Analysis of Phenotypic and Molecular Variability of Memory-like NK Cells for Cancer Adoptive Cell Therapy Screening
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. NK Cell Isolation
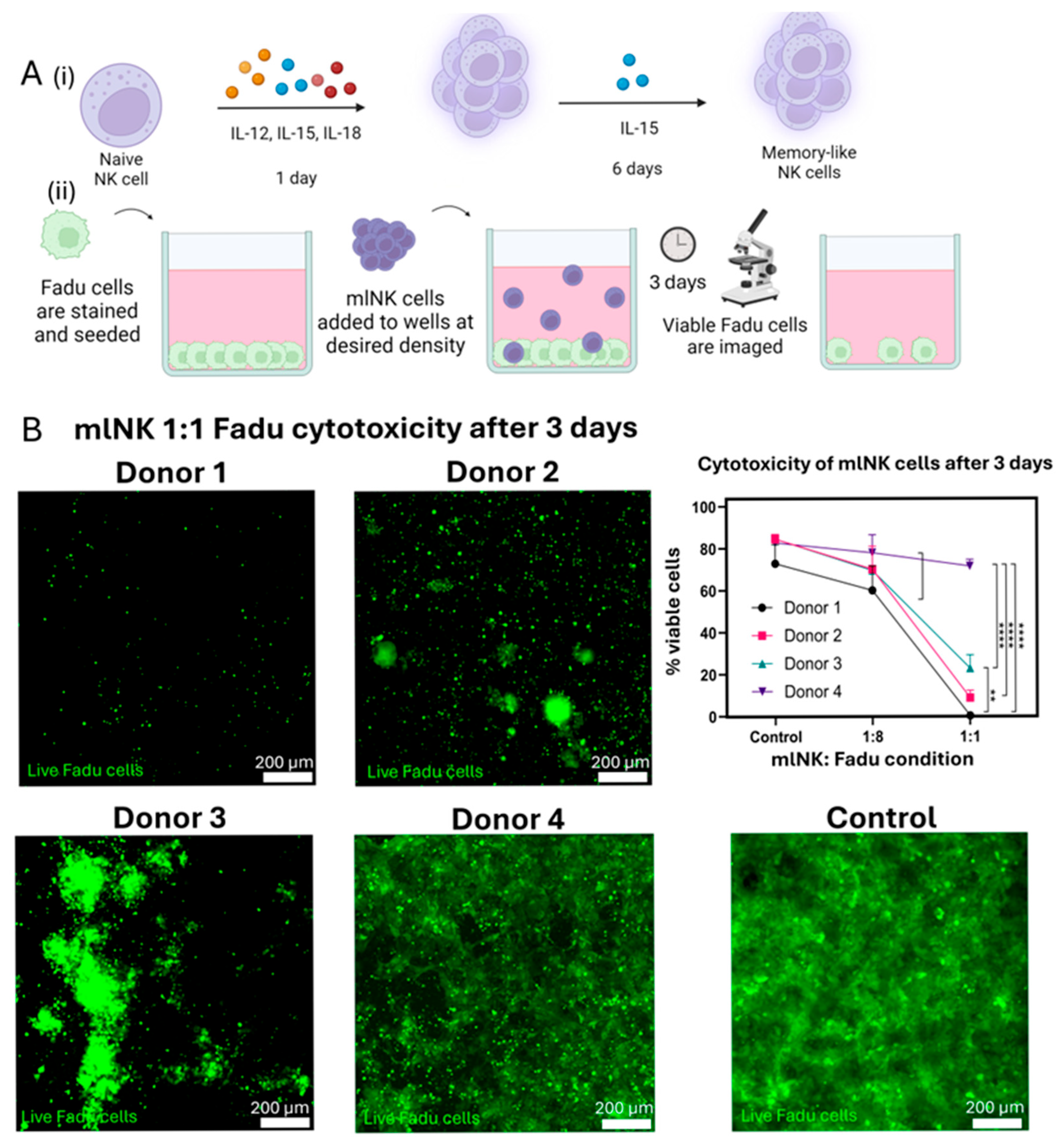
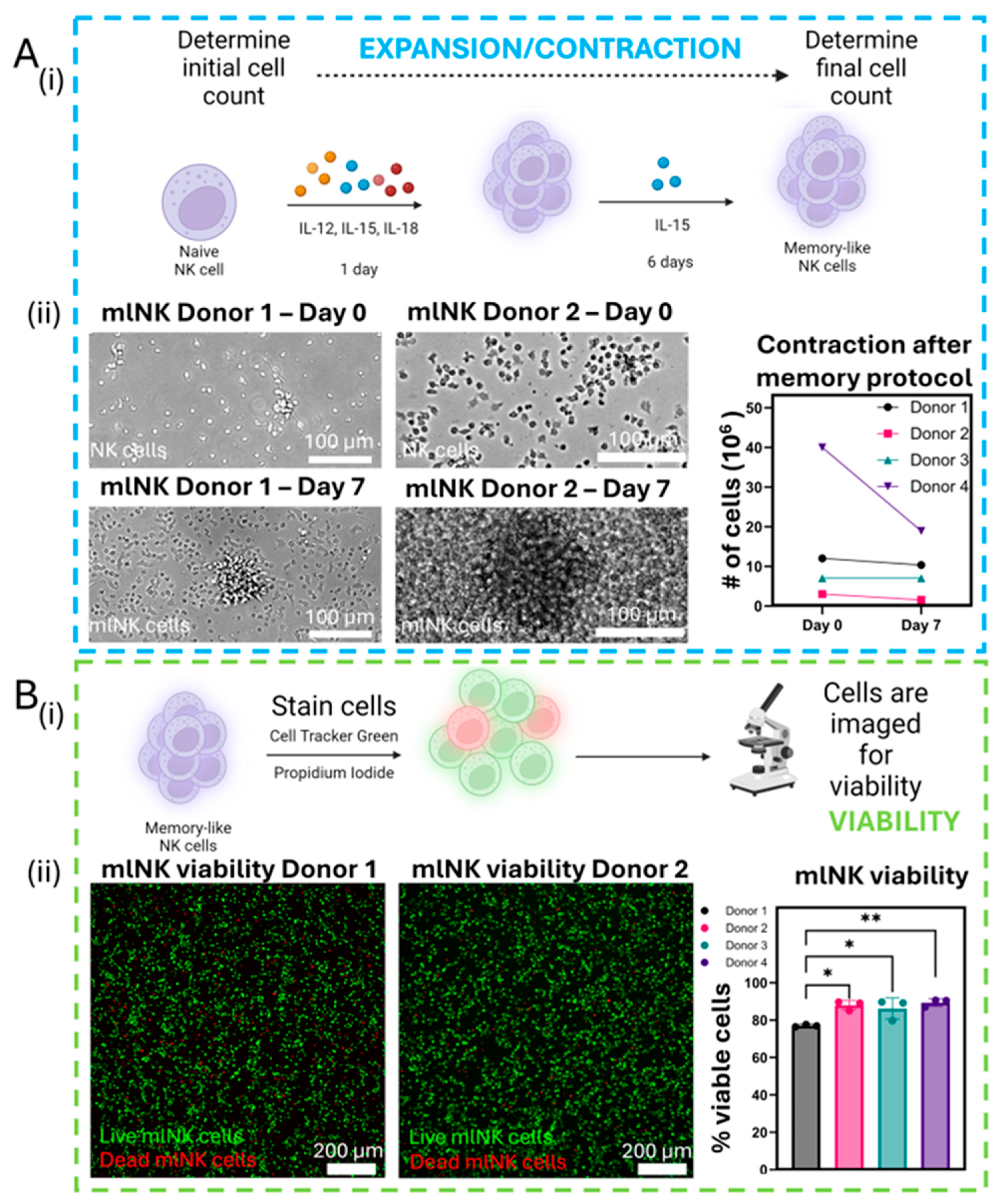
2.2. Memory-like NK Cell Generation and Culture
2.3. Flow Cytometry
2.4. Fadu Tumor Cell Culture
2.5. Cytotoxicity
2.6. Genotyping Analysis
2.7. mlNK Cell Viability
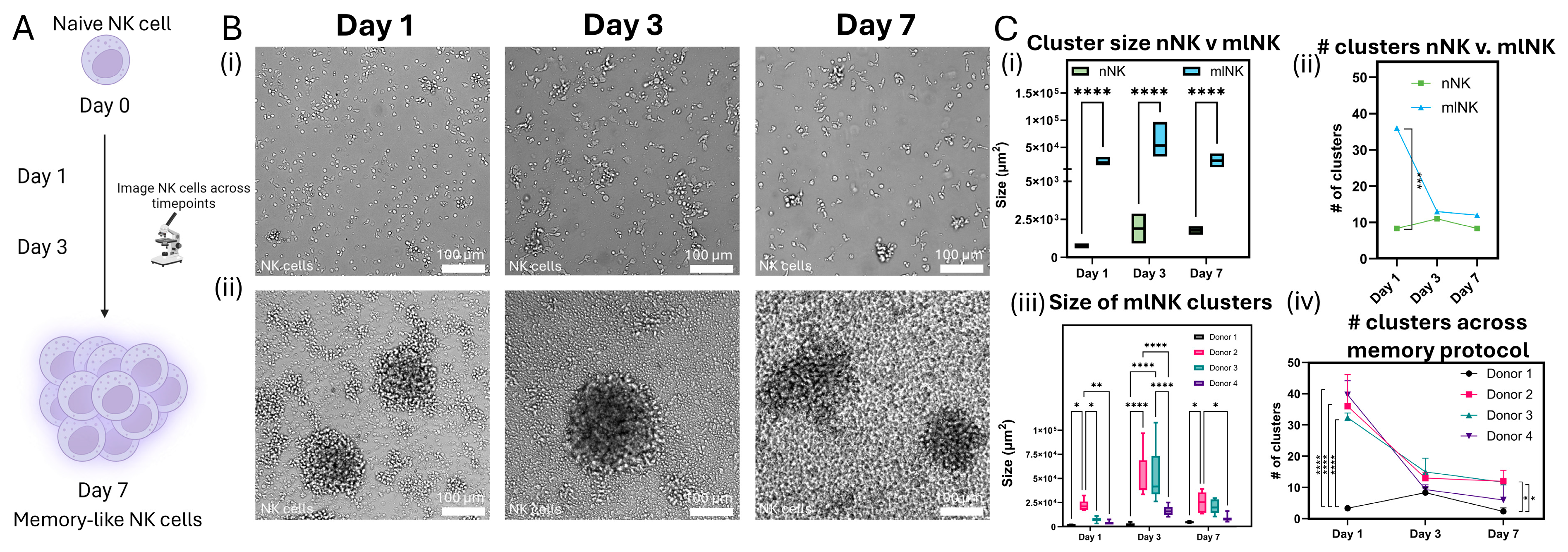
2.8. Clustering
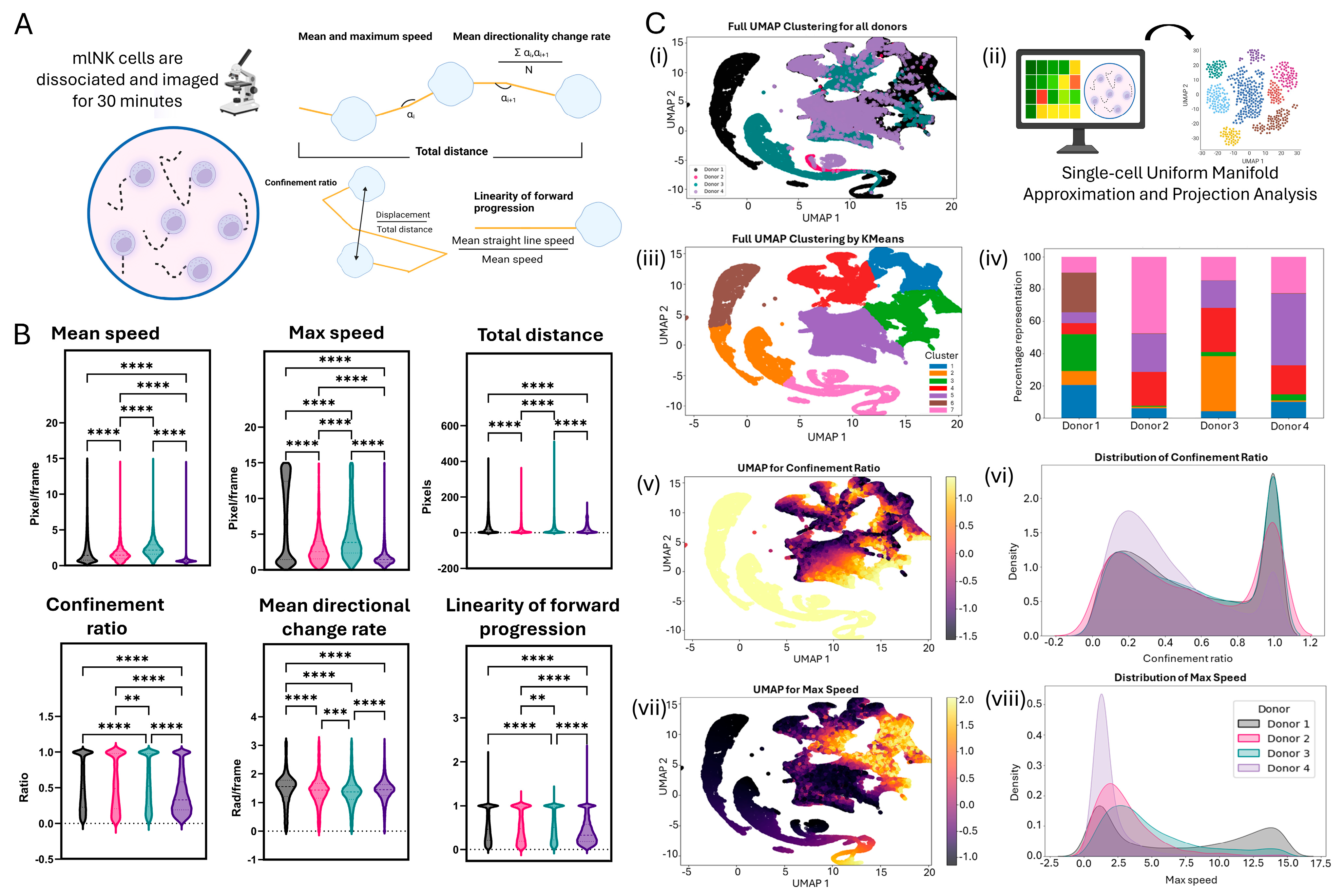
2.9. Motility Analysis
2.10. Single-Cell Computational Analysis
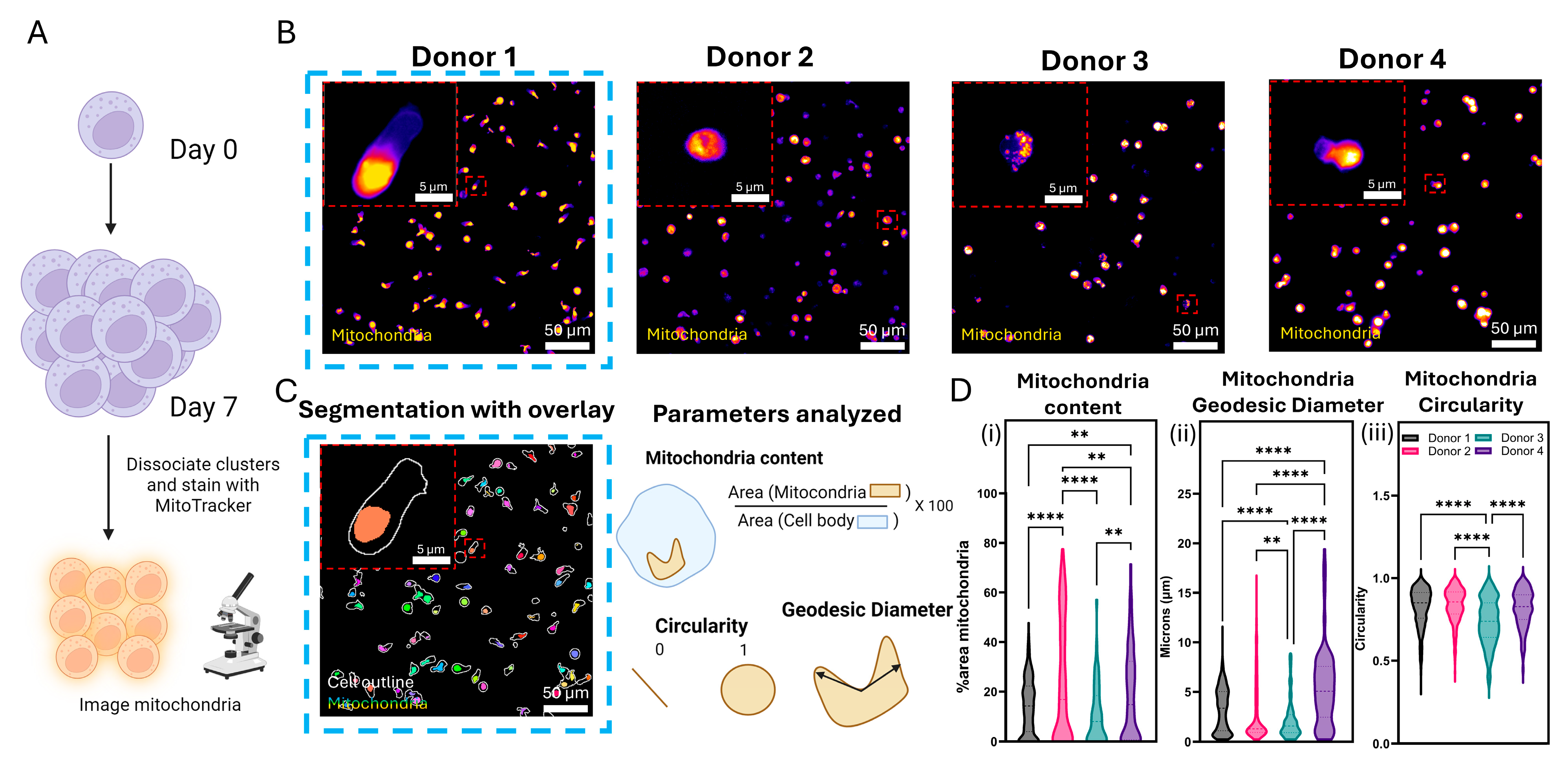
2.11. Mitochondrial Staining and Analysis
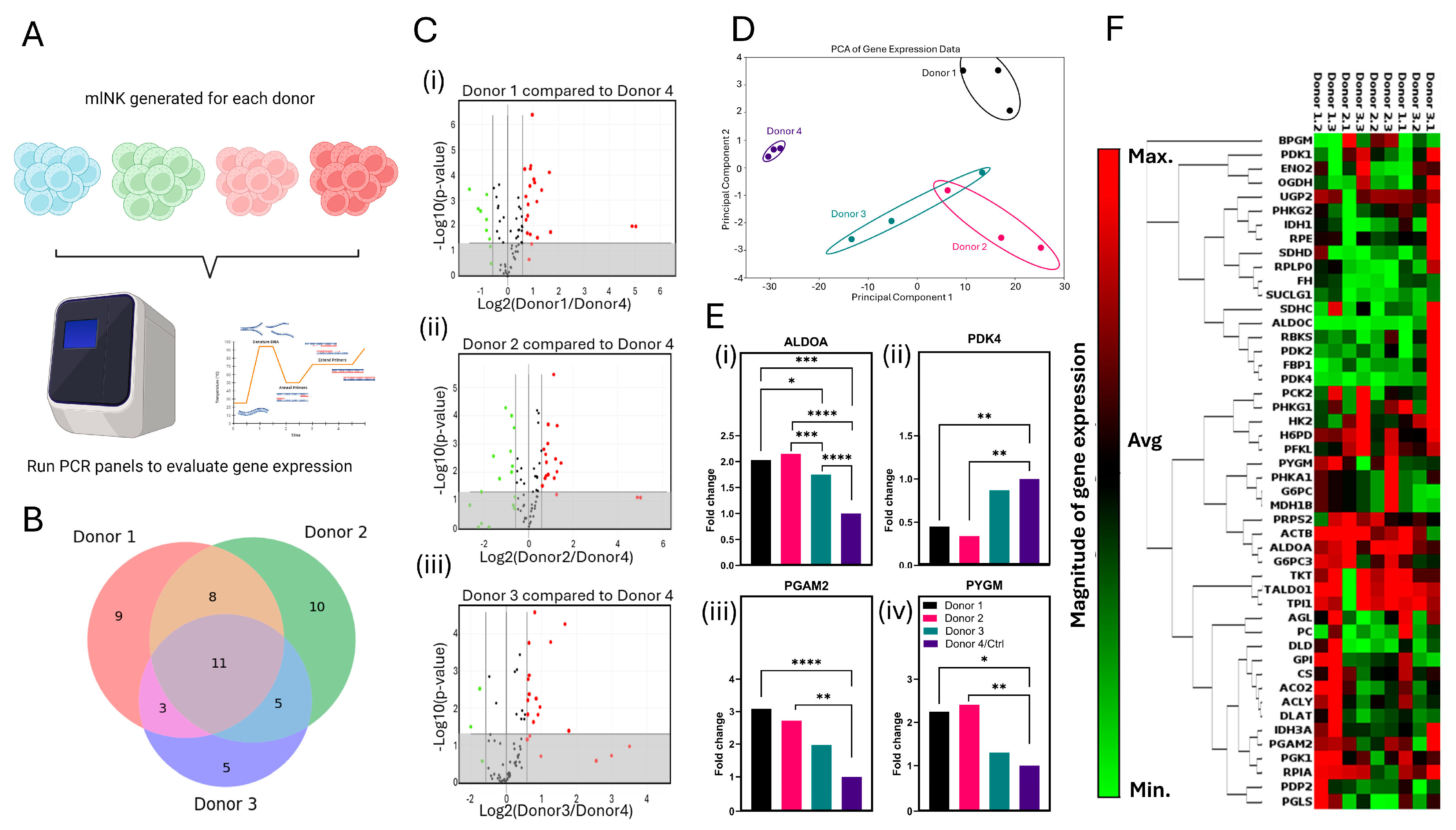
2.12. Gene Expression Analysis
2.13. Two-Dimensional and Oligomycin Exposure
2.14. Image Acquisition and Analysis
2.15. Equipment
2.16. Statistical Analysis
3. Results
3.1. Differences in Cytotoxic Potential Across mlNK Products Showcases the Need for Further Characterization
3.2. KIR/KIR-Ligand Genotyping Is Not Sufficient to Accurately Predict Phenotypic Heterogeneity
3.3. NK Viability Remains High After Memory Protocol Despite Drop in Total Count
3.4. mlNK Cells Self-Organize into Clusters of Different Sizes
3.5. Single-Cell Motility Analysis Identifies Subsets of mlNK Cells with Differences in Their Migration Potential
3.6. Mitochondrial Architecture Can Be an Indicator of Metabolic Activity of mlNK Cells
3.7. Gene Expression Analysis Suggests Disruption of Glycolytic Flux in mlNK Cells Could Affect Cytotoxicity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef]
- Lin, D.; Shen, Y.; Liang, T. Oncolytic virotherapy: Basic principles, recent advances and future directions. Signal Transduct. Target. Ther. 2023, 8, 156. [Google Scholar] [CrossRef] [PubMed]
- Van Der Bruggen, P.; Traversari, C.; Chomez, P.; Lurquin, C.; De Plaen, E.; Van Den Eynde, B.; Knuth, A.; Boon, T. A Gene Encoding an Antigen Recognized by Cytolytic T Lymphocytes on a Human Melanoma. Science 1991, 254, 1643–1647. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, J.M.; Strawderman, M.H.; Ernstoff, M.S.; Smith, T.J.; Borden, E.C.; Blum, R.H. Interferon alfa-2b adjuvant therapy of high-risk resected cutaneous melanoma: The Eastern Cooperative Oncology Group Trial EST 1684. J. Clin. Oncol. 1996, 14, 7–17. [Google Scholar] [CrossRef]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996, 271, 1734–1736. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Restifo, N.P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 2015, 348, 62–68. [Google Scholar] [CrossRef]
- Depil, S.; Duchateau, P.; Grupp, S.A.; Mufti, G.; Poirot, L. ‘Off-the-shelf’ allogeneic CAR T cells: Development and challenges. Nat. Rev. Drug Discov. 2020, 19, 185–199. [Google Scholar] [CrossRef]
- Mehta, R.S.; Rezvani, K. Can we make a better match or mismatch with KIR genotyping? Hematology 2016, 2016, 106–118. [Google Scholar] [CrossRef]
- Seiwert, T.Y.; Burtness, B.; Mehra, R.; Weiss, J.; Berger, R.; Eder, J.P.; Heath, K.; McClanahan, T.; Lunceford, J.; Gause, C.; et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): An open-label, multicentre, phase 1b trial. Lancet Oncol. 2016, 17, 956–965. [Google Scholar] [CrossRef]
- Mell, L.K.; Torres-Saavedra, P.A.; Wong, S.J.; Kish, J.A.; Chang, S.S.; Jordan, R.C.; Liu, T.; Truong, M.T.; Winquist, E.W.; Takiar, V.; et al. Radiotherapy with cetuximab or durvalumab for locoregionally advanced head and neck cancer in patients with a contraindication to cisplatin (NRG-HN004): An open-label, multicentre, parallel-group, randomised, phase 2/3 trial. Lancet Oncol. 2024, 25, 1576–1588. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, T.J.; Biederstädt, A.; Rezvani, K. Natural killer cells in antitumour adoptive cell immunotherapy. Nat. Rev. Cancer 2022, 22, 557–575. [Google Scholar] [CrossRef] [PubMed]
- Ayuso, J.M.; Truttschel, R.; Gong, M.M.; Humayun, M.; Virumbrales-Munoz, M.; Vitek, R.; Felder, M.; Gillies, S.D.; Sondel, P.; Wisinski, K.B.; et al. Evaluating natural killer cell cytotoxicity against solid tumors using a microfluidic model. Oncoimmunology 2019, 8, 1553477. [Google Scholar] [CrossRef]
- Schantz, S.P.; Shillitoe, E.J.; Brown, B.; Campbell, B. Natural killer cell activity and head and neck cancer: A clinical assessment. J. Natl. Cancer Inst. 1986, 77, 869–875. [Google Scholar]
- Schantz, S.P.; Campbell, B.H.; Guillamondegui, O.M. Pharyngeal carcinoma and natural killer cell activity. Am. J. Surg. 1986, 152, 467–474. [Google Scholar] [CrossRef]
- Charap, A.J.; Enokida, T.; Brody, R.; Sfakianos, J.; Miles, B.; Bhardwaj, N.; Horowitz, A. Landscape of natural killer cell activity in head and neck squamous cell carcinoma. J. Immunother. Cancer 2020, 8, 1523. [Google Scholar] [CrossRef]
- Berrien-Elliott, M.M.; Wagner, J.A.; Cashen, A.F.; Fehniger, T.A. Memory-Like Natural Killer Cells. Blood 2018, 132 (Suppl. S1), SCI-8. [Google Scholar] [CrossRef]
- Pahl, J.H.W.; Cerwenka, A.; Ni, J. Memory-Like NK cells: Remembering a previous activation by cytokines and NK cell receptors. Front. Immunol. 2018, 9, 2796. [Google Scholar] [CrossRef]
- Berrien-Elliott, M.M.; Cashen, A.F.; Cubitt, C.C.; Neal, C.C.; Wong, P.; Wagner, J.A.; Foster, M.; Schappe, T.; Desai, S.; McClain, E.; et al. Multidimensional anal-yses of donor memory-like NK cells reveal new associations with response after adoptive immunotherapy for leu-kemia. Cancer Discov. 2020, 10, 1854–1872. [Google Scholar] [CrossRef]
- Romee, R.; Rosario, M.; Berrien-Elliott, M.M.; Wagner, J.A.; Jewell, B.A.; Schappe, T.; Leong, J.W.; Abdel-Latif, S.; Schneider, S.E.; Willey, S.; et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci. Transl. Med. 2016, 8, 357ra123. [Google Scholar] [CrossRef]
- Erbe, A.K.; Wang, W.; Carmichael, L.; Kim, K.; Mendonça, E.A.; Song, Y.; Hess, D.; Reville, P.K.; London, W.B.; Naranjo, A.; et al. Neuroblastoma Patients’ KIR and KIR-Ligand Genotypes Influence Clinical Outcome for Dinutuximab-based Immunotherapy: A Report from the Children’s Oncology Group. Clin. Cancer Res. 2018, 24, 189–196. [Google Scholar] [CrossRef]
- Ayuso, J.M.; Farooqui, M.; Virumbrales-Muñoz, M.; Denecke, K.; Rehman, S.; Schmitz, R.; Guerrero, J.F.; Sanchez-De-Diego, C.; Campo, S.A.; Maly, E.M.; et al. Microphysiological model reveals the promise of memory-like natural killer cell immunotherapy for HIV ± cancer. Nat. Commun. 2023, 14, 6681. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.M.; Mahmoud, A.B.; Makrigiannis, A.P. Licensed and unlicensed NK cells: Differential roles in cancer and viral control. Front. Immunol. 2016, 7, 166. [Google Scholar] [CrossRef]
- Pende, D.; Falco, M.; Vitale, M.; Cantoni, C.; Vitale, C.; Munari, E.; Bertaina, A.; Moretta, F.; Del Zotto, G.; Pietra, G.; et al. Killer Ig-like receptors (KIRs): Their role in NK cell modulation and developments leading to their clinical exploitation. Front. Immunol. 2019, 10, 1179. [Google Scholar] [CrossRef]
- Moesta, A.K.; Norman, P.J.; Yawata, M.; Yawata, N.; Gleimer, M.; Parham, P. Synergistic Polymorphism at Two Positions Distal to the Ligand-Binding Site Makes KIR2DL2 a Stronger Receptor for HLA-C Than KIR2DL3. J. Immunol. 2008, 180, 3969–3979. [Google Scholar] [CrossRef]
- Almeida, C.R.; Ashkenazi, A.; Shahaf, G.; Kaplan, D.; Davis, D.M.; Mehr, R. Human NK cells differ more in their KIR2DL1-dependent thresholds for HLA-Cw6-mediated inhibition than in their maximal killing capacity. PLoS ONE 2011, 6, e24927. [Google Scholar] [CrossRef]
- Litwin, V.; Gumperz, J.; Parham, P.; Phillips, J.H.; Lanier, L.L. NKB1: A natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J. Exp. Med. 1994, 180, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, W.M.; Kim, S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol. Rev. 2006, 214, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, A.H.; Yokoyama, W.M. Natural killer cell tolerance licensing and other mechanisms. Adv. Immunol. 2009, 101, 27–79. [Google Scholar] [CrossRef]
- Kim, S.; Poursine-Laurent, J.; Truscott, S.M.; Lybarger, L.; Song, Y.J.; Yang, L.; French, A.R.; Sunwoo, J.B.; Lemieux, S.; Hansen, T.H.; et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 2005, 436, 709–713. [Google Scholar] [CrossRef]
- Kim, M.; Kim, T.J.; Kim, H.M.; Doh, J.; Lee, K.M. Multi-cellular natural killer (NK) cell clusters enhance NK cell activation through localizing IL-2 within the cluster. Sci. Rep. 2017, 7, 40623. [Google Scholar] [CrossRef]
- Palacios, D.; Majhi, R.K.; Szabo, E.K.; Clement, D.; Lachota, M.; Netskar, H.; Penna, L.; Krokeide, S.Z.; Vincenti, M.; Kveberg, L.; et al. The G Protein–Coupled Receptor GPR56 Is an Inhibitory Checkpoint for NK Cell Migration. J. Immunol. 2024, 213, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Tinevez, J.Y.; Perry, N.; Schindelin, J.; Hoopes, G.M.; Reynolds, G.D.; Laplantine, E.; Bednarek, S.Y.; Shorte, S.L.; Eliceiri, K.W. TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115, 80–90. [Google Scholar] [CrossRef]
- Xie, J.H.; Li, Y.Y.; Jin, J. The essential functions of mitochondrial dynamics in immune cells. Cell. Mol. Immunol. 2020, 17, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Surace, L.; Doisne, J.-M.; Escoll, P.; Marie, S.; Dardalhon, V.; Croft, C.; Thaller, A.; Topazio, D.; Sparaneo, A.; Cama, A.; et al. Polarized mitochondria as guardians of NK cell fitness. Blood Adv. 2021, 5, 26–38. [Google Scholar] [CrossRef]
- Chen, W.; Zhao, H.; Li, Y. Mitochondrial dynamics in health and disease: Mechanisms and potential targets. Signal Transduct. Target. Ther. 2023, 8, 333. [Google Scholar] [CrossRef]
- Legland, D.; Arganda-Carreras, I.; Andrey, P. MorphoLibJ: Integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 2016, 32, 3532–3534. [Google Scholar] [CrossRef]
- Gemmink, A.; Daemen, S.; Wefers, J.; Hansen, J.; Moorsel Dvan Astuti, P.; Jorgensen, J.A.; Kornips, E.; Schaart, G.; Hoeks, J.; Schrauwen, P.; et al. Twenty-four hour rhythmicity in mitochondrial network connectivity and mitochondrial respiration; a study in human skeletal muscle biopsies of young lean and older individuals with obesity. Mol. Metab. 2023, 72, 101727. [Google Scholar] [CrossRef]
- Zheng, X.; Qian, Y.; Fu, B.; Jiao, D.; Jiang, Y.; Chen, P.; Shen, Y.; Zhang, H.; Sun, R.; Tian, Z.; et al. Mitochondrial fragmentation limits NK cell-based tumor immunosurveillance. Nat. Immunol. 2019, 20, 1656–1667. [Google Scholar] [CrossRef]
- Haythorne, E.; Lloyd, M.; Walsby-Tickle, J.; Tarasov, A.I.; Sandbrink, J.; Portillo, I.; Exposito, R.T.; Sachse, G.; Cyranka, M.; Rohm, M.; et al. Altered glycolysis triggers impaired mitochondrial metabolism and mTORC1 activation in diabetic β-cells. Nat. Commun. 2022, 13, 6754. [Google Scholar] [CrossRef]
- Poznanski, S.M.; Ashkar, A.A. What defines NK cell functional fate: Phenotype or metabolism? Front. Immunol. 2019, 10, 1414. [Google Scholar] [CrossRef]
- Chang, Y.C.; Yang, Y.C.; Tien, C.P.; Yang, C.J.; Hsiao, M. Roles of Aldolase Family Genes in Human Cancers and Diseases. Trends Endocrinol. Metab. 2018, 29, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Sobanski, T.; Suraweera, A.; Burgess, J.T.; Richard, I.; Cheong, C.M.; Dave, K.; Rose, M.; Adams, M.N.; O’Byrne, K.J.; Richard, D.J.; et al. The fructose-bisphosphate, Aldolase A (ALDOA), facilitates DNA-PKcs and ATM kinase activity to regulate DNA double-strand break repair. Sci. Rep. 2023, 13, 15171. [Google Scholar] [CrossRef]
- Dou, X.; Fu, Q.; Long, Q.; Liu, S.; Zou, Y.; Fu, D.; Xu, Q.; Jiang, Z.; Ren, X.; Zhang, G.; et al. PDK4-dependent hypercatabolism and lactate production of senescent cells promotes cancer malignancy. Nat. Metab. 2023, 5, 1887–1910. [Google Scholar] [CrossRef]
- Flora, G.D.; Nayak, M.K.; Ghatge, M.; Kumskova, M.; Patel, R.B.; Chauhan, A.K. Mitochondrial pyruvate dehydrogenase kinases contribute to platelet function and thrombosis in mice by regulating aerobic glycolysis. Blood Adv. 2023, 7, 2347–2359. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.; Barciszewski, J.; Turlik, J.; Baran, K.; Duda, P.; Jaskolski, M.; Rakus, D. High-Resolution Crystal Structure of Muscle Phosphoglycerate Mutase Provides Insight into Its Nuclear Import and Role. Int. J. Mol. Sci. 2022, 23, 13198. [Google Scholar] [CrossRef]
- Migocka-Patrzałek, M.; Elias, M. Muscle glycogen phosphorylase and its functional partners in health and disease. Cells 2021, 10, 883. [Google Scholar] [CrossRef]
- PYGM Glycogen Phosphorylase, Muscle Associated—NIH Genetic Testing Registry (GTR)-NCBI. Available online: https://www.ncbi.nlm.nih.gov/gtr/genes/5837/ (accessed on 21 December 2024).
- Keppel, M.P.; Saucier, N.; Mah, A.Y.; Vogel, T.P.; Cooper, M.A. Activation-specific metabolic requirements for NK cell IFN-γ production. J. Immunol. 2015, 194, 1954. [Google Scholar] [CrossRef]
- Spiotto, M.T.; Taniguchi, C.M.; Klopp, A.H.; Colbert, L.E.; Lin, S.H.; Wang, L.; Frederick, M.J.; Osman, A.A.; Pickering, C.R.; Frank, S.J. Biology of the radio- and chemo-responsiveness in HPV malignancies. Semin. Radiat. Oncol. 2021, 31, 274–285. [Google Scholar] [CrossRef]
- Wang, W.; Erbe, A.K.; Alderson, K.A.; Phillips, E.; Gallenberger, M.; Gan, J.; Campana, D.; Hank, J.A.; Sondel, P.M. Human NK cells maintain licensing status and are subject to killer immunoglobulin-like receptor (KIR) and KIR-ligand inhibition following ex vivo expansion. Cancer Immunol. Immunother. CII 2016, 65, 1047–1059. [Google Scholar] [CrossRef]
- Wang, W.; Erbe, A.K.; Desantes, K.B.; Sondel, P.M. Donor selection for ex vivo-expanded natural killer cells as adoptive cancer immunotherapy. Future Oncol. 2017, 13, 1043–1047. [Google Scholar] [CrossRef]
- O’Sullivan, T.E.; Sun, J.C.; Lanier, L.L. Natural Killer Cell Memory. Immunity 2015, 43, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Vanherberghen, B.; Olofsson, P.E.; Forslund, E.; Sternberg-Simon, M.; Khorshidi, M.A.; Pacouret, S.; Guldevall, K.; Enqvist, M.; Malmberg, K.J.; Mehr, R.; et al. Classification of human natural killer cells based on migration behavior and cytotoxic response. Blood 2013, 121, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.L.; Finlay, D.K. Immunometabolism and natural killer cell responses. Nat. Rev. Immunol. 2019, 19, 282–290. [Google Scholar] [CrossRef]
- Lamb, R.; Bonuccelli, G.; Ozsvári, B.; Peiris-Pagès, M.; Fiorillo, M.; Smith, D.L.; Bevilacqua, G.; Mazzanti, C.M.; McDonnell, L.A.; Naccarato, A.G.; et al. Mitochondrial mass, a new metabolic biomarker for stem-like cancer cells: Understanding WNT/FGF-driven anabolic signaling. Oncotarget 2015, 6, 30453–30471. [Google Scholar] [CrossRef] [PubMed]
- Van Der Windt, G.J.; O’Sullivan, D.; Everts, B.; Huang, S.C.C.; Buck, M.D.; Curtis, J.D.; Chang, C.H.; Smith, A.M.; Ai, T.; Faubert, B.; et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl. Acad. Sci. USA 2013, 110, 14336–14341. [Google Scholar] [CrossRef]
- Duroux-Richard, I.; Roubert, C.; Ammari, M.; Présumey, J.; Grün, J.R.; Häupl, T.; Grützkau, A.; Lecellier, C.H.; Boitez, V.; Codogno, P.; et al. miR-125b controls monocyte adaptation to inflammation through mitochondrial metabolism and dynamics. Blood 2016, 128, 3125–3136. [Google Scholar] [CrossRef]
- Wang, Z.; Guan, D.; Wang, S.; Chai, L.Y.A.; Xu, S.; Lam, K.P. Glycolysis and Oxidative Phosphorylation Play Critical Roles in Natural Killer Cell Receptor-Mediated Natural Killer Cell Functions. Front. Immunol. 2020, 11, 202. [Google Scholar] [CrossRef]
- Reed-McBain, C.A.; Turaga, R.V.; Zima, S.R.T.; Abizanda Campo, S.; Riendeau, J.; Contreras Guzman, E.; Juang, T.D.; Juang, D.S.; Hampton, D.W.; Skala, M.C.; et al. Microfluidic device with reconfigurable spatial temporal gradients reveals plastic astrocyte response to stroke and reperfusion. Lab Chip 2023, 23, 3945–3960. [Google Scholar] [CrossRef]
- Reed-McBain, C.; Turaga, R.V.; Zima, S.R.T.; Patel, J.; Cunha, A.W.F.; Mixdorf, J.; Wehner, L.E.; Engle, J.W.; Hernandez, R.; Rehen, S.K.; et al. Non-destructive luminescence and PET imaging to monitor tissue microenvironment in microphysiological systems during brain metastasis using dissociated cerebral organoids. Biofabrication 2025. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turaga, R.V.; Zima, S.R.T.; Peterson, E.P.; Erbe, A.K.; Forsberg, M.H.; Capitini, C.M.; Cosper, P.F.; Sondel, P.M.; Ayuso, J.M. Analysis of Phenotypic and Molecular Variability of Memory-like NK Cells for Cancer Adoptive Cell Therapy Screening. Cancers 2025, 17, 2288. https://doi.org/10.3390/cancers17142288
Turaga RV, Zima SRT, Peterson EP, Erbe AK, Forsberg MH, Capitini CM, Cosper PF, Sondel PM, Ayuso JM. Analysis of Phenotypic and Molecular Variability of Memory-like NK Cells for Cancer Adoptive Cell Therapy Screening. Cancers. 2025; 17(14):2288. https://doi.org/10.3390/cancers17142288
Chicago/Turabian StyleTuraga, Rithvik V., Seth R. T. Zima, Ella P. Peterson, Amy K. Erbe, Matthew H. Forsberg, Christian M. Capitini, Pippa F. Cosper, Paul M. Sondel, and Jose M. Ayuso. 2025. "Analysis of Phenotypic and Molecular Variability of Memory-like NK Cells for Cancer Adoptive Cell Therapy Screening" Cancers 17, no. 14: 2288. https://doi.org/10.3390/cancers17142288
APA StyleTuraga, R. V., Zima, S. R. T., Peterson, E. P., Erbe, A. K., Forsberg, M. H., Capitini, C. M., Cosper, P. F., Sondel, P. M., & Ayuso, J. M. (2025). Analysis of Phenotypic and Molecular Variability of Memory-like NK Cells for Cancer Adoptive Cell Therapy Screening. Cancers, 17(14), 2288. https://doi.org/10.3390/cancers17142288





