Correction: Ye et al. The Mechanisms of lncRNA-Mediated Multidrug Resistance and the Clinical Application Prospects of lncRNAs in Breast Cancer. Cancers 2022, 14, 2101
Reference Updates:
Figure Revisions:
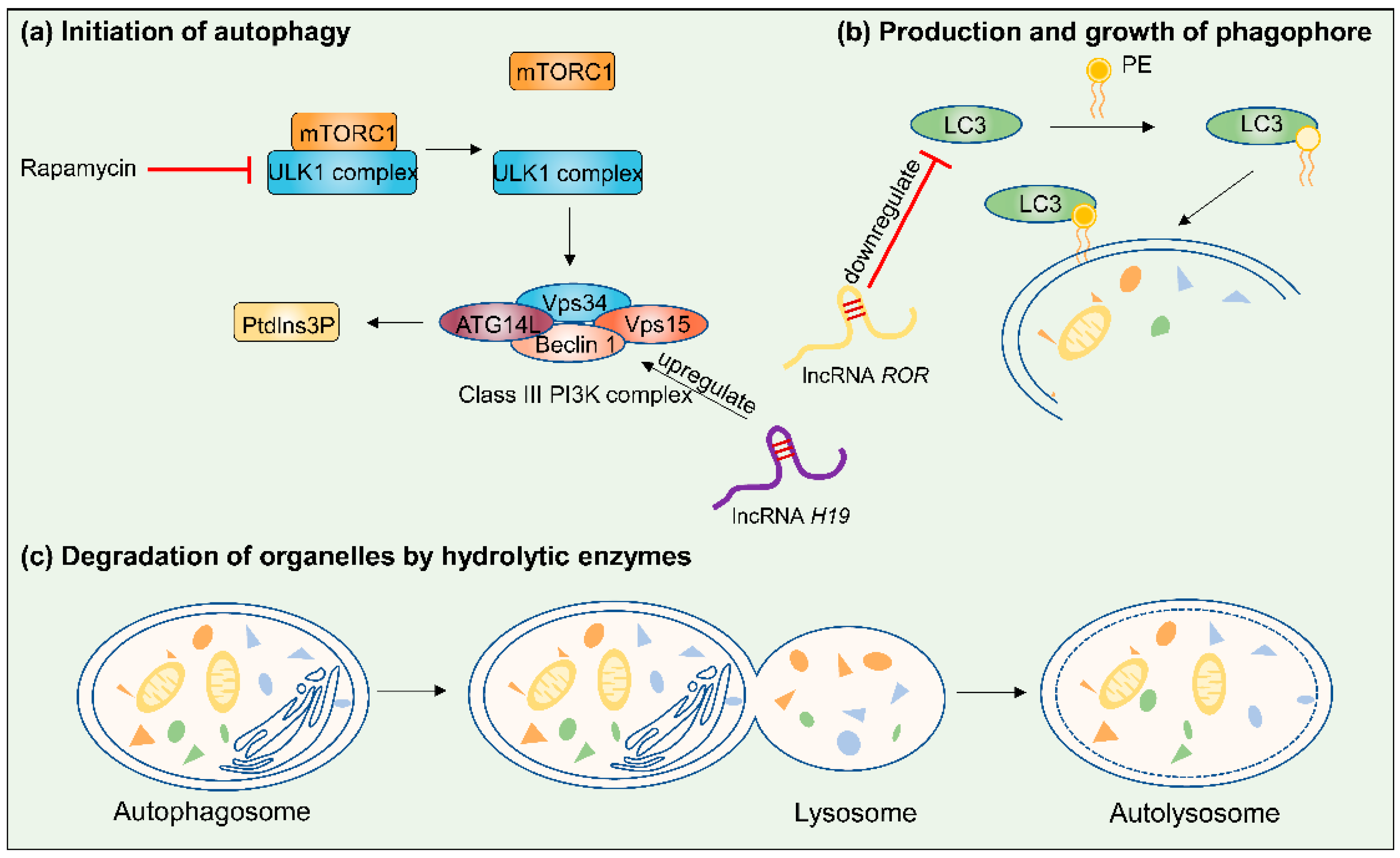
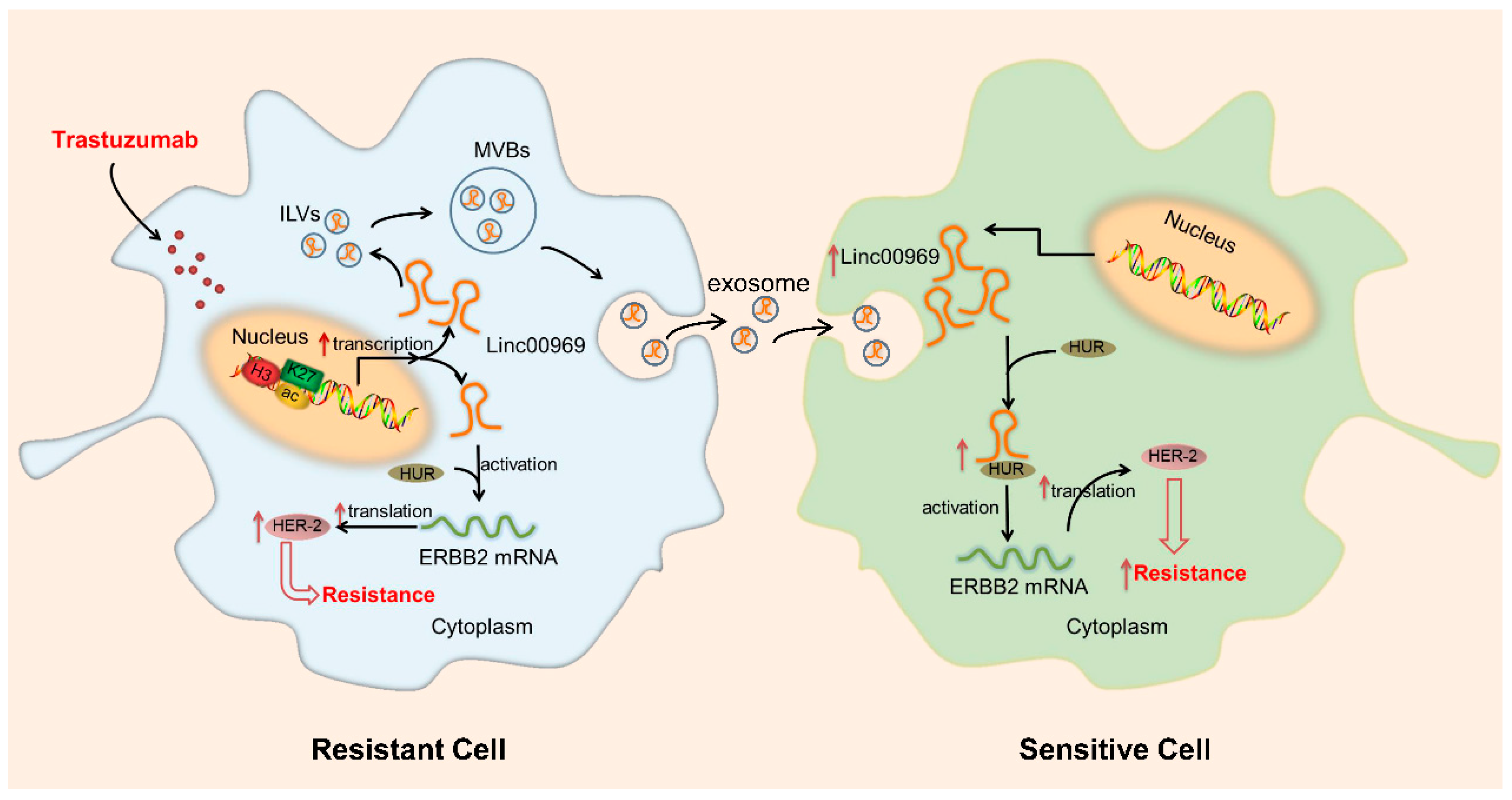
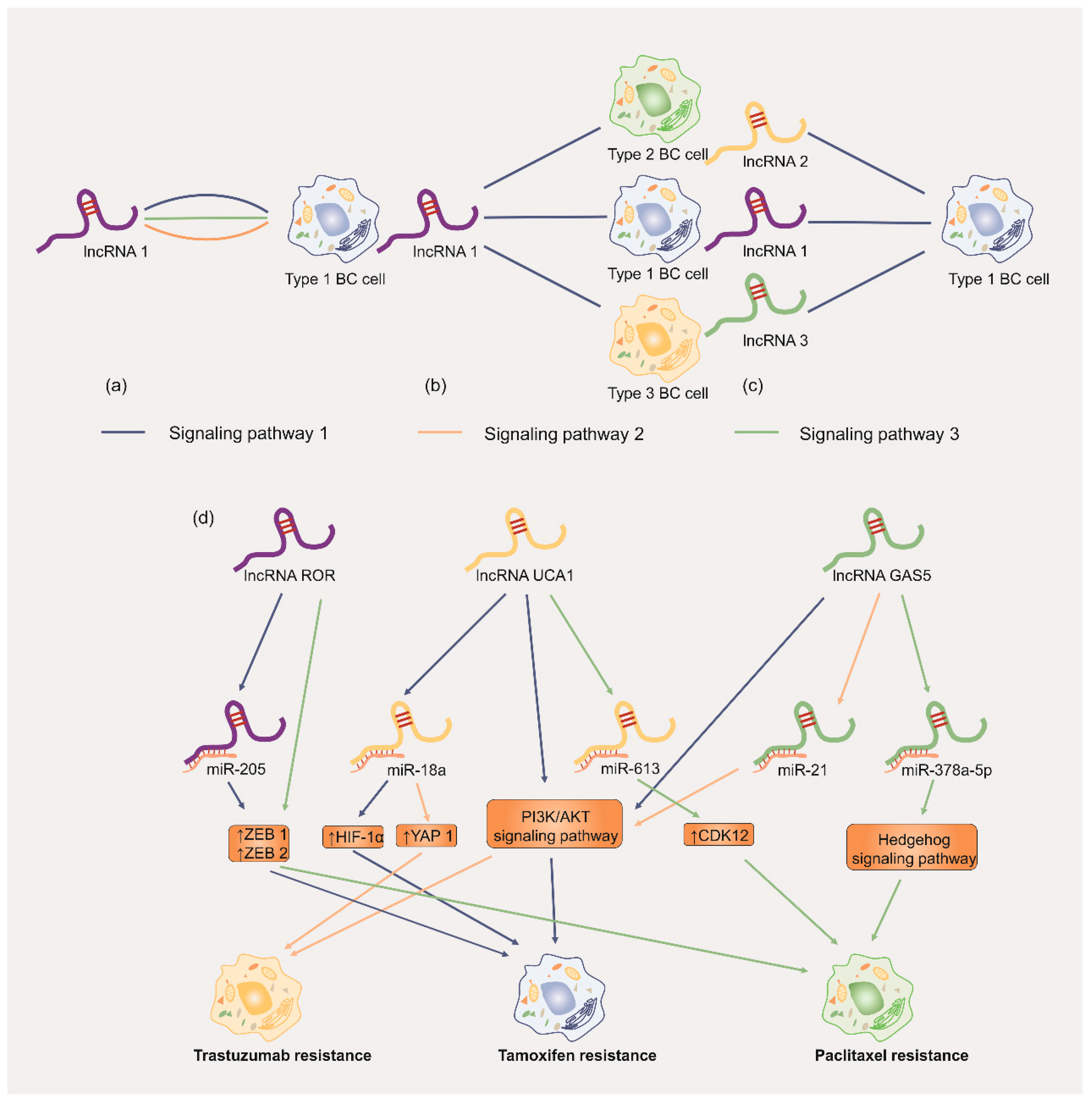
Table Revisions:
| Function | LncRNA | Type | Genomic Location | Expression Level * | Resistant Drugs | Cell Lines | Possible Mechanism § | References |
|---|---|---|---|---|---|---|---|---|
| Suppressing apoptosis | GAS5 | Tumor suppressor | chr1q25.1 | ↓ | paclitaxel; cisplatin | MDA-MB-231; BT549 | ↑ miR-378a-5p/↓ SUFU signaling | [49] |
| MEG3 | Tumor suppressor | chr14q32 | ↓ | doxorubicin; paclitaxel | Hs578T; MCF-7; MDA-MB-231 | ↑ TGF-β and N-cadherin protein; ↓ MMP 2, ZEB 1 and COL3A1 expression; ↓ miR-4513/↑ PBLD axis | [50,51] | |
| PTENP1 | Tumor suppressor | N/A ° | ↓ | adriamycin | MDA-MB-231; T-47D; MCF-7 | ↑ miR-20a/↓ PTEN axis; ↑ PI3K/AKT pathway | [52] | |
| UCA1 | Oncogene | chr19q13.12 | ↑ | tamoxifen | MCF-7; T-47D; LCC2; LCC9 | ↑ EZH2/↓ p21 axis; ↑ PI3K/AKT pathway; ↑ mTOR pathway | [53,54] | |
| H19 | Oncogene | chr11p15.5 | ↑ | paclitaxel | MDA-MB-453; MDA-MB-157; MDA-MB-231; ZR-75-1; MCF-7 | ↑ AKT pathway; ↓ BIK; ↓ NOXA | [47,48] | |
| PRLB | Oncogene | chr8p11.21 | ↑ | 5-fluorouracil | MDA-MB-231 | ↓ miR-4766-5p/↑ SIRT1 axis | [55] | |
| LINP1 | Oncogene | chr10 | ↑ | doxorubicin;5-fluorouracil | MDA-MB-231; MDA-MB-468; MCF-7 | ↓ p53; ↓ E-cadherin; ↑ N-cadherin; ↑ vimentin; ↓ caspase9/Bax | [56] | |
| LOC645166 | Oncogene | N/A | ↑ | adriamycin | MDA-MB-231; MCF-7 | ↑ NF-κB/GATA3 axis | [57] | |
| Autophagy | EGOT | Tumor suppressor | N/A | ↓ | paclitaxel | MCF-7; T-47D; UACC-812; SK-BR-3; HCC70; MDA-MB-453; MDA-MB-231; MDA-MB-468; BT549; Hs578T | ↑ ITPR1 | [58] |
| ROR | Oncogene | chr18q21.31 | ↑ | tamoxifen | BT474 | ↑ MDR1 and GST-π mRNA; ↓ LC3 and Beclin 1 | [59] | |
| H19 | Oncogene | chr11p15.5 | ↑ | tamoxifen | MCF-7 | H19/SAHH/DNMT3B axis; ↑ Beclin1 | [60] | |
| ZNF649-AS1 | Oncogene | chr19q13.41 | ↑ | trastuzumab | SK-BR-3; BT474 | ↑ ATG5 through associating with PTBP1 | [61] | |
| ASAH2B-2 | Oncogene | N/A | ↑ | everolimus | BT474; MCF-7 | ↑ mTOR pathway | [62] | |
| DNA repair | HCP5 | Oncogene | N/A | ↑ | cisplatin | MDA-MB-231 | ↓ PTEN | [63] |
| PTENP1 | Tumor suppressor | N/A | ↓ | adriamycin | MDA-MB-231; T-47D; MCF-7 | ↑ miR-20a/↓ PTEN axis; ↑ PI3K/AKT pathway | [52] | |
| GAS5 | Tumor suppressor | chr1q25.1 | ↓ | tamoxifen | MCF-7 | ↑ AKT/mTOR pathway; ↓ PTEN | [64] | |
| UCA1 | Oncogene | chr19q13.12 | ↑ | trastuzumab | SKBR-3 | ↓ miR-18a/↑ Yes-associated protein 1 (YAP1); ↓ PTEN; ↑ CD6 | [65] | |
| UCA1 | Oncogene | chr19q13.12 | ↑ | paclitaxel | MCF-7 | ↓ miR-613/↑ CDK12 axis | [66] | |
| GAS5 | Tumor suppressor | chr1q25.1 | ↓ | trastuzumab; lapatinib | SKBR-3 | ↑ miR-21; ↓ PTEN; ↑ mTOR; ↑ Ki-67 | [67] | |
| LINC-PINT | Tumor suppressor | N/A | ↓ | paclitaxel | MDA-MB-231; BT-20 | ↑ NONO | [68] | |
| H19 | Oncogene | chr11p15.5 | ↑ | doxorubicin | MCF-7 | ↓ PARP1 | [69] | |
| lncMat2B | Oncogene | N/A | ↑ | cisplatin | MDA-MB-231; MCF-7 | N/A | [70] | |
| ADAMTS9-AS2 | Tumor suppressor | N/A | ↓ | tamoxifen | MCF-7 | ↑ microRNA-130a-5p; ↓ PTEN | [71] |
| Function | LncRNA | Type | Genomic Location | Expression Level * | Resistant Drugs | Cell Lines | Possible Mechanism § | References |
|---|---|---|---|---|---|---|---|---|
| regulating cell cycle | TMPO-AS1 | Oncogene | N/A ° | ↑ | tamoxifen | MCF-7 | stabilize ESR1 mRNA | [109] |
| CASC2 | Oncogene | N/A | ↑ | paclitaxel | MDA-MB-231; MCF-7 | ↓ miR-18a-5p/↑ CDK19 axis | [110] | |
| LINC00511 | Oncogene | chr17q24.3 | ↑ | paclitaxel | MDA-MB-231; MCF-7; T-47D; Hs-578T | ↓ miR-29c/↑ CDK6 axis | [104] | |
| NEAT1 | Oncogene | N/A | ↑ | cisplatin/taxol | MDA-MB-231 | N/A | [111] | |
| LOL | Oncogene | N/A | ↑ | tamoxifen | MCF-7 | ↓ let-7 miRNA; ↓ ERα signaling | [112] | |
| UCA1 | Oncogene | chr19q13.12 | ↑ | tamoxifen | MCF-7; T-47D; LCC2; LCC9; BT474 | ↑ EZH2/↓ p21 axis; ↑ PI3K/AKT pathway; ↓ miR-18a/↑ HIF1α | [54,113] | |
| DSCAM-AS1 | Oncogene | chr21q22.3 | ↑ | tamoxifen | MCF-7; T-47D; SK-BR-3; MDA-MB-231 | ↑ epidermal growth factor receptor pathway substrate 8 (EPS8); ↑ ESR1; ↑ ERα; ↓ miR-137 | [114,115] | |
| FTH1P3 | Oncogene | N/A | ↑ | paclitaxel | MCF-7; MDA-MB-231; MDA-MB-468; MDA-MB-453 | ↓ miR-206/↑ ABCB1 | [116] | |
| MAFG-AS1 | Oncogene | N/A | ↑ | tamoxifen | MCF-7; BT474; T-47D; MCF10A | ↓ miR-339-5p/↑ CDK2 axis | [117] | |
| PRLB | Oncogene | chr8p11.21 | ↑ | 5-fluorouracil | MDA-MB-231 | ↓ miR-4766-5p/↑ SIRT1 axis | [55] | |
| UCA1 | Oncogene | chr19q13.12 | ↑ | trastuzumab | SKBR-3 | ↓ miR-18a/↑ Yes-associated protein 1 (YAP1); ↓ PTEN; ↑ CD6 | [65] | |
| LINP1 | Oncogene | chr10 | ↑ | doxorubicin; 5-fluorouracil | MDA-MB-231; MDA-MB-468; MCF-7 | ↓ p53; ↓ E-cadherin; ↑ N-cadherin; ↑ vimentin; ↓ caspase9/Bax | [56] | |
| TROJAN | Oncogene | N/A | ↑ | palbociclib | MCF7; T47D | ↑ NKRF/CDK2 axis | [5] | |
| DILA1 | Oncogene | N/A | ↑ | tamoxifen | MCF-7; 293-T; T47D | ↑ Cyclin D1 | [4] | |
| ARA | Oncogene | Xq23 | ↑ | adriamycin | MCF-7 | multiple signaling pathways | [118] | |
| drug efflux metabolism | GAS5 | Tumor suppressor | chr1q25.1 | ↓ | adriamycin | MCF-7 | ↑ miR-221-3p/↑ Dickkopf 2 (DKK2) axis; ↑ Wnt/b-catenin pathway | [119] |
| BC032585 | Tumor suppressor | chr9 | ↓ | taxane; anthracyclines | MDA-MB-231 | ↑ MDR1 | [120] | |
| Linc00518 | Oncogene | chr6 | ↑ | multidrugadriamycin; vincristine; paclitaxel | MCF-7 | ↓ miR-199a/↑ MRP1 axis | [121] | |
| FTH1P3 | Oncogene | N/A | ↑ | paclitaxel | MCF-7; MDA-MB-231; MDA-MB-468; MDA-MB-453 | ↓ miR-206/↑ ABCB1 | [116] | |
| ROR | Oncogene | chr18q21.31 | ↑ | tamoxifen | BT474 | ↑ MDR1 and GST-π mRNA; ↓ LC3 and Beclin 1 | [59] | |
| H19 | Oncogene | chr11p15.5 | ↑ | doxorubicin; anthracyclines | MCF-7 | ↑ CUL4A-ABCB1/MDR1 pathway | [122] | |
| RP11-770J1.3TMEM25 | Oncogene | N/A | ↑ | paclitaxel | MCF-7 | ↑ MRP, BCRP and MDR1/P-gp | [123] | |
| EMT | LINP1 | Oncogene | chr10 | ↑ | tamoxifen | MCF-7; T-47D | ↓ ER expression signaling pathway | [124] |
| MEG3 | Tumor suppressor | chr14q32 | ↓ | doxorubicin | Hs578T | ↑ TGF-β and N-cadherin protein; ↓ MMP 2, ZEB 1 and COL3A1 expression | [50] | |
| NONHSAT101069 | Oncogene | chr5 | ↑ | epirubicin | MCF-7 | ↓ miR-129-5p/↑ Twist1 axis | [125] | |
| NEAT1 | Oncogene | N/A | ↑ | cisplatin/taxol | MDA-MB-231 | N/A | [111] | |
| H19 | Oncogene | chr11p15.5 | ↑ | tamoxifen; paclitaxel | SK-BR-3; MCF-7 | ↑ Wnt pathway; ↓ miR-340-3p/YWHAZ axis | [126,127] | |
| PRLB | Oncogene | chr8p11.21 | ↑ | 5-fluorouracil | MDA-MB-231 | ↓ miR-4766-5p/↑ SIRT1 | [55] | |
| LINC00894002 | Tumor suppressor | X chromosome | ↓ | tamoxifen | MCF-7 | ↓ miR200/↑ TGFβ2 signaling pathway; ↑ ZEB1 | [128] | |
| LINP1 | Oncogene | chr10 | ↑ | doxorubicin; 5-fluorouracil | MDA-MB-231; MDA-MB-468; MCF-7 | ↓ p53; ↓ E-cadherin; ↑ N-cadherin; ↑ vimentin; ↓ caspase9/Bax | [56] | |
| NEAT1 | Oncogene | N/A | ↑ | 5-fluorouracil | MCF-7; T-47D; MDA-MB-231; ZR-75-1 | ↓ miR-211/↑ HMGA2 axis | [129] | |
| ROR | Oncogene | chr18q21.31 | ↑ | tamoxifen | MDA-MB-231; MCF-7 | ↓ microRNA-205; ↓ E-cadherin; ↑ vimentin; ↑ ZEB1 and ZEB2 | [130] | |
| DLX6-AS1 | Oncogene | N/A | ↑ | cisplatin | HCC1599; MDA-MB-231; HCC1806; Hs578T | ↓ miR-199b-5p/paxillin signaling | [131] | |
| ROR | Oncogene | chr18q21.31 | ↑ | 5-fluorouracil; paclitaxel | T-47D; MCF-7; SK-BR-3; Bcap-37; MDA-MB-231; MCF10A | ↓ E-cadherin; ↑ vimentin and N-cadherin | [132] | |
| ATB | Oncogene | chr14q11.2 | ↑ | trastuzumab | SKBR-3 | ↓ miR-200c; ↑ TGF-β signaling; ↑ ZEB1 and ZNF-217 | [133] | |
| SNHG7 | Oncogene | chr9q34.3 | ↑ | trastuzumab; adriamycin; paclitaxel | SKBR3; AU565; MDA-MB-231; MCF10A; MCF-7 | ↓ miR-186; ↓ miR-34a | [134,135] | |
| DCST1-AS1 | Oncogene | N/A | ↑ | doxorubicin; paclitaxel | MDA-MB-231; BT-549; T-47D; MCF-7 | ↑ TGF-β/Smad signaling through ANXA1 | [136] | |
| epigenetic alteration | LINC00969 | Oncogene | N/A | ↑ | trastuzumab | SKBR-3; BT474 | ↑ translation and stability of ERBB2 mRNA | [137] |
| TMPO-AS1 | Oncogene | N/A | ↑ | tamoxifen | MCF-7 | stabilize ESR1 mRNA | [109] | |
| ZNF649-AS1 | Oncogene | chr19q13.41 | ↑ | trastuzumab | SK-BR-3; BT474 | ↑ ATG5 through associating with PTBP1 | [61] | |
| MIR2052HG | Oncogene | N/A | ↑ | aromatase inhibitor | MDA-MB-231; CAMA-1; Au565; 293-T; MCF-7 | ↑ LMTK3; ↓ AKT/FOXO3-mediated ESR1 transcription; ↓ PKC/MEK/ERK/RSK1 pathway; ↓ ERα degradation | [138] | |
| LINC00472 | Tumor suppressor | N/A | ↓ | tamoxifen | MCF-7; T-47D; MDA-MB-231; Hs578T | ↑ phosphorylation NF-κB | [139] | |
| UCA1 | Oncogene | chr19q13.12 | ↑ | tamoxifen | MCF-7; T-47D; LCC2; LCC9 | ↑ EZH2/↓ p21 axis; ↑ PI3K/AKT pathway | [54] | |
| H19 | Oncogene | chr11p15.5 | ↑ | tamoxifen; fulverstrant | LCC2; LCC9; MCF-7 | ↑ ERα; ↑ Notch, HGF and c-MET signaling | [140] | |
| BORG | Oncogene | N/A | ↑ | doxorubicin | D2.OR; 67NR; 4T07; 4T1 | ↑ NF-κB signaling; ↑ RPA1 | [141] | |
| SNHG14 | Oncogene | chr15q11.2 | ↑ | trastuzumab | SKBR-3; BT474 | ↑ PABPC1; ↑ Nrf2 pathway | [142] | |
| MAPT-AS1 | Oncogene | chr17q21.31 | ↑ | paclitaxel | MDA-MB-231; MDA-MB-468 | ↑ MAPT mRNA | [143] | |
| Linc-RoR | Oncogene | N/A | ↑ | tamoxifen | MCF-7 | ↑ MAPK/ERK signaling; ↑ ER signaling; ↓ DUSP7 | [144] | |
| HOTAIR | Oncogene | chr12q13.13 | ↑ | tamoxifen; TNF-a | MCF-7; T-47D | ↑ ER signaling; ↑ SRC and p38MAPK kinases; ↑ EZH2 | [145,146] | |
| H19 | Oncogene | chr11p15.5 | ↑ | paclitaxel | ZR-75-1; MCF-7 | ↓ BIK; ↓ NOXA | [48] | |
| BDNF-AS | Oncogene | chr11p14.1 | ↑ | tamoxifen | MCF-7; T-47D; MDA-MB-231 | ↑ RNH1/TRIM21/mTOR | [147] | |
| BCAR4 | Oncogene | chr16p13.13 | ↑ | tamoxifen | ZR-75-1 | ↑ ERBB2/ERBB3 pathway; ↑ AKT | [148] |
| LncRNA | Type | Genomic Location | Expression Level * | Resistant Drugs | Cell Lines | Possible Mechanism § | Reference |
|---|---|---|---|---|---|---|---|
| LINC00969 | Oncogene | N/A | ↑ | trastuzumab | SKBR-3; BT474 | ↑ translation and stability of ERBB2 mRNA | [137] |
| H19 | Oncogene | chr11p15.5 | ↑ | doxorubicin | MCF-7; MDA-MB-231 | N/A ° | [192] |
| HISLA | Oncogene | chr14q31.3 | ↑ | docetaxel | MDA-MB-231; BT-474; MDA-MB-468; MCF-7 | inhibit the hydroxylation and degradation of HIF-1α | [193] |
| AGAP2-AS1 | Oncogene | chr12q14.1 | ↑ | trastuzumab | SKBR-3; BT474 | N/A | [194] |
| UCA1 | Oncogene | chr19q13.12 | ↑ | tamoxifen | MCF-7; LCC2 | ↓ cleaved caspase-3 | [190] |
Text Revisions:
5.1.3. Activating DNA Repair
5.3. Drug Efflux
5.5. Epigenetic Modification
5.6. Modifying the TME via Exosomal lncRNAs
7.1. Association of lncRNAs and Patients with BC
Reference
- Ye, P.; Feng, L.; Shi, S.; Dong, C. The Mechanisms of lncRNA-Mediated Multidrug Resistance and the Clinical Application Prospects of lncRNAs in Breast Cancer. Cancers 2022, 14, 2101. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, P.; Feng, L.; Shi, S.; Dong, C. Correction: Ye et al. The Mechanisms of lncRNA-Mediated Multidrug Resistance and the Clinical Application Prospects of lncRNAs in Breast Cancer. Cancers 2022, 14, 2101. Cancers 2025, 17, 2179. https://doi.org/10.3390/cancers17132179
Ye P, Feng L, Shi S, Dong C. Correction: Ye et al. The Mechanisms of lncRNA-Mediated Multidrug Resistance and the Clinical Application Prospects of lncRNAs in Breast Cancer. Cancers 2022, 14, 2101. Cancers. 2025; 17(13):2179. https://doi.org/10.3390/cancers17132179
Chicago/Turabian StyleYe, Pingting, Lei Feng, Shuo Shi, and Chunyan Dong. 2025. "Correction: Ye et al. The Mechanisms of lncRNA-Mediated Multidrug Resistance and the Clinical Application Prospects of lncRNAs in Breast Cancer. Cancers 2022, 14, 2101" Cancers 17, no. 13: 2179. https://doi.org/10.3390/cancers17132179
APA StyleYe, P., Feng, L., Shi, S., & Dong, C. (2025). Correction: Ye et al. The Mechanisms of lncRNA-Mediated Multidrug Resistance and the Clinical Application Prospects of lncRNAs in Breast Cancer. Cancers 2022, 14, 2101. Cancers, 17(13), 2179. https://doi.org/10.3390/cancers17132179





