Metastatic Melanoma Prognosis Prediction Using a TC Radiomic-Based Machine Learning Model: A Preliminary Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
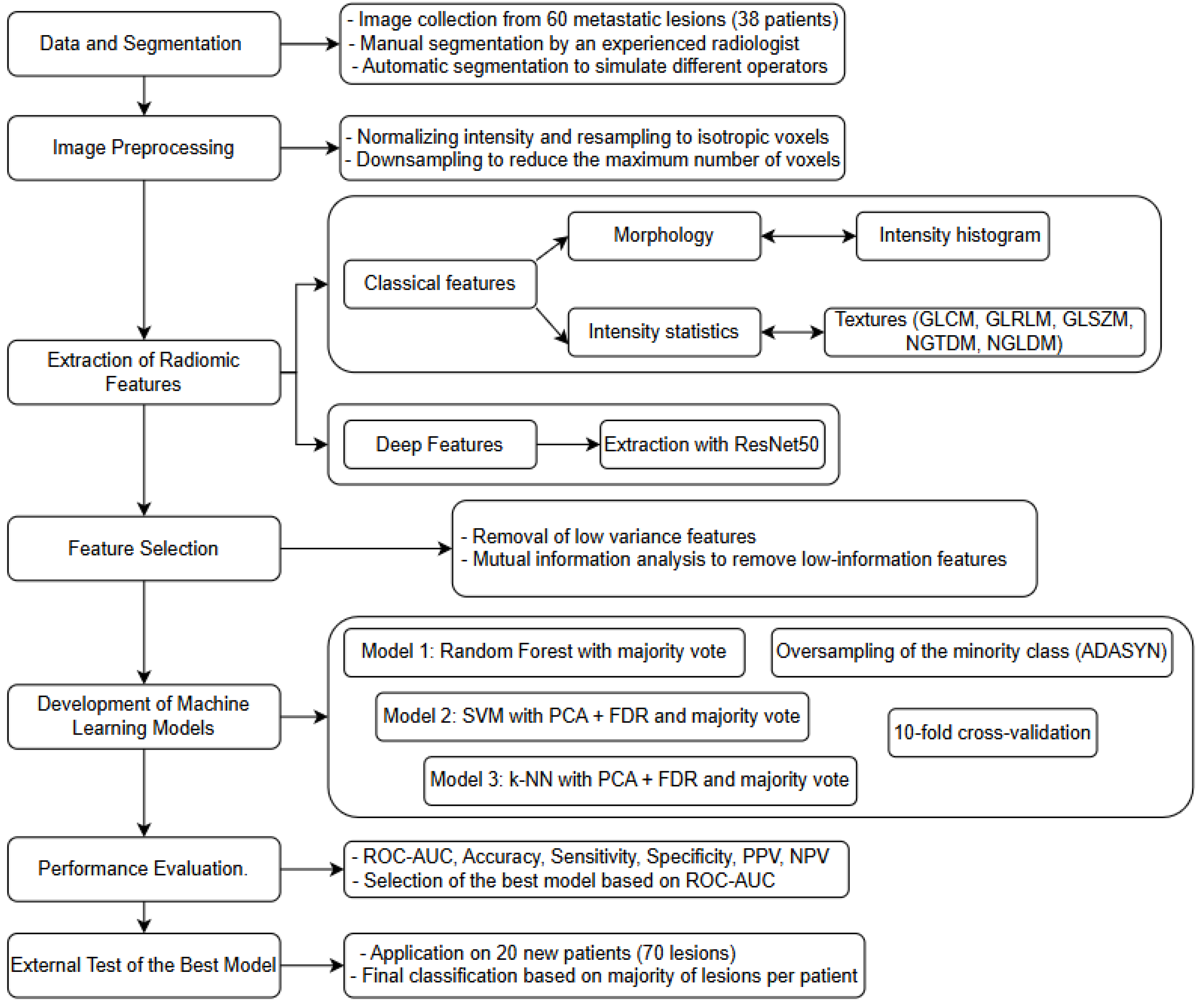
2.2. Radiomics Analysis and Model Construction
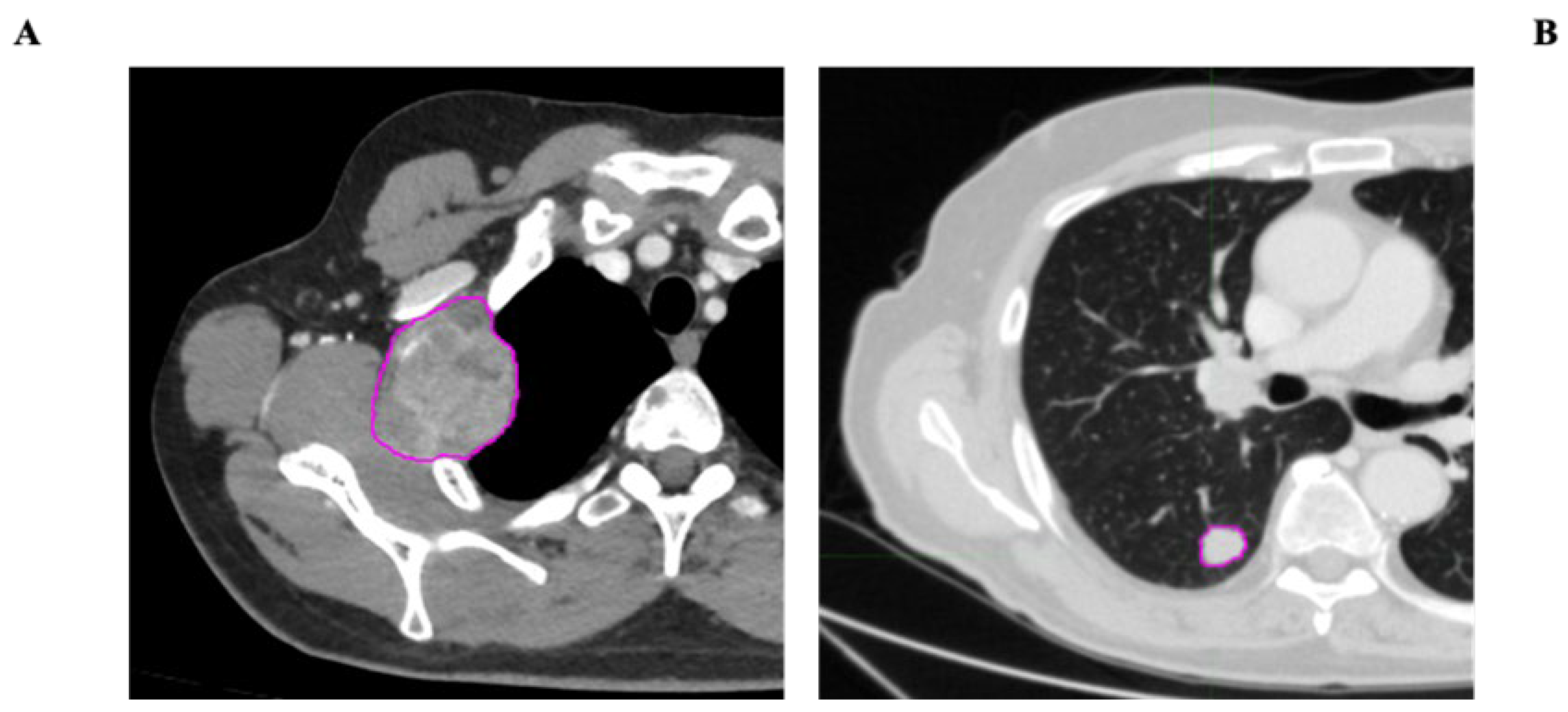
- VOI segmentation: Segmentation of the Volume of Interest (VOI) was performed manually on a slice-by-slice basis in consensus by two expert radiologists in CT imaging (15 and 10 years of experience) using the Trace4Research segmentation tool (named Manual segmentation by an experienced radiologist) (Figure 2).
- 2.
- Pre-processing of image intensities: To account for the potential variability introduced by the heterogeneous acquisition parameters, the intensity values within the segmented Volume of Interest (VOI) were pre-processed by resampling all CT series of each patient at an isotropic voxel spacing of 1 × 1 × 1 mm3, ensuring spatial consistency between datasets. Additionally, a voxel count limitation was applied to standardize computational load and reduce potential bias due to lesion volume differences: texture features were extracted from a maximum of 10 million voxels per VOI, while shape and first-order features were calculated on VOIs sampled up to a maximum of 1 million voxels. Voxel intensities within the segmented VOIs were quantized using fixed-width bins (64 bins for traditional radiomic features; 256 bins for deep features), enabling consistent intensity-based feature calculations in all cases. All preprocessing steps were carried out the Trace4Research platform, following a workflow aligned with Image Biomarker Standardization Initiative (IBSI) recommendations. This standard approach, commonly adopted for this type of data, helped to harmonize the feature space across cases, while also compensating for significant lesion size variations.
- 3.
- Radiomic feature extraction: Radiomic features were extracted from the segmented VOI across multiple feature families:
- Morphology.
- Intensity-based statistics.
- Intensity histogram (computed after discretizing the VOI intensities into 64 fixed bins).
- Texture features, including the following: Gray-Level Co-occurrence Matrix (GLCM), Gray-Level Run Length Matrix (GLRLM), Gray-Level Size Zone Matrix (GLSZM), Neighborhood Gray-Tone Difference Matrix (NGTDM), and Neighboring Gray-Level Dependence Matrix (NGLDM).
- Deep features: A set of 2048 deep features was extracted using the convolutional layers of a pre-trained ResNet50 model. The input images were resampled to dimensions of 224 × 224 × 16 voxels and discretized into 256 fixed intensity bins (these features are not included in the IBSI guidelines).
- 4.
- Feature selection: Low-variance features (variance < 0.1) were removed, followed by a mutual-information analysis to exclude features with low association with the class label (mutual information < 0.39). This selection pipeline ensured the inclusion of features with significant discriminant power while minimizing overfitting. All steps were performed within the Trace4Research platform and, where applicable, were aligned with the IBSI standard-compliant methodology.
- 5.
- Machine learning classification models: Three machine-learning classifiers were developed, validated, and tested for the binary classification task (FP vs. UP), using prognosis as the reference standard. A nested 10-fold cross-validation strategy was applied for all models:
- Model 1: Four ensembles of random forest classifiers, combined with the Gini index and a majority-vote rule.
- Model 2: Four ensembles of support vector machines (SVMs), combined with principal component analysis (PCA) and the Fisher Discriminant Ratio (FDR), using a majority-vote rule.
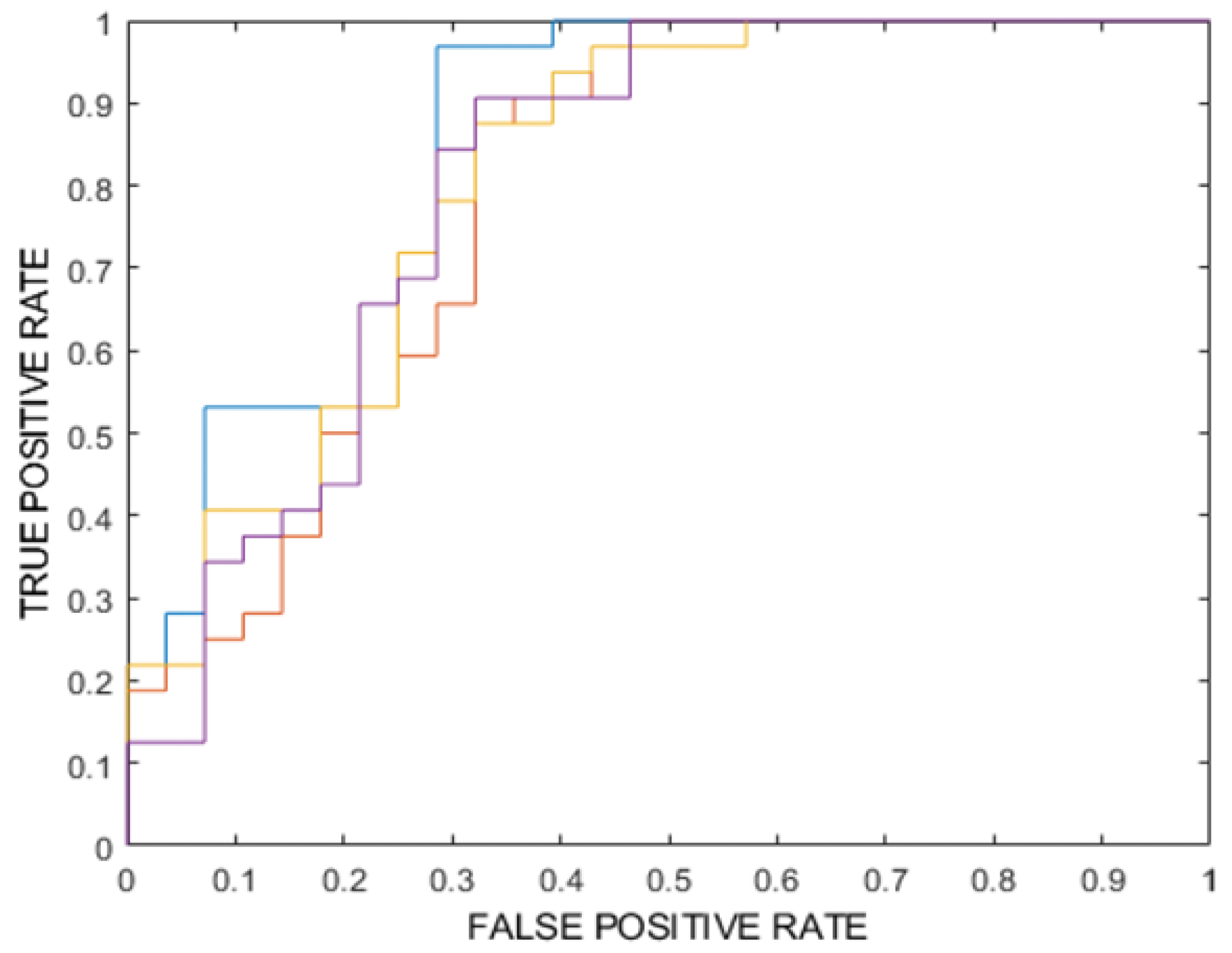
- Model 3: Four ensembles of k-nearest neighbor (k-NN) classifiers, also combined with PCA, FDR, and a majority-vote rule. To address potential bias due to class imbalance, the Adaptive Synthetic Sampling technique (ADASYN) was applied to oversample the minority (poor prognosis) class, ensuring a more balanced representation during model training. Model performance was evaluated using several metrics, including overall accuracy, area under the ROC curve (ROC-AUC), specificity, sensitivity, Negative Predictive Value (NPV) and Positive Predictive Value (PPV). For each of these metrics, 95% confidence intervals were calculated to provide a reliable estimate of their variability. Among all the models analyzed, the best performing binary classifier was selected based on the highest ROC-AUC value. The best-performing model, according to the ROC-AUC value of the internal testing, was then externally tested on the 20 MM patients of the external testing dataset (for a total of 70 lesions) using the most significant predictors. The classification of each patient’s prognosis was obtained as the one most frequently assigned by the classifier to the metastatic lesions of the same patient.
2.3. Statistical Analysis
3. Results
- -
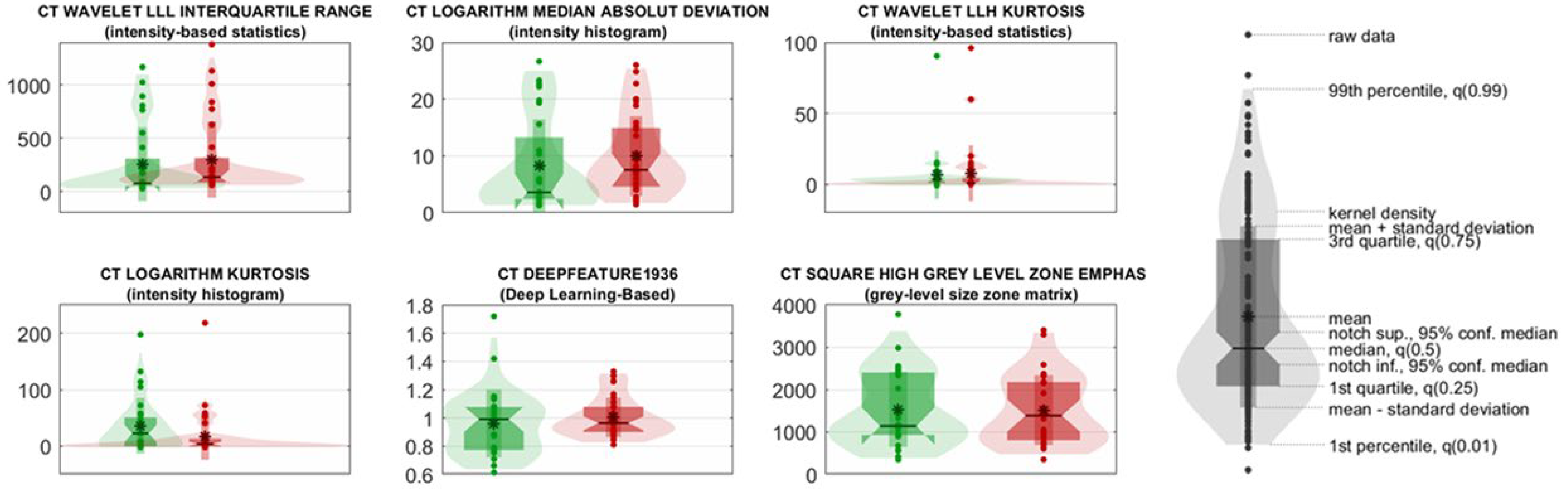
- CT wavelet LLL Interquartile Range and CT Logarithm Median Absolute Deviation reflect variability in lesion intensity, with higher values in UP patients indicating greater intratumoral heterogeneity;
- -
- CT Wavelet LLH Kurtosis and CT Logarithm Kurtosis measure the peaks and tails of the intensity distribution, with elevated kurtosis values suggesting more irregular and aggressive lesion profiles in UP cases;
- -
- CT DeepFeature1936, extracted from a ResNet50 convolutional neural network, captures abstract image patterns correlated with prognosis;
- -
- CT Square High-Gray-Level Zone Emphasis (GLSZM) quantifies the prominence of large, high-intensity zones within lesions, with higher values in UP patients suggesting the presence of denser, potentially more aggressive tumor regions.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADASYN | Adaptive Synthetic Sampling |
| AI | Artificial Intelligence |
| AUC | Area Under the Curve |
| CT | Computed Tomography |
| CI | Confidence Interval |
| FP | Favorable Prognosis |
| FDR | Fisher Discriminant Ratio |
| GLCM | Gray-Level Co-occurrence Matrix |
| GLRLM | Gray-Level Run Length Matrix |
| GLSZM | Gray-Level Size Zone Matrix |
| IBSI | Image Biomarker Standardisation Initiative |
| ICIs | Immune Checkpoint Inhibitors |
| LIME | Local Interpretable Model Diagnostic Explanation |
| k-NN | k-Nearest Neighbors |
| MM | Metastatic Melanoma |
| NGTDM | Neighborhood Gray Tone Difference Matrix |
| NGLDM | Neighboring Gray Level Dependence Matrix |
| NPV | Negative Predictive Value |
| OS | Overall Survival |
| PCA | Principal Component Analysis |
| PFS | Progression-Free Survival |
| PPV | Positive Predictive Value |
| ROC | Receiver Operating Characteristic |
| ROC-AUC | Receiver Operating Characteristic-Area Under the Curve |
| SHAP | SHapley Additive exPlanations |
| SVM | Support Vector Machine |
| UP | Unfavorable Prognosis |
| VOI | Volume of Interest |
References
- Grzywa, T.M.; Paskal, W.; Wlodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef] [PubMed]
- Chaharlashkar, Z.; Saeedi Honar, Y.; Abdollahpour-Alitappeh, M.; Parvizpour, S.; Barzegar, A.; Alizadeh, E. Metastatic melanoma: An integrated analysis to identify critical regulators associated with prognosis, pathogenesis and targeted therapies. PLoS ONE 2025, 20, e0312754. [Google Scholar] [CrossRef] [PubMed]
- Naseri, H.; Safaei, A.A. Diagnosis and prognosis of melanoma from dermoscopy images using machine learning and deep learning: A systematic literature review. BMC Cancer 2025, 25, 75. [Google Scholar] [CrossRef] [PubMed]
- Spain, L.; Coulton, A.; Lobon, I.; Rowan, A.; Schnidrig, D.; Shepherd, S.T.C.; Shum, B.; Byrne, F.; Goicoechea, M.; Piperni, E.; et al. Late-Stage Metastatic Melanoma Emerges through a Diversity of Evolutionary Pathways. Cancer Discov. 2023, 13, 1364–1385. [Google Scholar] [CrossRef]
- Xue, C.; Zhou, Q.; Xi, H.; Zhou, J. Radiomics: A review of current applications and possibilities in the assessment of tumor microenvironment. Diagn. Interv. Imaging 2023, 104, 113–122. [Google Scholar] [CrossRef]
- Savoia, P.; Zavattaro, E.; Cremona, O. Clinical Implications of Acquired BRAF Inhibitors Resistance in Melanoma. Int. J. Mol. Sci. 2020, 21, 9730. [Google Scholar] [CrossRef]
- Karami Fath, M.; Azargoonjahromi, A.; Soofi, A.; Almasi, F.; Hosseinzadeh, S.; Khalili, S.; Sheikhi, K.; Ferdousmakan, S.; Owrangi, S.; Fahimi, M.; et al. Current understanding of epigenetics role in melanoma treatment and resistance. Cancer Cell Int. 2022, 22, 313. [Google Scholar] [CrossRef]
- Krensel, M.; Schafer, I.; Augustin, M. Cost-of-illness of melanoma in Europe—A systematic review of the published literature. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 504–510. [Google Scholar] [CrossRef]
- Mosele, M.F.; Westphalen, C.B.; Stenzinger, A.; Barlesi, F.; Bayle, A.; Bieche, I.; Bonastre, J.; Castro, E.; Dienstmann, R.; Kramer, A.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with advanced cancer in 2024: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2024, 35, 588–606. [Google Scholar] [CrossRef]
- Carvajal, R.D.; Schwartz, G.K.; Tezel, T.; Marr, B.; Francis, J.H.; Nathan, P.D. Metastatic disease from uveal melanoma: Treatment options and future prospects. Br. J. Ophthalmol. 2017, 101, 38–44. [Google Scholar] [CrossRef]
- Higgins, H.; Nakhla, A.; Lotfalla, A.; Khalil, D.; Doshi, P.; Thakkar, V.; Shirini, D.; Bebawy, M.; Ammari, S.; Lopci, E.; et al. Recent Advances in the Field of Artificial Intelligence for Precision Medicine in Patients with a Diagnosis of Metastatic Cutaneous Melanoma. Diagnostics 2023, 13, 3483. [Google Scholar] [CrossRef] [PubMed]
- Dercle, L.; Zhao, B.; Gonen, M.; Moskowitz, C.S.; Firas, A.; Beylergil, V.; Connors, D.E.; Yang, H.; Lu, L.; Fojo, T.; et al. Early Readout on Overall Survival of Patients With Melanoma Treated With Immunotherapy Using a Novel Imaging Analysis. JAMA Oncol. 2022, 8, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Brendlin, A.S.; Peisen, F.; Almansour, H.; Afat, S.; Eigentler, T.; Amaral, T.; Faby, S.; Calvarons, A.F.; Nikolaou, K.; Othman, A.E. A Machine learning model trained on dual-energy CT radiomics significantly improves immunotherapy response prediction for patients with stage IV melanoma. J. Immunother. Cancer 2021, 9, e003261. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Mao, L.L.; Zhou, Z.G.; Si, L.; Zhu, H.T.; Chen, X.; Zhou, M.J.; Sun, Y.S.; Guo, J. Pilot Study of CT-Based Radiomics Model for Early Evaluation of Response to Immunotherapy in Patients With Metastatic Melanoma. Front. Oncol. 2020, 10, 1524. [Google Scholar] [CrossRef]
- Peisen, F.; Gerken, A.; Hering, A.; Dahm, I.; Nikolaou, K.; Gatidis, S.; Eigentler, T.K.; Amaral, T.; Moltz, J.H.; Othman, A.E. Can Whole-Body Baseline CT Radiomics Add Information to the Prediction of Best Response, Progression-Free Survival, and Overall Survival of Stage IV Melanoma Patients Receiving First-Line Targeted Therapy: A Retrospective Register Study. Diagnostics 2023, 13, 3210. [Google Scholar] [CrossRef]
- Ter Maat, L.S.; van Duin, I.A.J.; Elias, S.G.; Leiner, T.; Verhoeff, J.J.C.; Arntz, E.; Troenokarso, M.F.; Blokx, W.A.M.; Isgum, I.; de Wit, G.A.; et al. CT radiomics compared to a clinical model for predicting checkpoint inhibitor treatment outcomes in patients with advanced melanoma. Eur. J. Cancer 2023, 185, 167–177. [Google Scholar] [CrossRef]
- Guerrisi, A.; Russillo, M.; Loi, E.; Ganeshan, B.; Ungania, S.; Desiderio, F.; Bruzzaniti, V.; Falcone, I.; Renna, D.; Ferraresi, V.; et al. Exploring CT Texture Parameters as Predictive and Response Imaging Biomarkers of Survival in Patients With Metastatic Melanoma Treated With PD-1 Inhibitor Nivolumab: A Pilot Study Using a Delta-Radiomics Approach. Front. Oncol. 2021, 11, 704607. [Google Scholar] [CrossRef]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef]
- Robert, C.; Grob, J.J.; Stroyakovskiy, D.; Karaszewska, B.; Hauschild, A.; Levchenko, E.; Chiarion Sileni, V.; Schachter, J.; Garbe, C.; Bondarenko, I.; et al. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. N. Engl. J. Med. 2019, 381, 626–636. [Google Scholar] [CrossRef]
- Wolchok, J.D.; Chiarion-Sileni, V.; Rutkowski, P.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Queirolo, P.; Dummer, R.; Butler, M.O.; Hill, A.G.; et al. Final, 10-Year Outcomes with Nivolumab plus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2025, 392, 11–22. [Google Scholar] [CrossRef]
- Dummer, R.; Ascierto, P.A.; Gogas, H.J.; Arance, A.; Mandala, M.; Liszkay, G.; Garbe, C.; Schadendorf, D.; Krajsova, I.; Gutzmer, R.; et al. Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2018, 19, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Dummer, R.; Ascierto, P.A.; Gogas, H.J.; Arance, A.; Mandala, M.; Liszkay, G.; Garbe, C.; Schadendorf, D.; Krajsova, I.; Gutzmer, R.; et al. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): A multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2018, 19, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Schadendorf, D.; Dummer, R.; Flaherty, K.T.; Robert, C.; Arance, A.; de Groot, J.W.B.; Garbe, C.; Gogas, H.J.; Gutzmer, R.; Krajsova, I.; et al. COLUMBUS 7-year update: A randomized, open-label, phase III trial of encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF V600E/K-mutant melanoma. Eur. J. Cancer 2024, 204, 114073. [Google Scholar] [CrossRef] [PubMed]
- Bomar, L.; Senithilnathan, A.; Ahn, C. Systemic Therapies for Advanced Melanoma. Dermatol. Clin. 2019, 37, 409–423. [Google Scholar] [CrossRef]
- Falcone, I.; Conciatori, F.; Bazzichetto, C.; Ferretti, G.; Cognetti, F.; Ciuffreda, L.; Milella, M. Tumor Microenvironment: Implications in Melanoma Resistance to Targeted Therapy and Immunotherapy. Cancers 2020, 12, 2870. [Google Scholar] [CrossRef]
- Nikolaou, V.; Tsimpidakis, A.; Stratigos, A. Cutaneous Adverse Reactions of Immunotherapy in Patients with Advanced Melanoma. Cancers 2023, 15, 2084. [Google Scholar] [CrossRef]
- Ascierto, P.A.; Bastholt, L.; Hersey, P.; Cinat, G.; Eggermont, A.M.; Hauschild, A.; Espinosa, E.; Robert, C. Side effects and toxicities of targeted therapies in stage IV melanoma. Am. J. Ther. 2015, 22, 44–53. [Google Scholar] [CrossRef]
- Li, J.; Dan, K.; Ai, J. Machine learning in the prediction of immunotherapy response and prognosis of melanoma: A systematic review and meta-analysis. Front. Immunol. 2024, 15, 1281940. [Google Scholar] [CrossRef]
- Valenti, F.; Falcone, I.; Ungania, S.; Desiderio, F.; Giacomini, P.; Bazzichetto, C.; Conciatori, F.; Gallo, E.; Cognetti, F.; Ciliberto, G.; et al. Precision Medicine and Melanoma: Multi-Omics Approaches to Monitoring the Immunotherapy Response. Int. J. Mol. Sci. 2021, 22, 3837. [Google Scholar] [CrossRef]
- McGale, J.; Hama, J.; Yeh, R.; Vercellino, L.; Sun, R.; Lopci, E.; Ammari, S.; Dercle, L. Artificial Intelligence and Radiomics: Clinical Applications for Patients with Advanced Melanoma Treated with Immunotherapy. Diagnostics 2023, 13, 3065. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5, 13087. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Brush, J.; Boyd, K.; Chappell, F.; Crawford, F.; Dozier, M.; Fenwick, E.; Glanville, J.; McIntosh, H.; Renehan, A.; Weller, D.; et al. The value of FDG positron emission tomography/computerised tomography (PET/CT) in pre-operative staging of colorectal cancer: A systematic review and economic evaluation. Health Technol. Assess. 2011, 15, 1–192, iii–iv. [Google Scholar] [CrossRef]
- Ger, R.B.; Zhou, S.; Chi, P.M.; Lee, H.J.; Layman, R.R.; Jones, A.K.; Goff, D.L.; Fuller, C.D.; Howell, R.M.; Li, H.; et al. Comprehensive Investigation on Controlling for CT Imaging Variabilities in Radiomics Studies. Sci. Rep. 2018, 8, 13047. [Google Scholar] [CrossRef]
- Guan, H.; Yap, P.T.; Bozoki, A.; Liu, M. Federated learning for medical image analysis: A survey. Pattern Recognit. 2024, 151, 110424. [Google Scholar] [CrossRef]
- Vetto, J.T. Clinical and Imaging Follow-Up for High-Risk Cutaneous Melanoma: Current Evidence and Guidelines. Cancers 2024, 16, 2572. [Google Scholar] [CrossRef]
- Valente, D.; Gentileschi, M.P.; Valenti, A.; Burgio, M.; Soddu, S.; Bruzzaniti, V.; Guerrisi, A.; Verdina, A. Cumulative Dose from Recurrent CT Scans: Exploring the DNA Damage Response in Human Non-Transformed Cells. Int. J. Mol. Sci. 2024, 25, 7064. [Google Scholar] [CrossRef]
| Internal Testing | Internal Testing | |||
|---|---|---|---|---|
| FP | UP | FP | UP | |
| % Male | 70.6% | 60.0% | 66.7% | 50.0% |
| % Female | 29.4% | 40.0% | 33.3% | 50.0% |
| Median Number of Patients Lesion | 1.88 | 2.8 | 2.0 | 2.6 |
| Median Age | 64.9 | 66.7 | 73.7 | 66.9 |
| Median PFS (months) | 33.8 | 2.2 | 44.3 | 6.0 |
| Median OS (months) | 56.7 | 20.8 | 96.7 | 32.5 |
| Metric | Training (%) [95% CI] | Validation (%) [95% CI] | Internal Testing (Mean) (%) [95% CI] | Internal Testing (Majority Vote) (%) [95% CI] | |
|---|---|---|---|---|---|
| 1 | ROC-AUC | 100 * [99–100] | 80 ** [79–81] | 82 ** [78–85] | 82 |
| Accuracy | 100 * [99–100] | 74 ** [72–76] | 75 ** [74–77] | 77 | |
| Sensitivity | 100 * [99–100] | 80 ** [78–82] | 80 ** [78–83] | 84 | |
| Specificity | 100 * [99–100] | 67 ** [64–70] | 70 ** [66–73] | 68 | |
| PPV | 100 * [99–100] | 77 ** [75–78] | 75 ** [73–77] | 75 | |
| NPV | 100 * [99–100] | 77 ** [74–80] | 76 ** [74–78] | 79 | |
| 2 | ROC-AUC | 67 ** [66–68] | 56 ** [56–57] | 49 ** [42–56] | 47 |
| Accuracy | 65 ** [64–66] | 56 ** [54–59] | 54 ** [62–83] | 55 | |
| Sensitivity | 77 ** [76–78] | 69 ** [63–75] | 73 ** [62–83] | 75 | |
| Specificity | 51 ** [48–54] | 42 ** [37–48] | 32 ** [28–37] | 32 | |
| PPV | 64 ** [63–65] | 59 ** [57–61] | 55 ** [51–59] | 56 | |
| NPV | 66 ** [65–67] | 55 ** [49–61] | 51 ** [41–61] | 53 | |
| 3 | ROC-AUC | 81 ** [80–82] | 51 ** [48–54] | 67 ** [65–69] | 70 |
| Accuracy | 74 ** [73–75] | 52 ** [50–54] | 60 ** [56–64] | 67 | |
| Sensitivity | 79 ** [77–81] | 61 ** [59–62] | 76 ** [71–81] | 81 | |
| Specificity | 68 ** [68–68] | 42 ** [37–48] | 43 ** [35–51] | 50 | |
| PPV | 74 ** [74–75] | 55 ** [49–60] | 60 ** [57–64] | 65 | |
| NPV | 74 ** [72–76] | 48 ** [46–51] | 61 ** [55–66] | 70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerrisi, A.; Maccallini, M.T.; Falcone, I.; Valenti, A.; Miseo, L.; Ungania, S.; Dolcetti, V.; Valenti, F.; Cerro, M.; Desiderio, F.; et al. Metastatic Melanoma Prognosis Prediction Using a TC Radiomic-Based Machine Learning Model: A Preliminary Study. Cancers 2025, 17, 2304. https://doi.org/10.3390/cancers17142304
Guerrisi A, Maccallini MT, Falcone I, Valenti A, Miseo L, Ungania S, Dolcetti V, Valenti F, Cerro M, Desiderio F, et al. Metastatic Melanoma Prognosis Prediction Using a TC Radiomic-Based Machine Learning Model: A Preliminary Study. Cancers. 2025; 17(14):2304. https://doi.org/10.3390/cancers17142304
Chicago/Turabian StyleGuerrisi, Antonino, Maria Teresa Maccallini, Italia Falcone, Alessandro Valenti, Ludovica Miseo, Sara Ungania, Vincenzo Dolcetti, Fabio Valenti, Marianna Cerro, Flora Desiderio, and et al. 2025. "Metastatic Melanoma Prognosis Prediction Using a TC Radiomic-Based Machine Learning Model: A Preliminary Study" Cancers 17, no. 14: 2304. https://doi.org/10.3390/cancers17142304
APA StyleGuerrisi, A., Maccallini, M. T., Falcone, I., Valenti, A., Miseo, L., Ungania, S., Dolcetti, V., Valenti, F., Cerro, M., Desiderio, F., Calabrò, F., Ferraresi, V., & Russillo, M. (2025). Metastatic Melanoma Prognosis Prediction Using a TC Radiomic-Based Machine Learning Model: A Preliminary Study. Cancers, 17(14), 2304. https://doi.org/10.3390/cancers17142304







