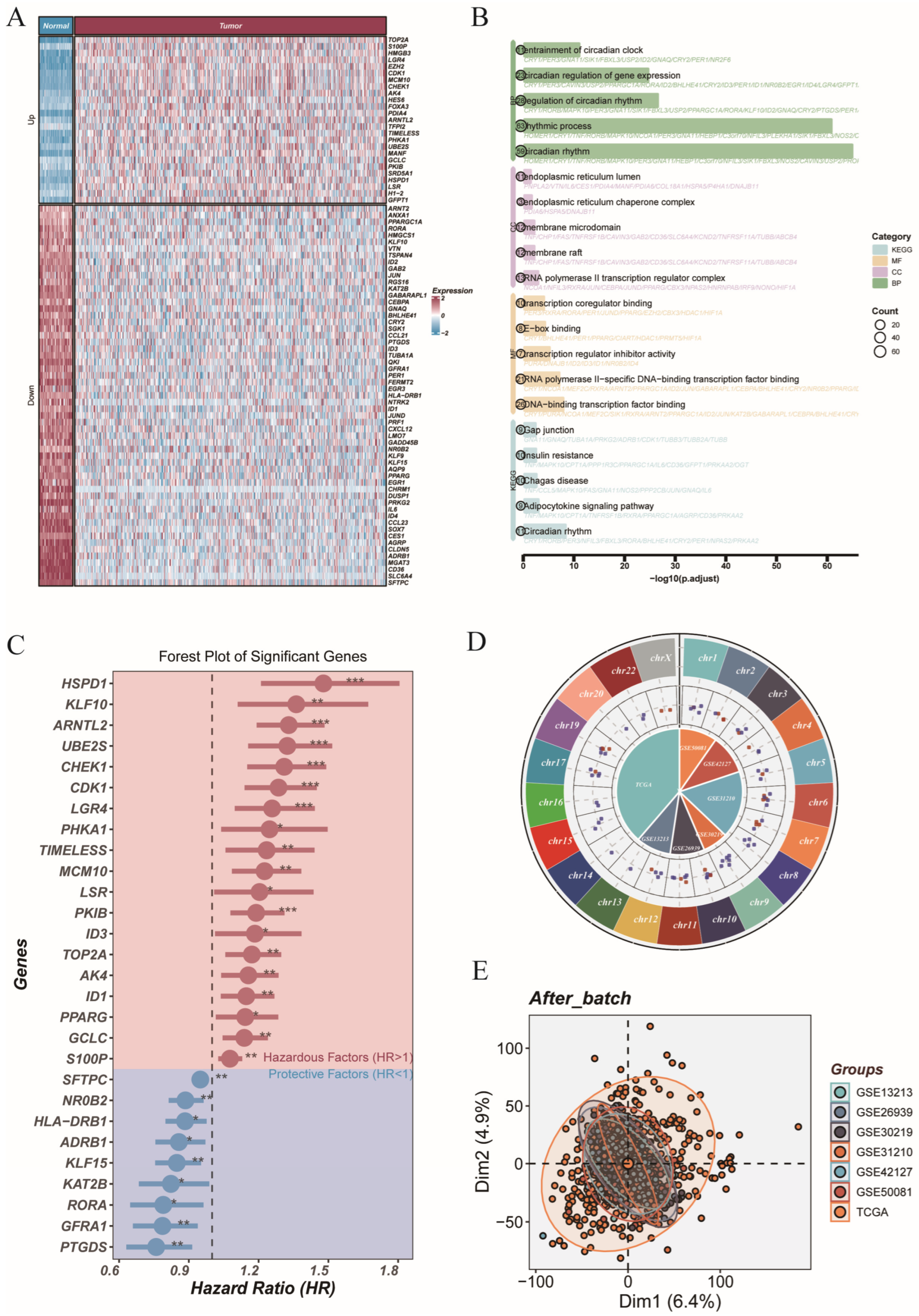
3.1. Genome-Wide Profiling of Circadian Genes Reveals Dysregulation and Survival Association in LUAD
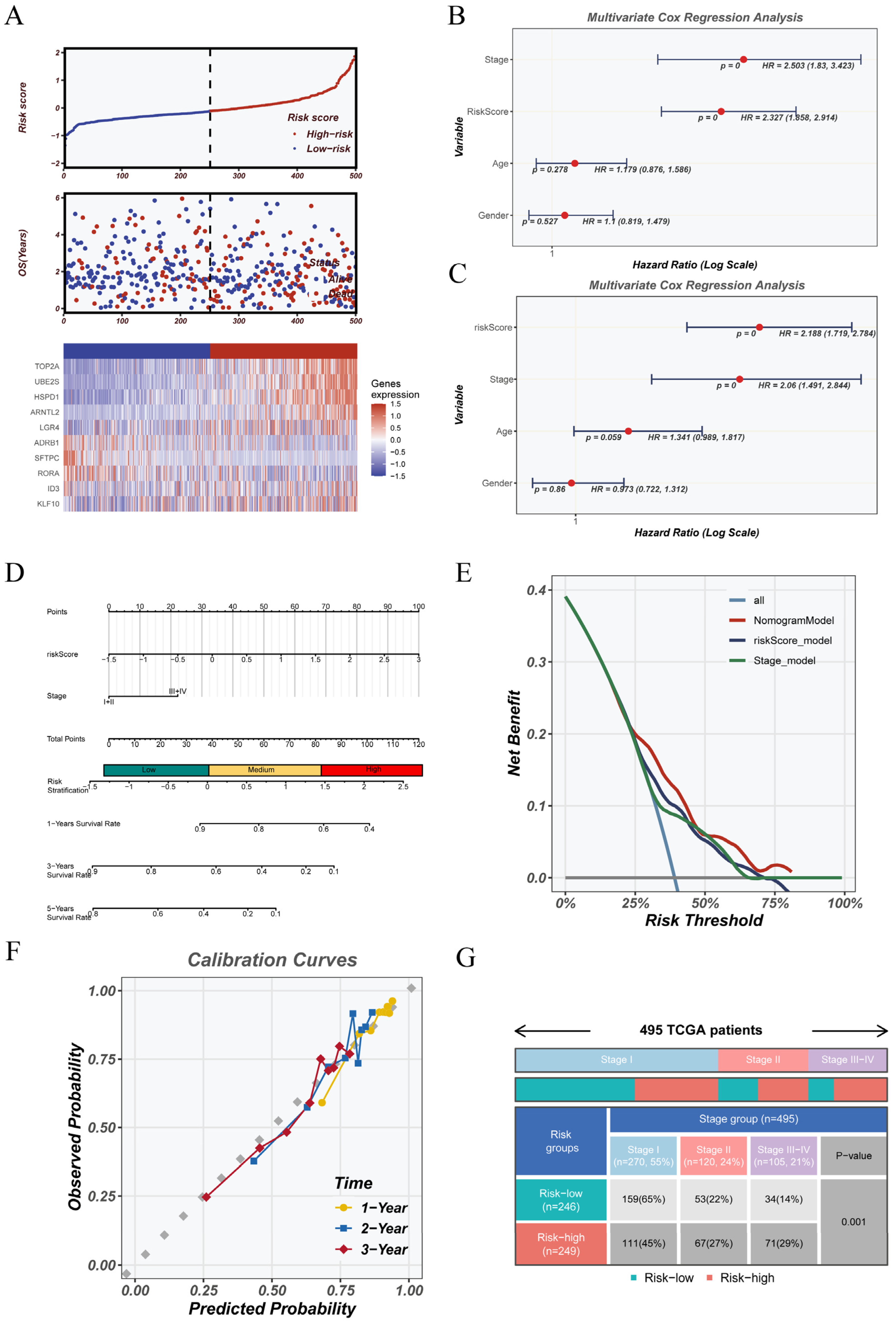
3.2. Machine Learning-Based CRGs Risk Score Enables Prognostic Stratification
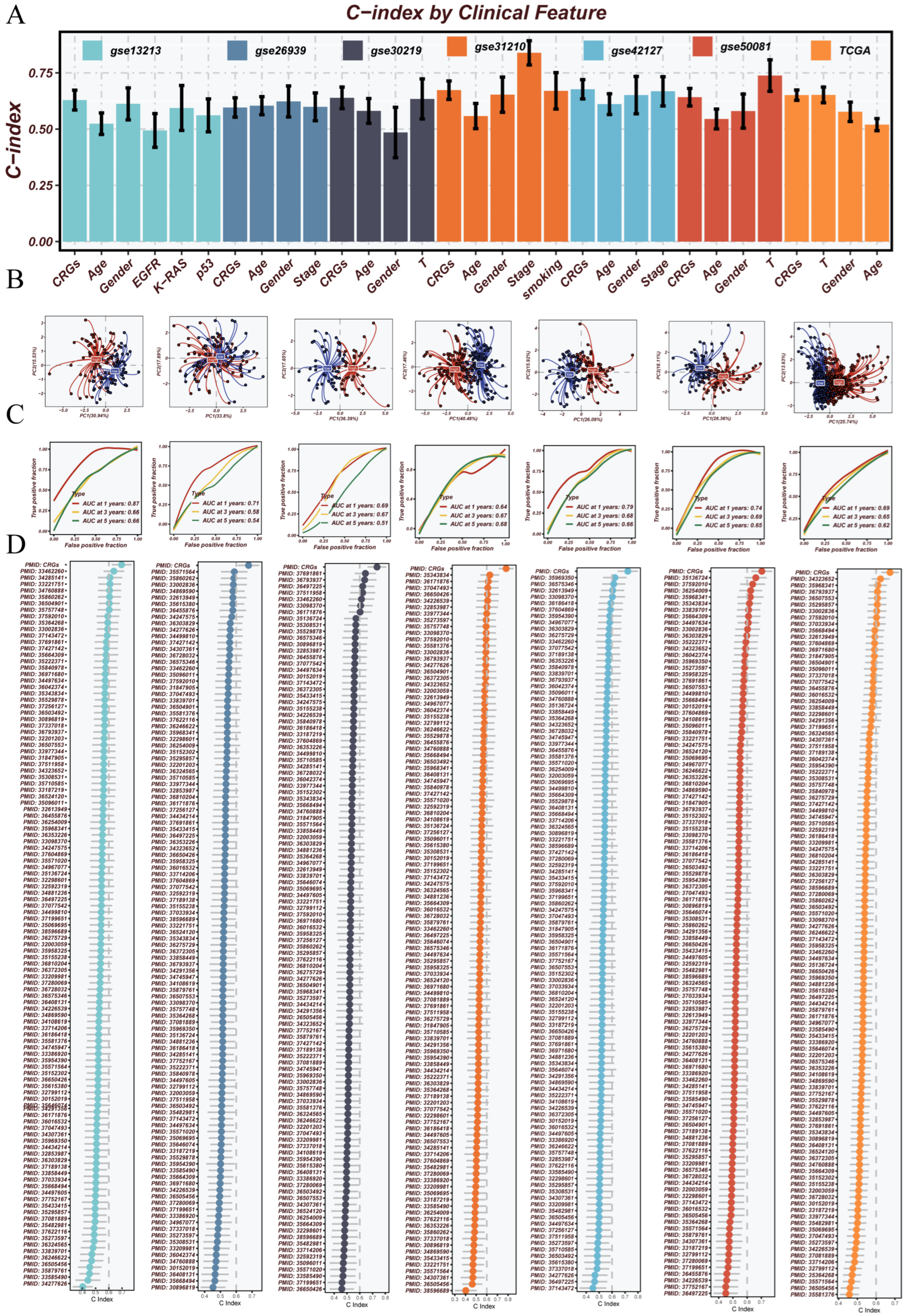
3.3. Robust Validation and Independent Prognostic Value of the CRGs Model Across Clinical and External Cohorts
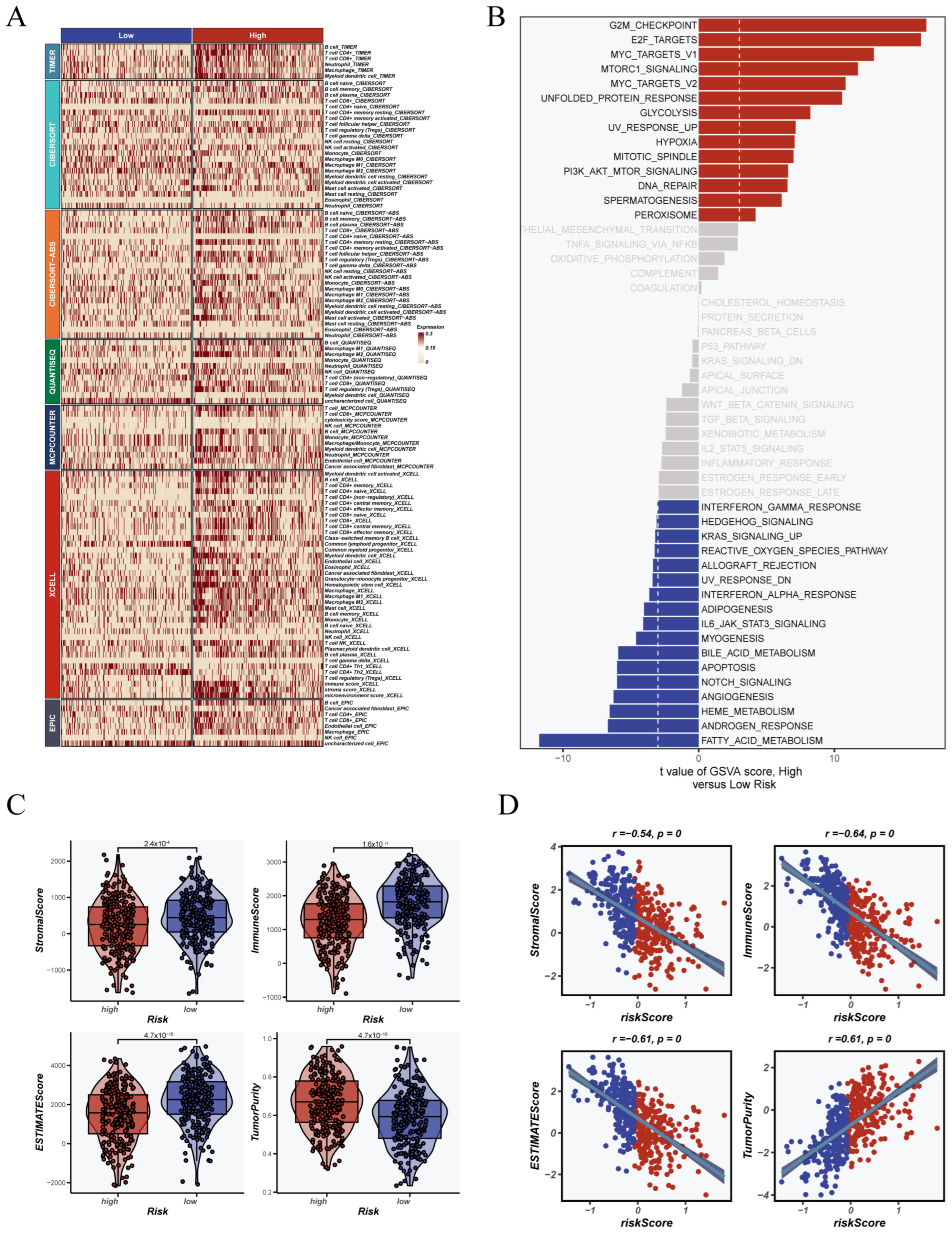
3.4. Association Between CRGs Score and Immune Landscape
3.5. Multi-Algorithm Scoring Identifies Distinct Metabolic Profiles Across Single-Cell Types
3.6. Metabolic Activity and Cell–Cell Communication in CRGs-Based Subtypes
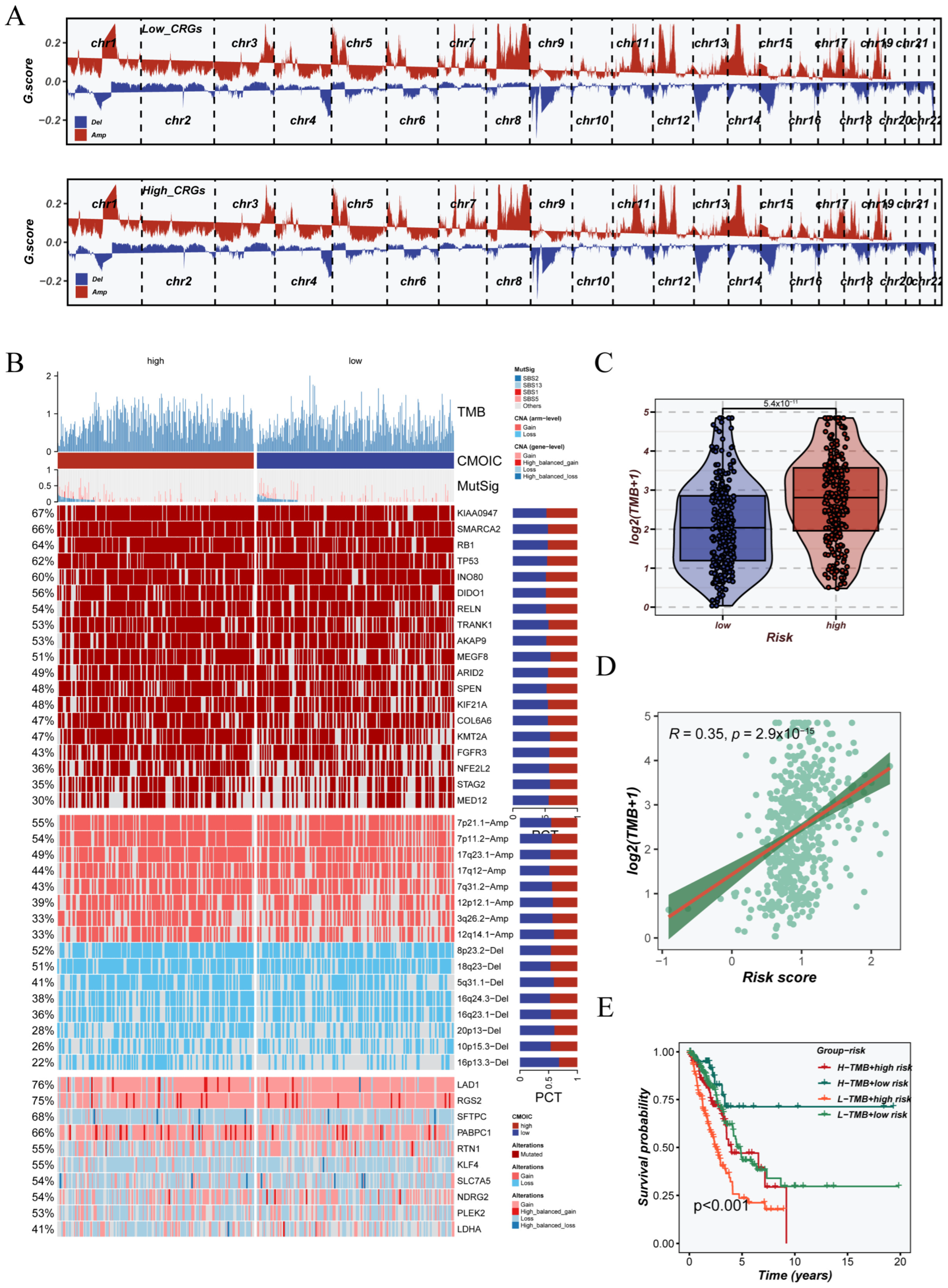
3.7. Genomic Instability Landscape Associated with CRGs Signature
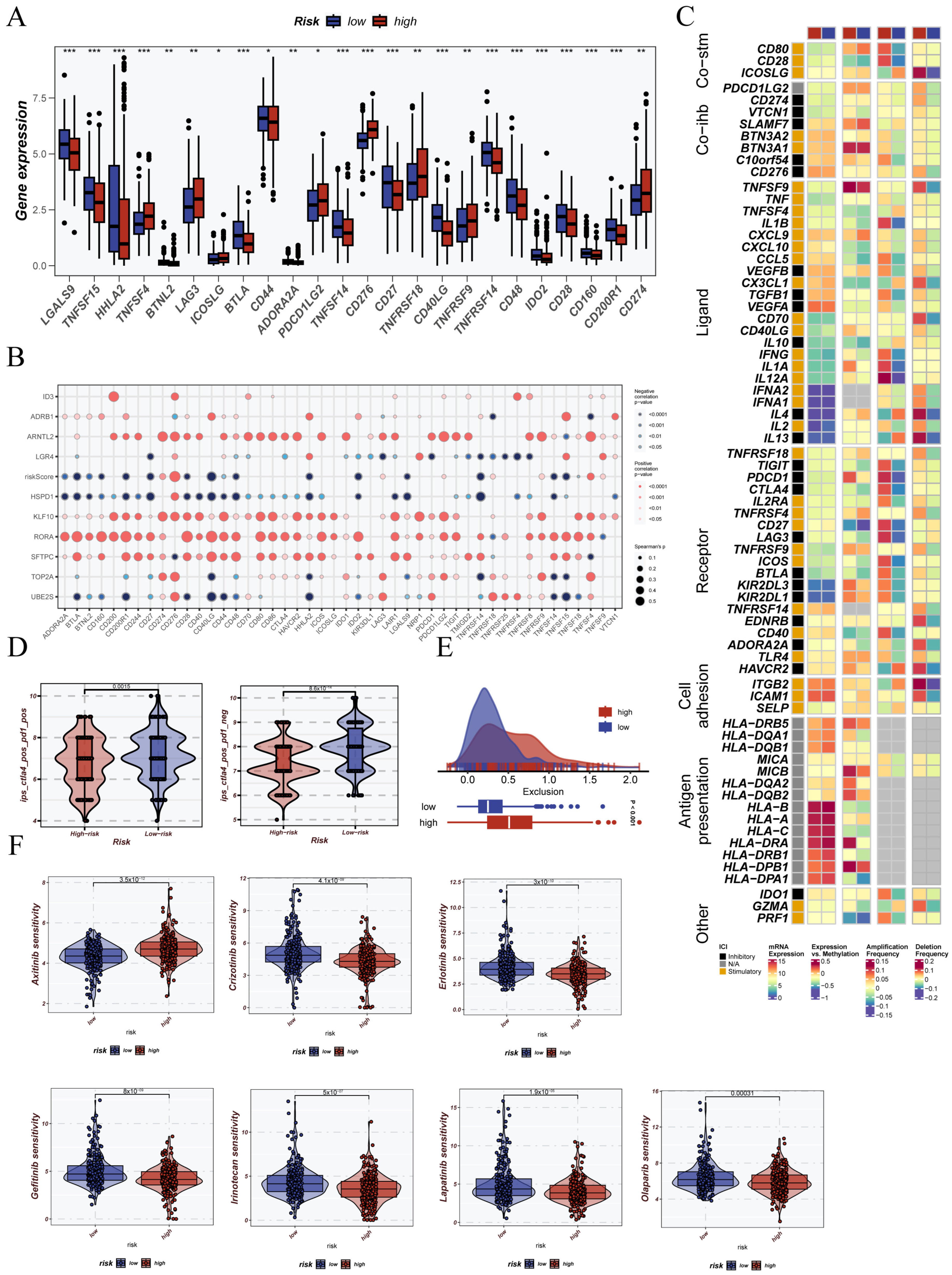
3.8. Immunological Status Assessment and Immunotherapy Prediction Based on CRG Stratification
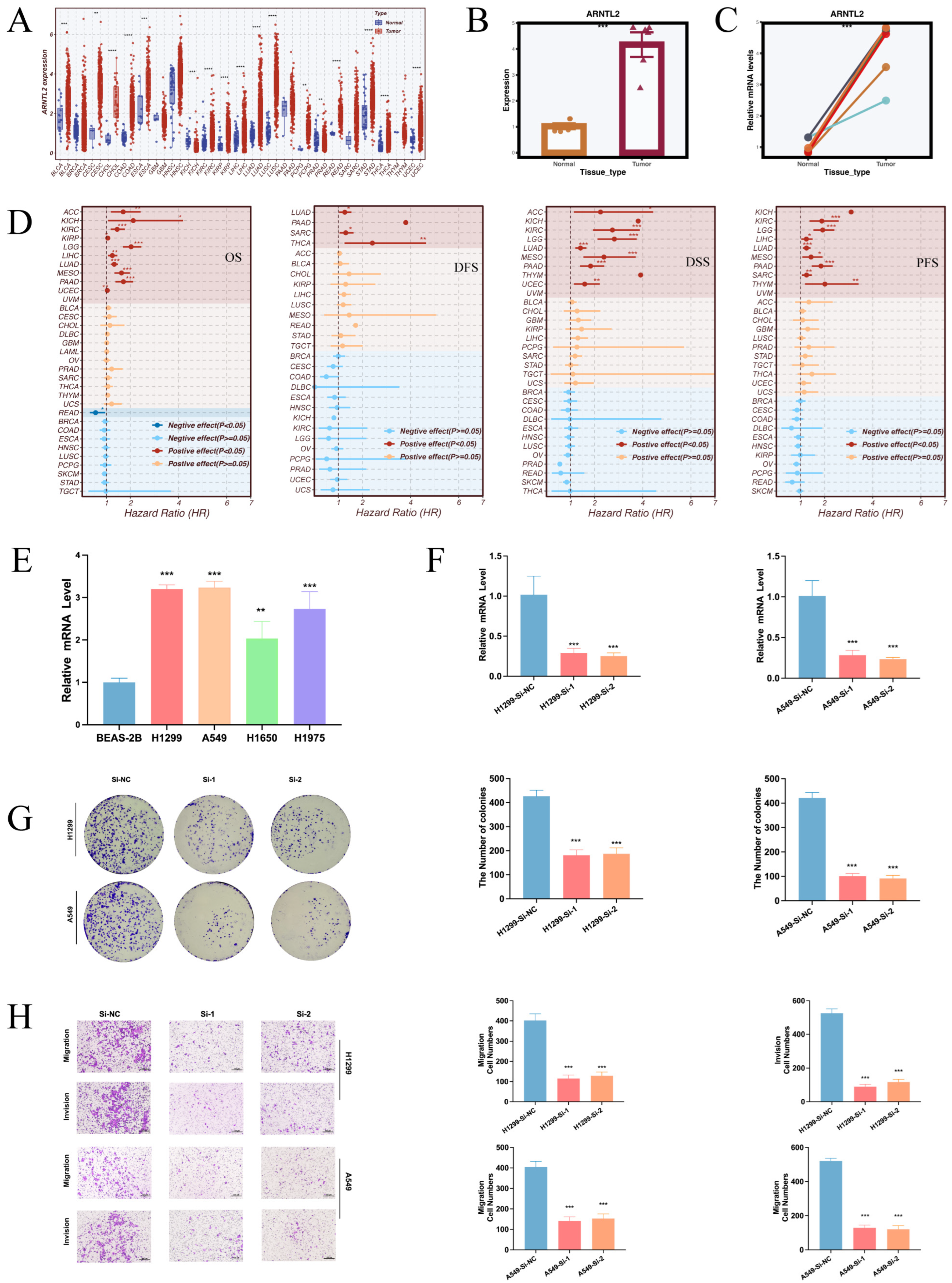
3.9. Pan-Cancer Prognostic Significance and Functional Impact of ARNTL2 in LUAD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LUAD | lung adenocarcinoma |
| GSVA | gene set variation analysis |
| TCGA | The Cancer Genome Atlas |
| qRT-PCR | quantitative real-time PCR |
| TME | tumor microenvironment |
| IPS | immunophenoscore |
| TCIA | The Cancer Immunome Atlas |
| TIDE | tumor immune dysfunction and exclusion |
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [CrossRef]
- de Groot, P.M.; Wu, C.C.; Carter, B.W.; Munden, R.F. The epidemiology of lung cancer. Transl. Lung Cancer Res. 2018, 7, 220–233. [Google Scholar] [CrossRef]
- Han, G.; Sinjab, A.; Rahal, Z.; Lynch, A.M.; Treekitkarnmongkol, W.; Liu, Y.; Serrano, A.G.; Feng, J.; Liang, K.; Khan, K.; et al. An atlas of epithelial cell states and plasticity in lung adenocarcinoma. Nature 2024, 627, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Gao, J.; Wang, J.; Fan, F.; Cheng, C.; Qian, D.; Guo, R.; Zhang, Y.; Ye, T.; Augustine, M.; et al. Genomic and immune heterogeneity of multiple synchronous lung adenocarcinoma at different developmental stages. Nat. Commun. 2024, 15, 7928. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.; Han, Y.; Dong, D.; Cao, Y.; Chen, X.; Liu, D.; Cheng, X.; Sun, D.; Li, H.; et al. Immune microenvironment heterogeneity of concurrent adenocarcinoma and squamous cell carcinoma in multiple primary lung cancers. NPJ Precis. Oncol. 2024, 8, 55. [Google Scholar] [CrossRef]
- Lee, Y. Roles of circadian clocks in cancer pathogenesis and treatment. Exp. Mol. Med. 2021, 53, 1529–1538. [Google Scholar] [CrossRef]
- Qu, M. Molecular crosstalk between circadian clock and cancer and therapeutic implications. Front. Nutr. 2023, 10, 1143001. [Google Scholar] [CrossRef]
- Vainshelbaum, N.M.; Salmina, K.; Gerashchenko, B.I.; Lazovska, M.; Zayakin, P.; Cragg, M.S.; Pjanova, D.; Erenpreisa, J. Role of the Circadian Clock “Death-Loop” in the DNA Damage Response Underpinning Cancer Treatment Resistance. Cells 2022, 11, 880. [Google Scholar] [CrossRef]
- Papagiannakopoulos, T.; Bauer, M.R.; Davidson, S.M.; Heimann, M.; Subbaraj, L.; Bhutkar, A.; Bartlebaugh, J.; Vander Heiden, M.G.; Jacks, T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016, 24, 324–331. [Google Scholar] [CrossRef]
- Wang, Z.H.; Zhang, P.; Du, Y.H.; Wei, X.S.; Ye, L.L.; Niu, Y.R.; Xiang, X.; Peng, W.B.; Su, Y.; Zhou, Q. High-risk early-stage lung adenocarcinoma patients are identified by an immune-related circadian clock gene signature. J. Thorac. Dis. 2022, 14, 3748–3761. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Yuan, X.; Eltzschig, H.K. Circadian rhythm as a therapeutic target. Nat. Rev. Drug Discov. 2021, 20, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Fan, J.; He, Y.; Xiong, A.; Yu, J.; Li, Y.; Zhang, Y.; Zhao, W.; Zhou, F.; Li, W.; et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 2021, 12, 2540. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Carbonetto, P.; Willwerscheid, J.; Oakes, S.A.; Macleod, K.F.; Stephens, M. Dissecting tumor transcriptional heterogeneity from single-cell RNA-seq data by generalized binary covariance decomposition. Nat. Genet. 2025, 57, 263–273. [Google Scholar] [CrossRef]
- Ho, D.W.; Tsui, Y.M.; Chan, L.K.; Sze, K.M.; Zhang, X.; Cheu, J.W.; Chiu, Y.T.; Lee, J.M.; Chan, A.C.; Cheung, E.T.; et al. Single-cell RNA sequencing shows the immunosuppressive landscape and tumor heterogeneity of HBV-associated hepatocellular carcinoma. Nat. Commun. 2021, 12, 3684. [Google Scholar] [CrossRef]
- Tian, Y.; Li, Q.; Yang, Z.; Zhang, S.; Xu, J.; Wang, Z.; Bai, H.; Duan, J.; Zheng, B.; Li, W.; et al. Single-cell transcriptomic profiling reveals the tumor heterogeneity of small-cell lung cancer. Signal Transduct. Target. Ther. 2022, 7, 346. [Google Scholar] [CrossRef]
- Bischoff, P.; Trinks, A.; Obermayer, B.; Pett, J.P.; Wiederspahn, J.; Uhlitz, F.; Liang, X.; Lehmann, A.; Jurmeister, P.; Elsner, A.; et al. Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene 2021, 40, 6748–6758. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Zhang, Y.; Parmigiani, G.; Johnson, W.E. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020, 2, lqaa078. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, L.; Weng, S.; Guo, C.; Dang, Q.; Xu, H.; Wang, L.; Lu, T.; Zhang, Y.; Sun, Z.; et al. Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nat. Commun. 2022, 13, 816. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef]
- Petitprez, F.; Levy, S.; Sun, C.M.; Meylan, M.; Linhard, C.; Becht, E.; Elarouci, N.; Tavel, D.; Roumenina, L.T.; Ayadi, M.; et al. The murine Microenvironment Cell Population counter method to estimate abundance of tissue-infiltrating immune and stromal cell populations in murine samples using gene expression. Genome Med. 2020, 12, 86. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Racle, J.; de Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife 2017, 6, e26476. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Shao, X.; Liao, J.; Li, C.; Lu, X.; Cheng, J.; Fan, X. CellTalkDB: A manually curated database of ligand-receptor interactions in humans and mice. Brief. Bioinform. 2021, 22, bbaa269. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Munteanu, C.; Turti, S.; Achim, L.; Muresan, R.; Souca, M.; Prifti, E.; Mârza, S.M.; Papuc, I. The Relationship between Circadian Rhythm and Cancer Disease. Int. J. Mol. Sci. 2024, 25, 5846. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, Q.; Zhang, H.; Wang, K.; Tan, L.; Li, X.; Sun, D. Application of Single-Cell Sequencing and Machine Learning in Prognosis and Immune Profiling of Lung Adenocarcinoma: Exploring Disease Mechanisms and Treatment Strategies Based on Circadian Rhythm Gene Signatures. Cancers 2025, 17, 2911. https://doi.org/10.3390/cancers17172911
Mu Q, Zhang H, Wang K, Tan L, Li X, Sun D. Application of Single-Cell Sequencing and Machine Learning in Prognosis and Immune Profiling of Lung Adenocarcinoma: Exploring Disease Mechanisms and Treatment Strategies Based on Circadian Rhythm Gene Signatures. Cancers. 2025; 17(17):2911. https://doi.org/10.3390/cancers17172911
Chicago/Turabian StyleMu, Qiuqiao, Han Zhang, Kai Wang, Lin Tan, Xin Li, and Daqiang Sun. 2025. "Application of Single-Cell Sequencing and Machine Learning in Prognosis and Immune Profiling of Lung Adenocarcinoma: Exploring Disease Mechanisms and Treatment Strategies Based on Circadian Rhythm Gene Signatures" Cancers 17, no. 17: 2911. https://doi.org/10.3390/cancers17172911
APA StyleMu, Q., Zhang, H., Wang, K., Tan, L., Li, X., & Sun, D. (2025). Application of Single-Cell Sequencing and Machine Learning in Prognosis and Immune Profiling of Lung Adenocarcinoma: Exploring Disease Mechanisms and Treatment Strategies Based on Circadian Rhythm Gene Signatures. Cancers, 17(17), 2911. https://doi.org/10.3390/cancers17172911





