Metabolic Signature in Combination with Fecal Immunochemical Test as a Non-Invasive Tool for Advanced Colorectal Neoplasia Diagnosis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Clinical Samples and Study Population
2.3. Metabolite Extraction
2.4. UHPLC-MS Metabolic Profiling
2.5. Data Pre-Processing
2.6. Data Normalization and Quality Control
2.7. Statistical Analysis
3. Results
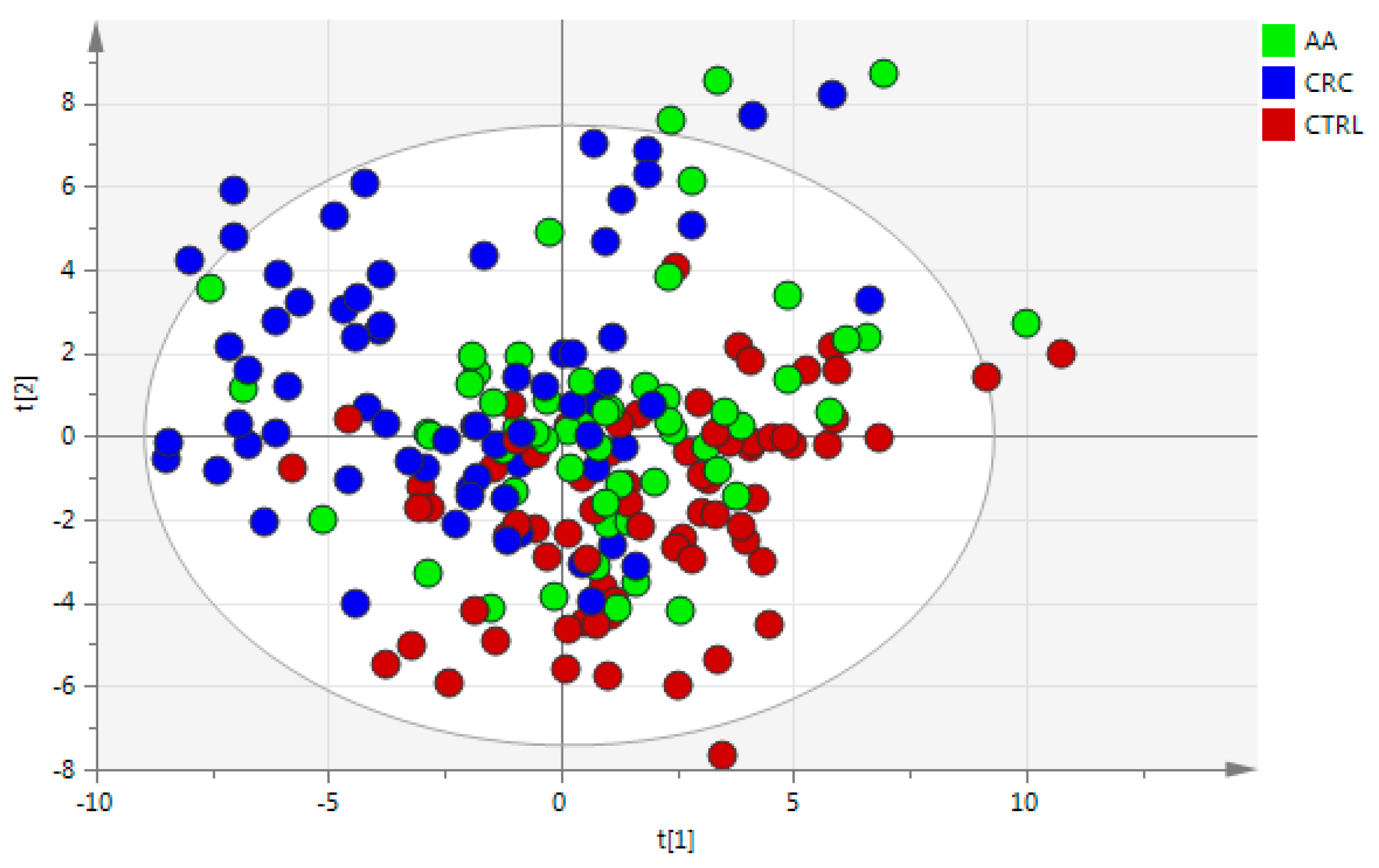
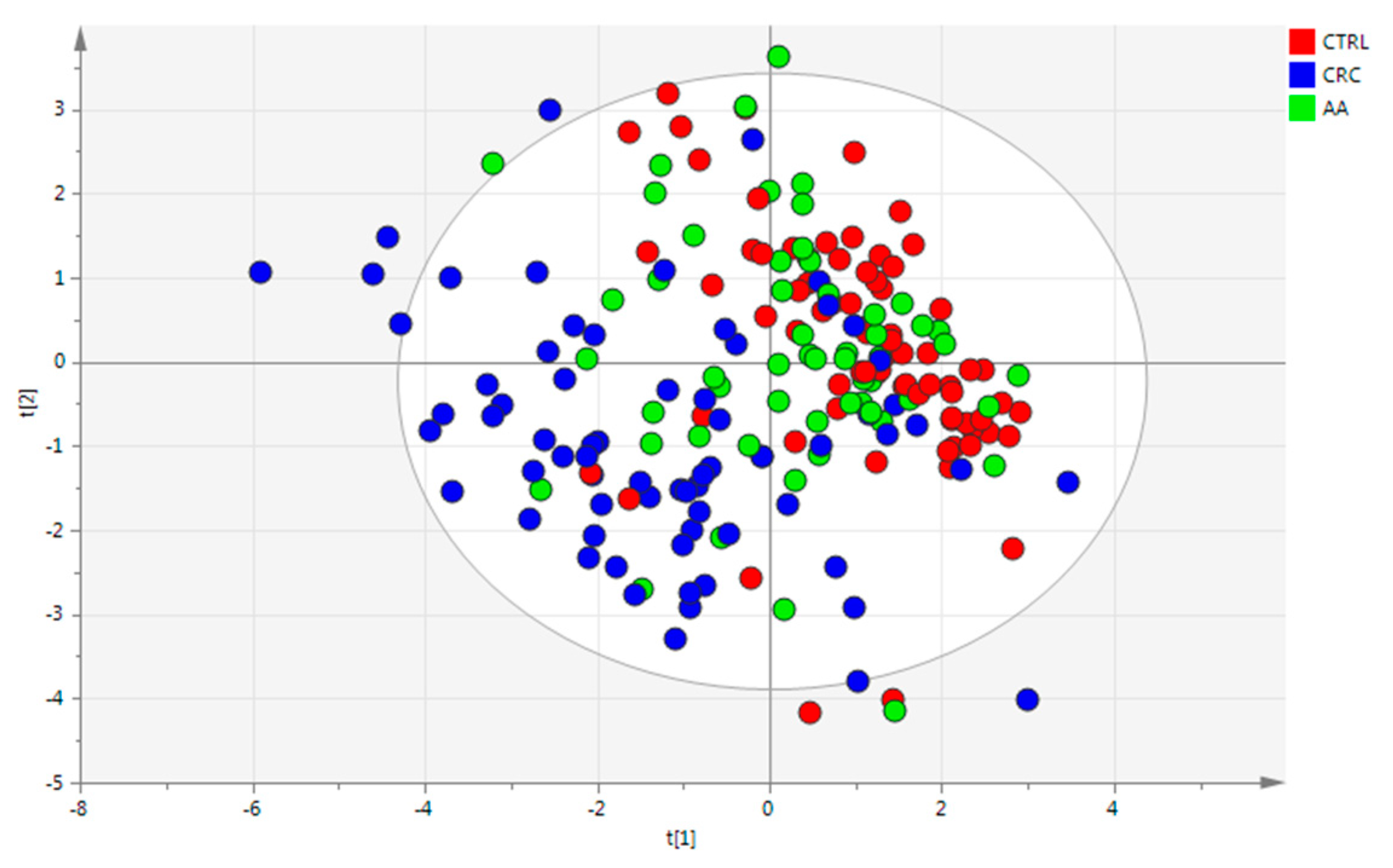
3.1. Data
3.2. Cohort Description
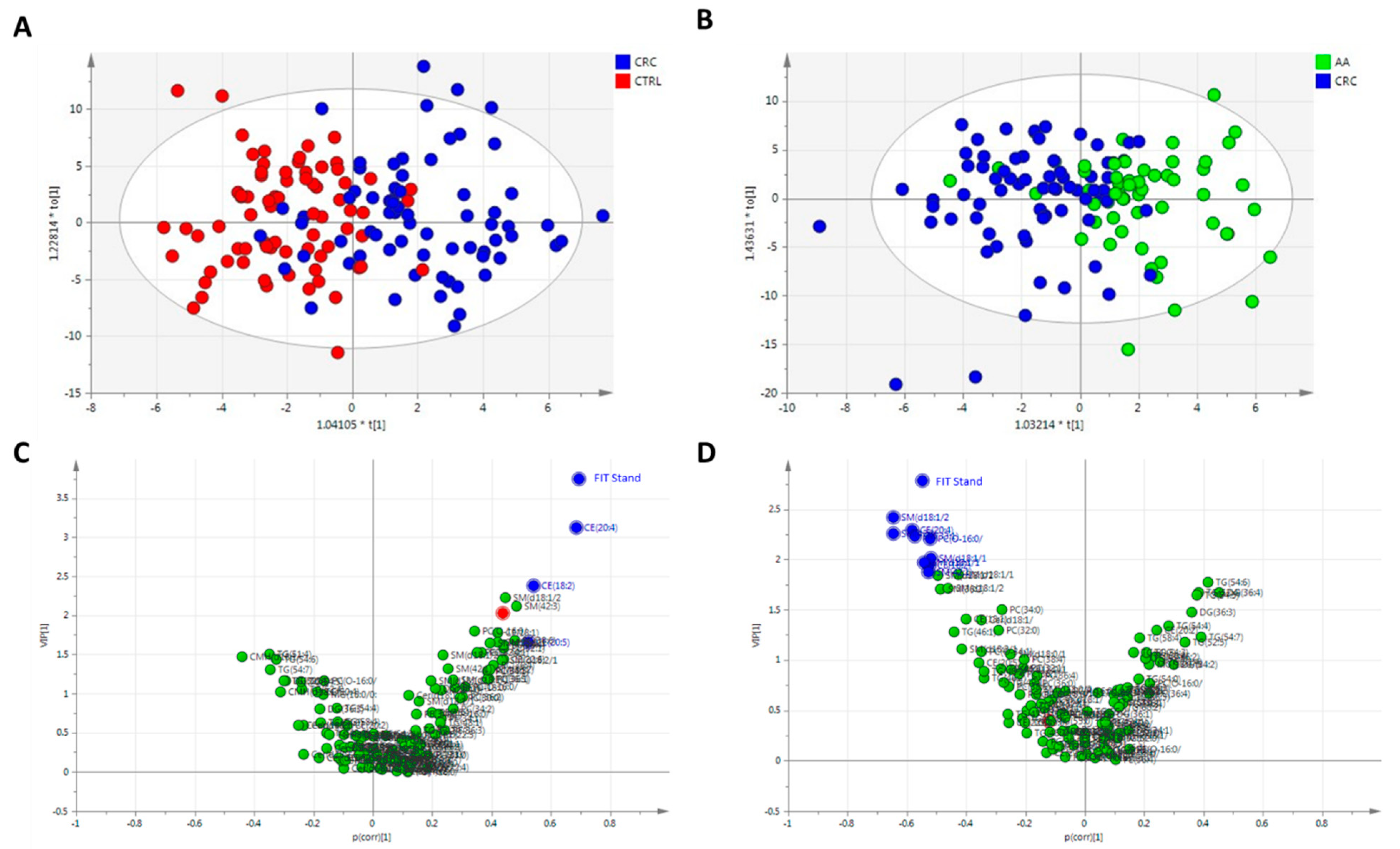
3.3. Statistical Analysis
3.4. Cholesteryl Esters as Targets in CRC Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- World Health Organisation. Colorectal Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/colorectal-cancer (accessed on 11 July 2023).
- Menon, G.; Recio-Boiles, A.; Lotfollahzadeh, S.; Cagir, B. Colon Cancer. In StatPearls; StatPearls Publishing: Tampa, FL, USA, 2025. [Google Scholar]
- Ballinger, A.B.; Anggiansah, C. Colorectal Cancer. BMJ 2007, 335, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Tao, S. Superior Diagnostic Performance of Faecal Immunochemical Tests for Haemoglobin in a Head-to-Head Comparison with Guaiac Based Faecal Occult Blood Test among 2235 Participants of Screening Colonoscopy. Eur. J. Cancer 2013, 49, 3049–3054. [Google Scholar] [CrossRef]
- Hoseini, S.H.; Enayati, P.; Nazari, M.; Babakhanzadeh, E.; Rastgoo, M.; Sohrabi, N.B. Biomarker Profile of Colorectal Cancer: Current Findings and Future Perspective. J. Gastrointest. Cancer 2024, 55, 497–510. [Google Scholar] [CrossRef]
- Kim, B.S.M. Diagnosis of Gastrointestinal Bleeding: A Practical Guide for Clinicians. World J. Gastrointest. Pathophysiol. 2014, 5, 467. [Google Scholar] [CrossRef] [PubMed]
- Avram, L.; Crișan, D.; Moldovan, R.-C.; Bogos, L.-G.; Iuga, C.-A.; Andraș, D.; Crișan, S.; Bodolea, C.; Nemeş, A.; Donca, V. Metabolomic Exploration of Colorectal Cancer Through Amino Acids and Acylcarnitines Profiling of Serum Samples. Cancers 2025, 17, 427. [Google Scholar] [CrossRef] [PubMed]
- Gold, A.; Choueiry, F.; Jin, N.; Mo, X.; Zhu, J. The Application of Metabolomics in Recent Colorectal Cancer Studies: A State-of-the-Art Review. Cancers 2022, 14, 725. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, Y.; Zhao, W.; Deng, K.; Wang, Z.; Yang, C.; Ma, L.; Openkova, M.S.; Hou, Y.; Li, K. Metabolomics for Biomarker Discovery in the Diagnosis, Prognosis, Survival and Recurrence of Colorectal Cancer: A Systematic Review. Oncotarget 2017, 8, 35460–35472. [Google Scholar] [CrossRef]
- Cubiella, J.; Clos-Garcia, M.; Alonso, C.; Martinez-Arranz, I.; Perez-Cormenzana, M.; Barrenetxea, Z.; Berganza, J.; Rodríguez-Llopis, I.; D’Amato, M.; Bujanda, L.; et al. Targeted UPLC-MS Metabolic Analysis of Human Faeces Reveals Novel Low-Invasive Candidate Markers for Colorectal Cancer. Cancers 2018, 10, 300. [Google Scholar] [CrossRef]
- Albóniga, O.E.; Cubiella, J.; Bujanda, L.; Blanco, M.E.; Lanza, B.; Alonso, C.; Nafría, B.; Falcón-Pérez, J.M. A Novel Approach on the Use of Samples from Faecal Occult Blood Screening Kits for Metabolomics Analysis: Application in Colorectal Cancer Population. Metabolites 2023, 13, 321. [Google Scholar] [CrossRef]
- Ni, Y.; Xie, G.; Jia, W. Metabonomics of Human Colorectal Cancer: New Approaches for Early Diagnosis and Biomarker Discovery. J. Proteome Res. 2014, 13, 3857–3870. [Google Scholar] [CrossRef]
- Duportet, X.; Aggio, R.B.M.; Carneiro, S.; Villas-Bôas, S.G. The Biological Interpretation of Metabolomic Data Can Be Misled by the Extraction Method Used. Metabolomics 2012, 8, 410–421. [Google Scholar] [CrossRef]
- Baker, M. Metabolomics: From Small Molecules to Big Ideas. Nat. Methods 2011, 8, 117–121. [Google Scholar] [CrossRef]
- Barr, J.; Caballería, J.; Martínez-Arranz, I.; Domínguez-Díez, A.; Alonso, C.; Muntané, J.; Pérez-Cormenzana, M.; García-Monzón, C.; Mayo, R.; Martín-Duce, A.; et al. Obesity-Dependent Metabolic Signatures Associated with Nonalcoholic Fatty Liver Disease Progression. J. Proteome Res. 2012, 11, 2521–2532. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Arranz, I.; Mayo, R.; Pérez-Cormenzana, M.; Mincholé, I.; Salazar, L.; Alonso, C.; Mato, J.M. Enhancing Metabolomics Research through Data Mining. J. Proteomics 2015, 127, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on Univariate and Multivariate Analysis of Metabolomics Data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Godzien, J.; Ciborowski, M.; Angulo, S.; Barbas, C. From Numbers to a Biological Sense: How the Strategy Chosen for Metabolomics Data Treatment May Affect Final Results. A Practical Example Based on Urine Fingerprints Obtained by LC-MS. Electrophoresis 2013, 34, 2812–2826. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, Å.M.; Wheelock, C.E. Trials and Tribulations of ‘omics Data Analysis: Assessing Quality of SIMCA-Based Multivariate Models Using Examples from Pulmonary Medicine. Mol. Biosyst. 2013, 9, 2589. [Google Scholar] [CrossRef]
- Box, G.E.P.; Cox, D.R. An Analysis of Transformations. J. R. Stat. Soc. Ser. B Stat. Methodol. 1964, 26, 211–243. [Google Scholar] [CrossRef]
- Goldstein, J.L.; DeBose-Boyd, R.A.; Brown, M.S. Protein Sensors for Membrane Sterols. Cell 2006, 124, 35–46. [Google Scholar] [CrossRef]
- Jiang, W.; Jin, W.-L.; Xu, A.-M. Cholesterol Metabolism in Tumor Microenvironment: Cancer Hallmarks and Therapeutic Opportunities. Int. J. Biol. Sci. 2024, 20, 2044–2071. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gomez, C.R.; Espinoza, I.; Le, T.P.T.; Shenoy, V.; Zhou, X. Correlation of Cholesteryl Ester Metabolism to Pathogenesis, Progression and Disparities in Colorectal Cancer. Lipids Health Dis. 2022, 21, 22. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-Y.; Li, B.-L.; Chang, C.C.Y.; Urano, Y. Acyl-Coenzyme A:Cholesterol Acyltransferases. Am. J. Physiol.-Endocrinol. Metab. 2009, 297, E1–E9. [Google Scholar] [CrossRef]
- Kopecka, J.; Godel, M.; Riganti, C. Cholesterol Metabolism: At the Cross Road between Cancer Cells and Immune Environment. Int. J. Biochem. Cell Biol. 2020, 129, 105876. [Google Scholar] [CrossRef]
- Huang, B.; Song, B.; Xu, C. Cholesterol Metabolism in Cancer: Mechanisms and Therapeutic Opportunities. Nat. Metab. 2020, 2, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Guillaumond, F.; Bidaut, G.; Ouaissi, M.; Servais, S.; Gouirand, V.; Olivares, O.; Lac, S.; Borge, L.; Roques, J.; Gayet, O.; et al. Cholesterol Uptake Disruption, in Association with Chemotherapy, Is a Promising Combined Metabolic Therapy for Pancreatic Adenocarcinoma. Proc. Natl. Acad. Sci. USA 2015, 112, 2473–2478. [Google Scholar] [CrossRef]
- Chen, X.; Liang, H.; Song, Q.; Xu, X.; Cao, D. Insulin Promotes Progression of Colon Cancer by Upregulation of ACAT1. Lipids Health Dis. 2018, 17, 122. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; López-Vilaró, L.; Nasarre, L.; Perez-Olabarria, M.; Vázquez, T.; Escuin, D.; Badimon, L.; Barnadas, A.; Lerma, E.; Llorente-Cortés, V. Intratumor Cholesteryl Ester Accumulation Is Associated with Human Breast Cancer Proliferation and Aggressive Potential: A Molecular and Clinicopathological Study. BMC Cancer 2015, 15, 460. [Google Scholar] [CrossRef]
- Yue, S.; Li, J.; Lee, S.-Y.; Lee, H.J.; Shao, T.; Song, B.; Cheng, L.; Masterson, T.A.; Liu, X.; Ratliff, T.L.; et al. Cholesteryl Ester Accumulation Induced by PTEN Loss and PI3K/AKT Activation Underlies Human Prostate Cancer Aggressiveness. Cell Metab. 2014, 19, 393–406. [Google Scholar] [CrossRef]
- Sun, T.; Xiao, X. Targeting ACAT1 in Cancer: From Threat to Treatment. Front. Oncol. 2024, 14, 1395192. [Google Scholar] [CrossRef]
- Ye, K.; Wu, Y.; Sun, Y.; Lin, J.; Xu, J. TLR4 SiRNA Inhibits Proliferation and Invasion in Colorectal Cancer Cells by Downregulating ACAT1 Expression. Life Sci. 2016, 155, 133–139. [Google Scholar] [CrossRef]
- Kuldeep, S.; Soni, S.; Srivastava, A.; Mishra, A.; Sharma, L.K.; Mandal, C.C. Dysregulated Cholesterol Regulatory Genes as a Diagnostic Biomarker for Cancer. J. Gene Med. 2023, 25, e3475. [Google Scholar] [CrossRef]
- Guan, C.; Niu, Y.; Chen, S.-C.; Kang, Y.; Wu, J.-X.; Nishi, K.; Chang, C.C.Y.; Chang, T.-Y.; Luo, T.; Chen, L. Structural Insights into the Inhibition Mechanism of Human Sterol O-Acyltransferase 1 by a Competitive Inhibitor. Nat. Commun. 2020, 11, 2478. [Google Scholar] [CrossRef]
- Wu, C.; Wang, M.; Shi, H. Cholesterol Promotes Colorectal Cancer Growth by Activating the PI3K/AKT Pathway. J. Oncol. 2022, 2022, 1515416. [Google Scholar] [CrossRef]
- Vara, J.Á.F.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. PI3K/Akt Signalling Pathway and Cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef]
- Li, J.; Gu, D.; Lee, S.S.-Y.; Song, B.; Bandyopadhyay, S.; Chen, S.; Konieczny, S.F.; Ratliff, T.L.; Liu, X.; Xie, J.; et al. Abrogating Cholesterol Esterification Suppresses Growth and Metastasis of Pancreatic Cancer. Oncogene 2016, 35, 6378–6388. [Google Scholar] [CrossRef]
- Tu, T.; Zhang, H.; Xu, H. Targeting Sterol-O-Acyltransferase 1 to Disrupt Cholesterol Metabolism for Cancer Therapy. Front. Oncol. 2023, 13, 1197502. [Google Scholar] [CrossRef]
- Hughes-Fulford, M.; Li, C.-F.; Boonyaratanakornkit, J.; Sayyah, S. Arachidonic Acid Activates Phosphatidylinositol 3-Kinase Signaling and Induces Gene Expression in Prostate Cancer. Cancer Res. 2006, 66, 1427–1433. [Google Scholar] [CrossRef]
- Ghosh, J.; Myers, C.E. Arachidonic Acid Stimulates Prostate Cancer Cell Growth: Critical Role of 5-Lipoxygenase. Biochem. Biophys. Res. Commun. 1997, 235, 418–423. [Google Scholar] [CrossRef]
- Llaverías, G.; Laguna, J.C.; Alegret, M. Pharmacology of the AC AT Inhibitor Avasimibe (CI-1011). Cardiovasc. Drug Rev. 2003, 21, 33–50. [Google Scholar] [CrossRef]
- Bemlih, S.; Poirier, M.-D.; Andaloussi, A. El Acyl-Coenzyme A: Cholesterol Acyltransferase Inhibitor Avasimibe Affect Survival and Proliferation of Glioma Tumor Cell Lines. Cancer Biol. Ther. 2010, 9, 1025–1032. [Google Scholar] [CrossRef]
- Antalis, C.J.; Arnold, T.; Rasool, T.; Lee, B.; Buhman, K.K.; Siddiqui, R.A. High ACAT1 Expression in Estrogen Receptor Negative Basal-like Breast Cancer Cells Is Associated with LDL-Induced Proliferation. Breast Cancer Res. Treat. 2010, 122, 661–670. [Google Scholar] [CrossRef]
- Hornbuckle, W.E.; Simpson, K.W.; Tennant, B.C. Gastrointestinal Function. In Clinical Biochemistry of Domestic Animals; Elsevier: Amsterdam, The Netherlands, 2008; pp. 413–457. [Google Scholar]
- Mercadante, S. Nutrition in Cancer Patients. Support. Care Cancer 1996, 4, 10–20. [Google Scholar] [CrossRef]
- Arends, J. Malnutrition in Cancer Patients: Causes, Consequences and Treatment Options. Eur. J. Surg. Oncol. 2024, 50, 107074. [Google Scholar] [CrossRef]
- Record, M.; Poirot, M.; Silvente-Poirot, S. Emerging Concepts on the Role of Exosomes in Lipid Metabolic Diseases. Biochimie 2014, 96, 67–74. [Google Scholar] [CrossRef]
- Carayon, K.; Chaoui, K.; Ronzier, E.; Lazar, I.; Bertrand-Michel, J.; Roques, V.; Balor, S.; Terce, F.; Lopez, A.; Salomé, L.; et al. Proteolipidic Composition of Exosomes Changes during Reticulocyte Maturation. J. Biol. Chem. 2011, 286, 34426–34439. [Google Scholar] [CrossRef]
- Paillasse, M.R.; de Medina, P.; Amouroux, G.; Mhamdi, L.; Poirot, M.; Silvente-Poirot, S. Signaling through Cholesterol Esterification: A New Pathway for the Cholecystokinin 2 Receptor Involved in Cell Growth and Invasion. J. Lipid. Res. 2009, 50, 2203–2211. [Google Scholar] [CrossRef]
- Bowden, K.L.; Bilbey, N.J.; Bilawchuk, L.M.; Boadu, E.; Sidhu, R.; Ory, D.S.; Du, H.; Chan, T.; Francis, G.A. Lysosomal Acid Lipase Deficiency Impairs Regulation of ABCA1 Gene and Formation of High Density Lipoproteins in Cholesteryl Ester Storage Disease. J. Biol. Chem. 2011, 286, 30624–30635. [Google Scholar] [CrossRef]
- Azparren-Angulo, M.; Mleczko, J.; Alboniga, O.E.; Kruglik, S.; Guigner, J.; Gonzalez, E.; Garcia-Vallicrosa, C.; Llop, J.; Simó, C.; Alonso, C.; et al. Lipidomics and Biodistribution of Extracellular Vesicles-secreted by Hepatocytes from Zucker Lean and Fatty Rats. J. Extracell. Biol. 2024, 3, e140. [Google Scholar] [CrossRef]
- Yang, J.S.; Lee, J.C.; Byeon, S.K.; Rha, K.H.; Moon, M.H. Size Dependent Lipidomic Analysis of Urinary Exosomes from Patients with Prostate Cancer by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. Anal. Chem. 2017, 89, 2488–2496. [Google Scholar] [CrossRef]
- Glover, S.C.; Nouri, M.; Tuna, K.M.; Mendoza Alvarez, L.B.; Ryan, L.K.; Shirley, J.F.; Tang, Y.; Denslow, N.D.; Alli, A.A. Lipidomic Analysis of Urinary Exosomes from Hereditary A-tryptasemia Patients and Healthy Volunteers. FASEB Bioadv. 2019, 1, 624–638. [Google Scholar] [CrossRef] [PubMed]
- Clos-Garcia, M.; Loizaga-Iriarte, A.; Zuñiga-Garcia, P.; Sánchez-Mosquera, P.; Rosa Cortazar, A.; González, E.; Torrano, V.; Alonso, C.; Pérez-Cormenzana, M.; Ugalde-Olano, A.; et al. Metabolic Alterations in Urine Extracellular Vesicles Are Associated to Prostate Cancer Pathogenesis and Progression. J. Extracell. Vesicles 2018, 7, 1470442. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S.; et al. Metabolomics Workbench: An International Repository for Metabolomics Data and Metadata, Metabolite Standards, Protocols, Tutorials and Training, and Analysis Tools. Nucleic Acids. Res. 2016, 44, D463–D470. [Google Scholar] [CrossRef] [PubMed]
| Diagnosis | ANOVA p-Value | |||
|---|---|---|---|---|
| Control (CTRL) | Adenoma (AA) | Colorectal Cancer (CRC) | ||
| N | 78 | 58 | 75 | Not applicable |
| Gender (% women) | 50 | 34 | 29 | 0.024 |
| Age (years; mean ± sd) | 68.40 ± 14.85 | 76.69 ± 9.95 | 77.81 ± 10.74 | <0.001 |
| FIT (+) | 21 | 38 | 65 | <0.001 |
| FIT (−) | 57 | 20 | 10 | |
| Variable | p (corr) | VIP a | |
|---|---|---|---|
| CRC vs. CTRL | Standardized fecal Hb | 0.6948 | 3.7443 |
| CE (20:4) | 0.6846 | 3.1189 | |
| CE (18:2) | 0.5408 | 2.3798 | |
| CE (20:5) | 0.5250 | 1.6608 | |
| CRC vs. AA | Standardized fecal Hb | −0.5484 | 2.7858 |
| SM (d18:1/24:1) + SM (d18:2/24:0) b | −0.6451 | 2.4180 | |
| CE (20:4) | −0.5825 | 2.2940 | |
| SM (d18:2/24:1) + SM (d18:1/24:2) b | −0.6459 | 2.2555 | |
| SM (d17:1/16:0) + SM (d18:1/15:0) b | −0.5728 | 2.2294 | |
| PC (O-16:0/16:0) | −0.5203 | 2.2089 | |
| SM (d18:1/16:0) | −0.5186 | 2.0138 | |
| SM (d18:1/17:0) | −0.5421 | 1.9714 | |
| CE (18:2) | −0.5229 | 1.9402 | |
| SM (42:1) | −0.5280 | 1.8786 |
| Metabolite | Log2(Robust Fold-Change) | q-Value |
|---|---|---|
| CE (18:1) | 0.7931 | 5.70 × 10−3 |
| CE (18:2) | 1.8460 | 4.10 × 10−6 |
| CE (20:4) | 2.5633 | 7.70 × 10−10 |
| CE (20:5) | 1.2566 | 1.50 × 10−2 |
| CE (22:6) | 1.0574 | 4.70 × 10−3 |
| PC (16:0/16:0) | 0.6417 | 6.50 × 10−3 |
| PC (16:0/16:1) + PC (14:1/18:0) + PC (14:0/18:1) b | 0.8143 | 5.90 × 10−3 |
| PC (16:0/18:1) | 0.7270 | 1.10 × 10−2 |
| PC (18:0/18:1) | 0.7239 | 1.80 × 10−2 |
| PC (16:0/20:4) | 0.4835 | 1.50 × 10−2 |
| PC (18:0/20:4) | 0.4735 | 7.40 × 10−3 |
| PC (16:0/22:6) | 1.1738 | 4.40 × 10−3 |
| DG (32:1) | 0.4271 | 4.50 × 10−2 |
| PC (O-16:0/16:0) | 0.6236 | 8.20 × 10−3 |
| PC (O-16:0/18:1) + PC (18:1e/16:0) b | 0.8781 | 1.60 × 10−2 |
| PC (O-16:0/18:2) | 1.1600 | 7.40 × 10−3 |
| PC (O-18:0/18:2) | 0.5693 | 4.80 × 10−2 |
| SM (d17:1/16:0) + SM (d18:1/15:0) b | 0.9500 | 7.40 × 10−3 |
| SM (d18:1/16:0) | 0.9900 | 5.70 × 10−3 |
| SM (d18:2/16:0) | 0.7225 | 8.00 × 10−3 |
| SM (d18:1/23:0) | 1.1646 | 3.90 × 10−2 |
| SM (d18:1/24:1) + SM (d18:2/24:0) | 1.3371 | 1.20 × 10−3 |
| SM (d18:2/24:1) + SM (d18:1/24:2) b | 0.9671 | 1.40 × 10−2 |
| SM (42:1) | 0.8341 | 3.50 × 10−2 |
| TG (18:2_18:2_15:0) + TG (17:1_18:2_16:1) + TG (17:1_18:3_16:0) b | −1.0903 | 5.90 × 10−3 |
| TG (16:0_18:2_18:3) b | −1.3093 | 4.50 × 10−2 |
| TG (18:2_18:3_18:1) b | −1.8290 | 2.10 × 10−2 |
| TG (54:7) | −1.7835 | 4.50 × 10−2 |
| Metabolites | Log2(Robust Fold-Change) | q-Value |
|---|---|---|
| CE (18:1) | 0.6281 | 5.00 × 10−2 |
| CE (18:2) | 1.4507 | 1.70 × 10−3 |
| CE (20:2) | −0.7016 | 5.00 × 10−2 |
| CE (20:4) | 1.8026 | 1.90 × 10−3 |
| PC (16:0/16:0) | 0.5055 | 4.40 × 10−2 |
| PC (16:0/18:0) | 0.1995 | 2.00 × 10−2 |
| DG (18:1_18:2_0:0) b | −1.4547 | 2.60 × 10−2 |
| DG (18:2_18:2_0:0) b | −1.5312 | 1.50 × 10−2 |
| PC (O-16:0/16:0) | 0.9914 | 1.90 × 10−3 |
| PC (O-16:0/18:1) + PC (18:1e/16:0) b | 0.8829 | 4.30 × 10−2 |
| PC (O-16:0/18:2) | 0.9022 | 1.90 × 10−2 |
| SM (d17:1/16:0) + SM (d18:1/15:0) b | 0.9711 | 2.60 × 10−3 |
| SM (d18:1/16:0) | 1.2231 | 4.30 × 10−3 |
| SM (d18:1/17:0) | 1.0336 | 3.50 × 10−2 |
| SM (d18:1/18:0) | 1.6791 | 5.70 × 10−3 |
| SM (d18:1/18:1) + SM (d18:2/18:0) b | 1.1187 | 2.60 × 10−2 |
| SM (d18:1/22:0) | 1.3192 | 1.80 × 10−2 |
| SM (d18:1/23:0) | 1.3881 | 1.40 × 10−2 |
| SM (d18:1/24:1) + SM (d18:2/24:0) | 1.7776 | 1.70 × 10−3 |
| SM (d18:2/24:1) + SM (d18:1/24:2) b | 0.9900 | 2.60 × 10−2 |
| SM (42:1) | 1.1880 | 1.40 × 10−2 |
| TG (16:0_18:2_18:2) b | −1.4691 | 2.00 × 10−2 |
| TG (18:2_18:1_18:1) + TG (18:2_18:2_18:0) b | −1.4075 | 4.40 × 10−2 |
| TG (18:2_18:2_18:1) b | −1.6763 | 1.40 × 10−2 |
| TG (18:2_18:3_18:1) b | −1.8445 | 3.30 × 10−3 |
| TG (18:2_18:3_18:2) b | −1.1997 | 3.60 × 10−2 |
| 2-by-2 Comparison | Variable(s) | OOB Estimate Error Rate (%) | Accuracy | Precision | Recall | F1-Score | AUC |
|---|---|---|---|---|---|---|---|
| CRC vs. CTRL | FIT | 22.45 | 83.64 | 85.71 | 82.76 | 84.21 | 90 |
| FIT + CEs | 19.39 | 89.09 | 89.65 | 89.65 | 89.65 | 91 | |
| CRC vs. AA | FIT | 33.72 | 63.83 | 77.78 | 65.62 | 71.19 | 69 |
| FIT + CEs | 32.56 | 74.47 | 75.00 | 93.75 | 83.33 | 81 | |
| AA vs. CTRL | FIT | 39.08 | 69.39 | 69.05 | 93.55 | 79.45 | 76 |
| FIT + CEs | 45.98 | 65.31 | 64.58 | 100 | 78.48 | 70 | |
| CTRL vs. AA + CTRL | FIT | 32.33 | 82.05 | 78.12 | 78.12 | 78.12 | 84 |
| FIT + CEs | 27.07 | 74.36 | 63.04 | 90.62 | 74.36 | 84 |
| Samples Correctly Classified (YpredPS > 0.65) | Samples in the Borderline (0.35 > YpredPS < 0.65) c | Samples Not Classified (YpredPS < 0.35) | ||
|---|---|---|---|---|
| CRC vs. CTRL | CRC | 48 | 11 (4) | 10 |
| CTRL | 61 | 7 (1) | 6 | |
| CRC vs. AA | CRC | 47 | 14 (3) | 8 |
| AA | 25 | 24 (7) | 7 | |
| CTRL vs. AA | CTRL | 32 | 37 (13) | 5 |
| AA | 7 | 37 (5) | 12 |
| AA vs. CTRL | CRC vs. CTRL | CRC vs. AA | ||||
|---|---|---|---|---|---|---|
| Metabolites | log2 (Robust FC) | q-Value | log2 (Robust FC) | q-Value | log2 (Robust FC) | q-Value |
| Cholesterol and derivatives | −0.045 | 0.9201 | 0.012 | 0.6467 | 0.058 | 0.8154 |
| CE (18:1) | 0.165 | 0.8436 | 0.793 | 0.0057 | 0.628 | 0.0498 |
| CE (18:2) | 0.395 | 0.8436 | 1.846 | 4.10 × 10−6 | 1.451 | 0.0017 |
| CE (20:2) | 0.383 | 0.8436 | −0.319 | 0.1909 | −0.702 | 0.0498 |
| CE (20:4) | 0.761 | 0.1739 | 2.563 | 7.72 × 10−10 | 1.803 | 0.0019 |
| CE (20:5) | 0.809 | 0.8436 | 1.257 | 0.0154 | 0.448 | 0.3486 |
| CE (22:4) | 0.054 | 0.9201 | −0.052 | 0.9551 | −0.106 | 0.8732 |
| CE (22:5) | 0.212 | 0.8436 | −0.136 | 0.8465 | −0.348 | 0.6582 |
| CE (22:6) | 0.783 | 0.7589 | 1.057 | 0.0047 | 0.275 | 0.5992 |
| 2-by-2 Comparison | OOB Estimate Error Rate (%) | Accuracy | Precision | Recall | F1-Score | AUC |
|---|---|---|---|---|---|---|
| CRC vs. CTRL | 21.43 | 89.09 | 89.65 | 89.65 | 89.65 | 89 |
| CRC vs. AA | 29.07 | 68.08 | 69.77 | 93.75 | 80.00 | 79 |
| AA vs. CTRL | 36.78 | 71.43 | 71.80 | 90.32 | 80.00 | 70 |
| CTRL vs. AA + CRC | 31.58 | 71.79 | 62.50 | 78.12 | 69.44 | 80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albóniga, O.E.; Cubiella, J.; Bujanda, L.; Aspichueta, P.; Blanco, M.E.; Lanza, B.; Alonso, C.; Falcón-Pérez, J.M. Metabolic Signature in Combination with Fecal Immunochemical Test as a Non-Invasive Tool for Advanced Colorectal Neoplasia Diagnosis. Cancers 2025, 17, 2339. https://doi.org/10.3390/cancers17142339
Albóniga OE, Cubiella J, Bujanda L, Aspichueta P, Blanco ME, Lanza B, Alonso C, Falcón-Pérez JM. Metabolic Signature in Combination with Fecal Immunochemical Test as a Non-Invasive Tool for Advanced Colorectal Neoplasia Diagnosis. Cancers. 2025; 17(14):2339. https://doi.org/10.3390/cancers17142339
Chicago/Turabian StyleAlbóniga, Oihane E., Joaquín Cubiella, Luis Bujanda, Patricia Aspichueta, María Encarnación Blanco, Borja Lanza, Cristina Alonso, and Juan Manuel Falcón-Pérez. 2025. "Metabolic Signature in Combination with Fecal Immunochemical Test as a Non-Invasive Tool for Advanced Colorectal Neoplasia Diagnosis" Cancers 17, no. 14: 2339. https://doi.org/10.3390/cancers17142339
APA StyleAlbóniga, O. E., Cubiella, J., Bujanda, L., Aspichueta, P., Blanco, M. E., Lanza, B., Alonso, C., & Falcón-Pérez, J. M. (2025). Metabolic Signature in Combination with Fecal Immunochemical Test as a Non-Invasive Tool for Advanced Colorectal Neoplasia Diagnosis. Cancers, 17(14), 2339. https://doi.org/10.3390/cancers17142339








