Challenges and Advantages of Using Spatially Resolved Lipidomics to Assess the Pathological State of Human Lung Tissue
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
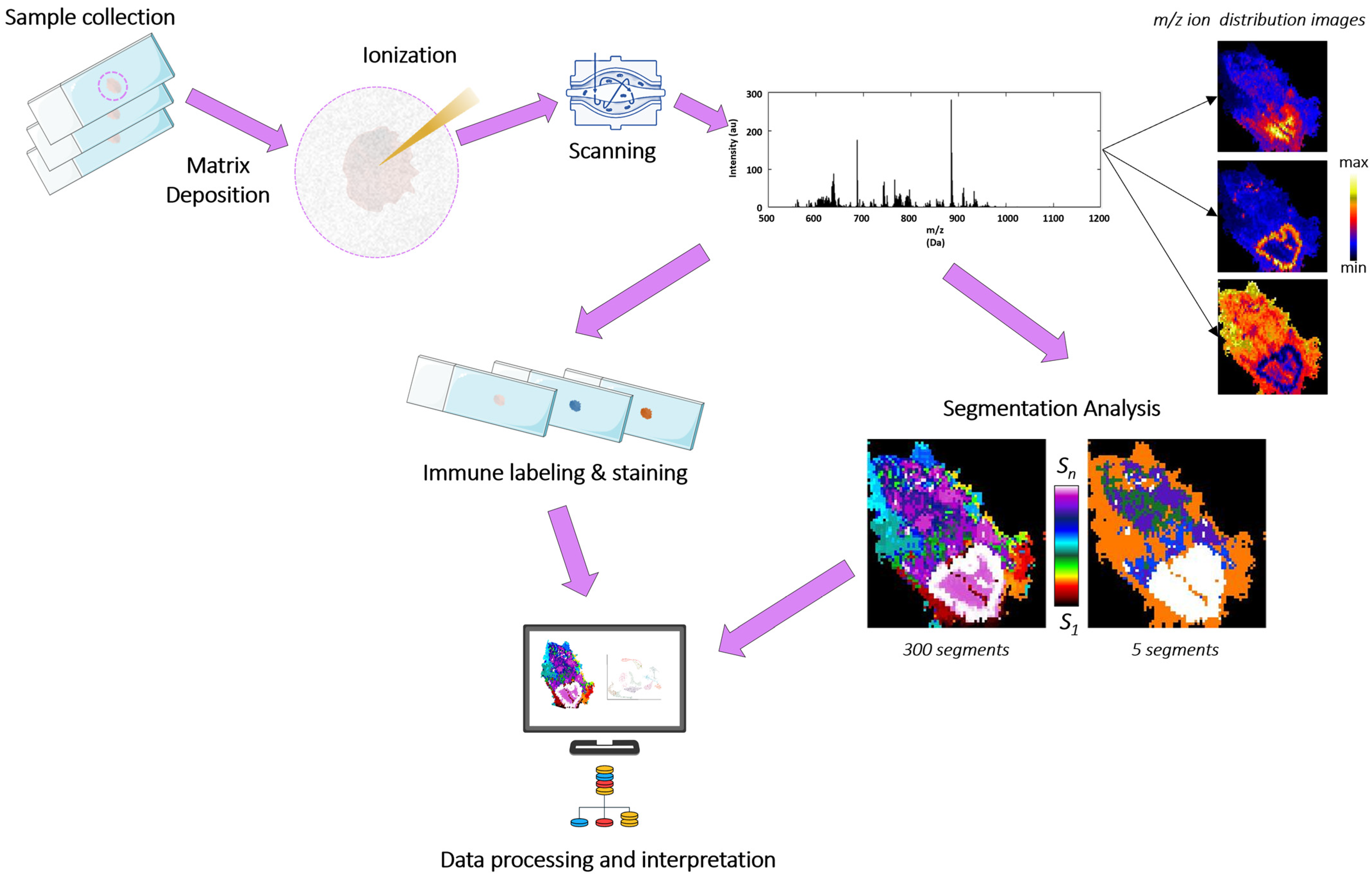
2.2. Sample Preparation for MALDI-Imaging, Measurement Conditions, and Data Analysis
2.3. Gene Set Enrichment Analysis
3. Results
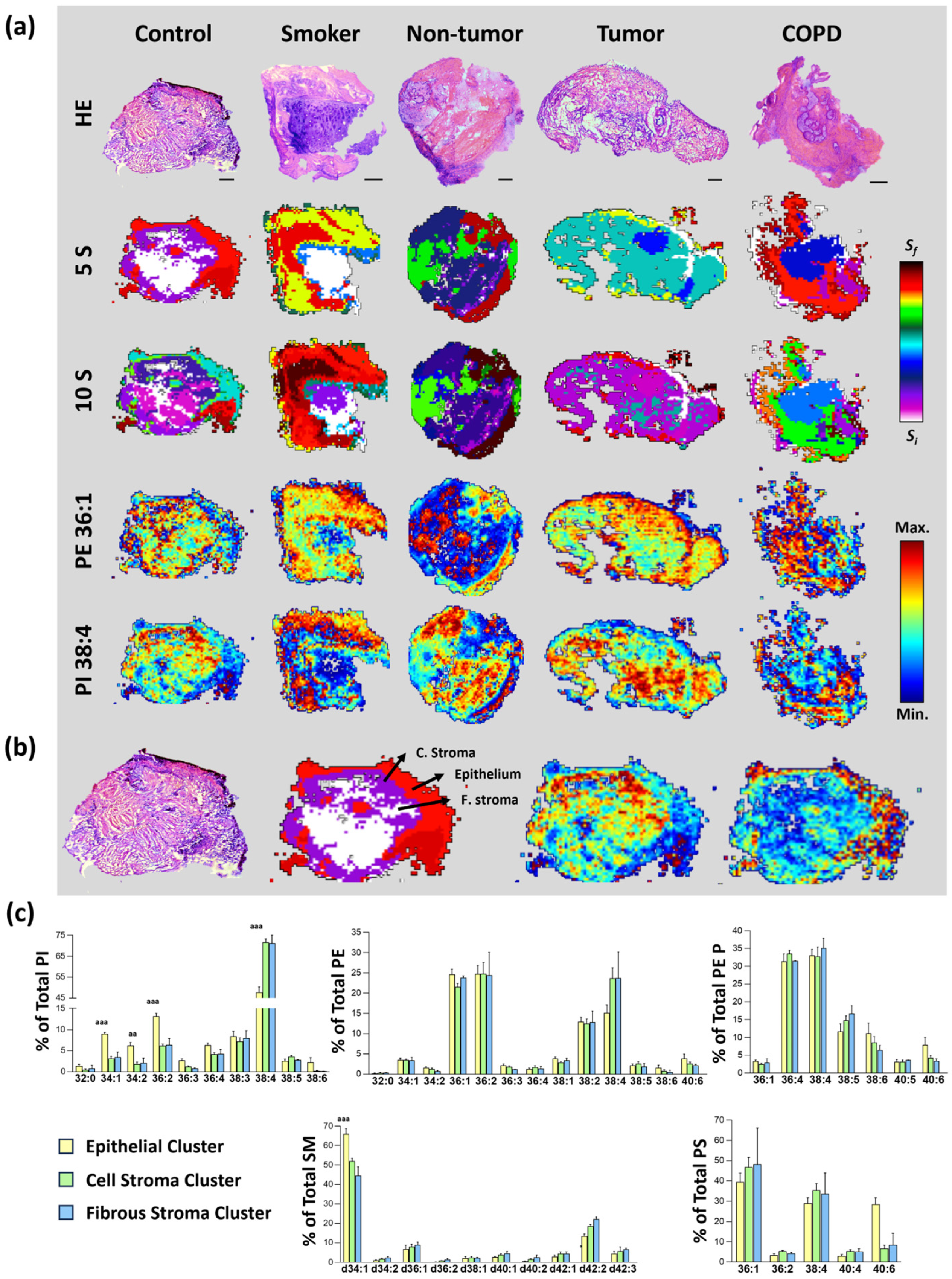
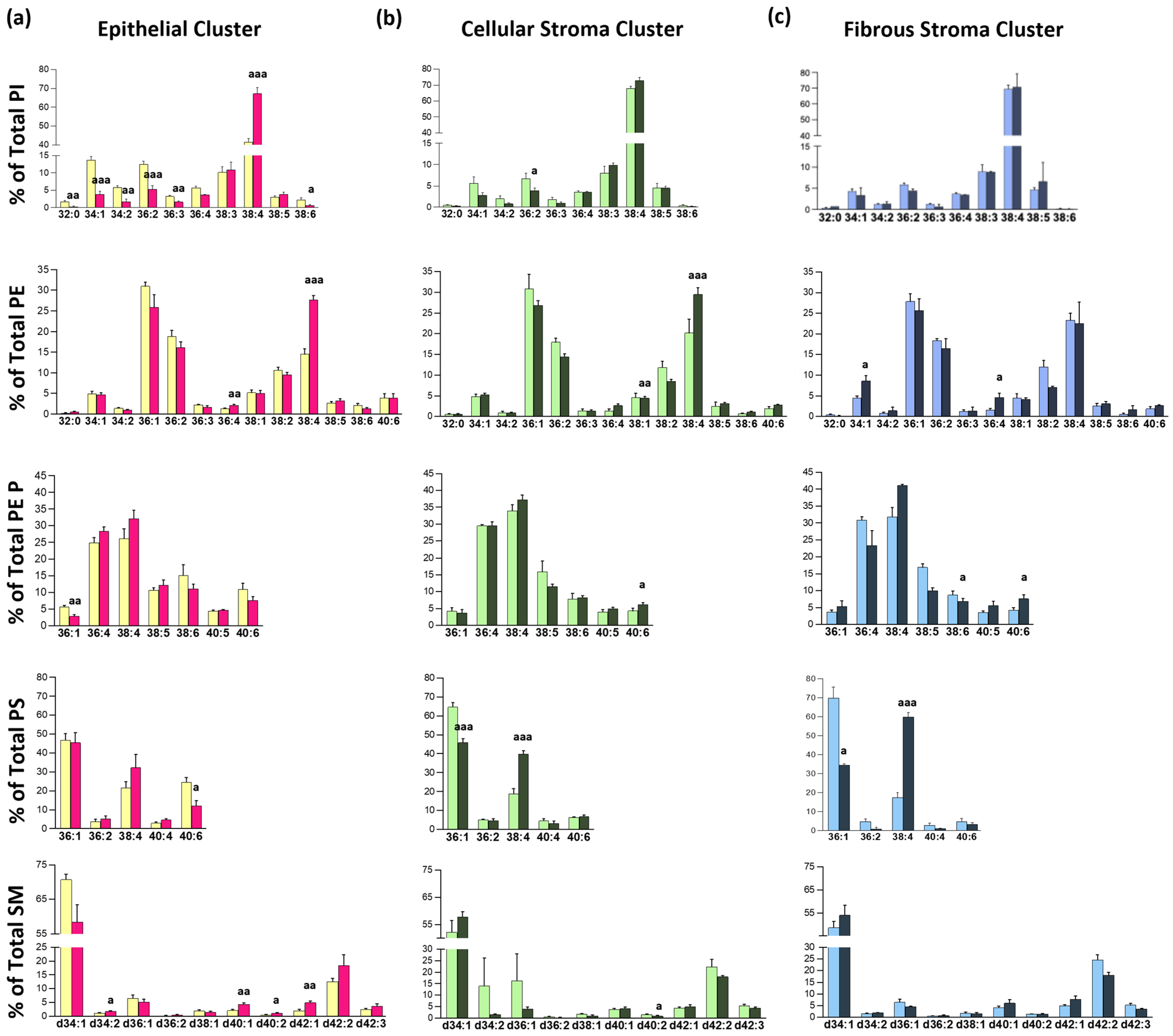
3.1. MALDI-MSI Lipid Segments Established Accurately Described the Tissue Architecture Found in Bronchoscopic Biopsies
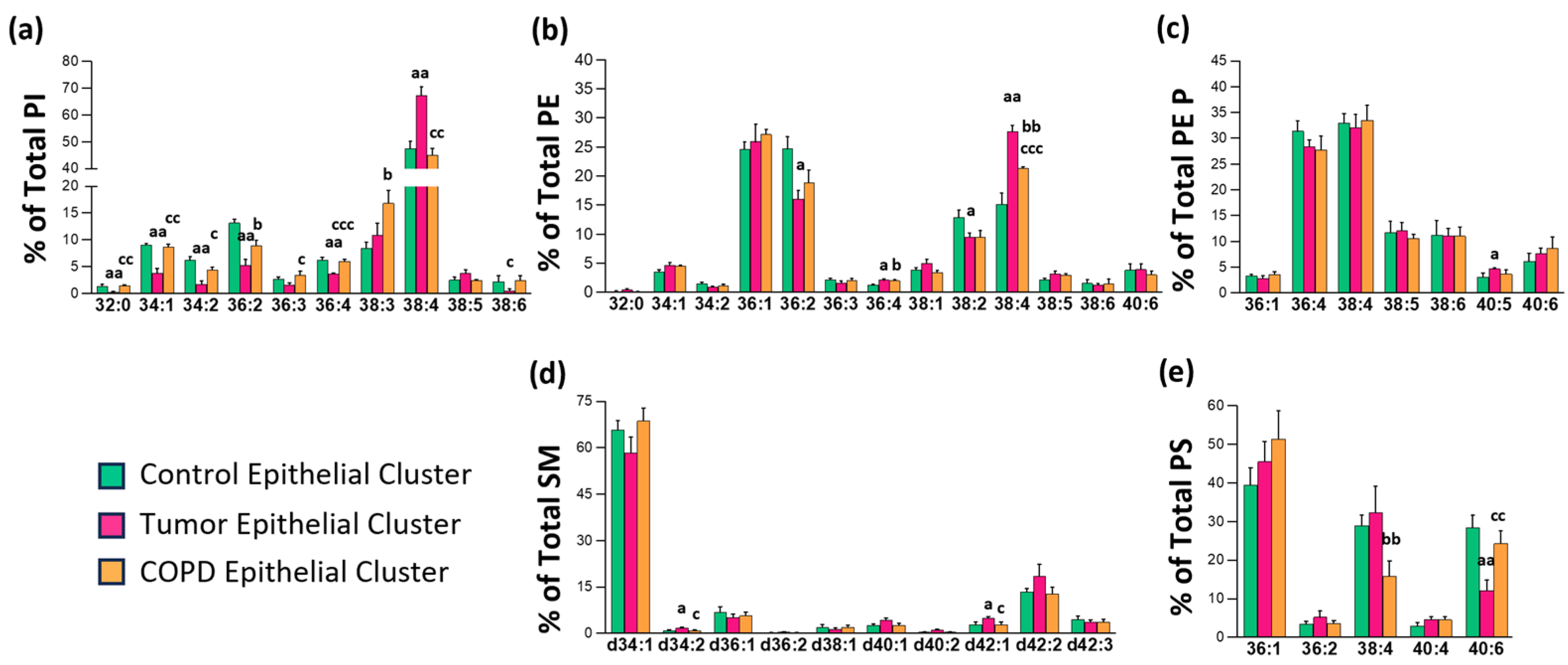
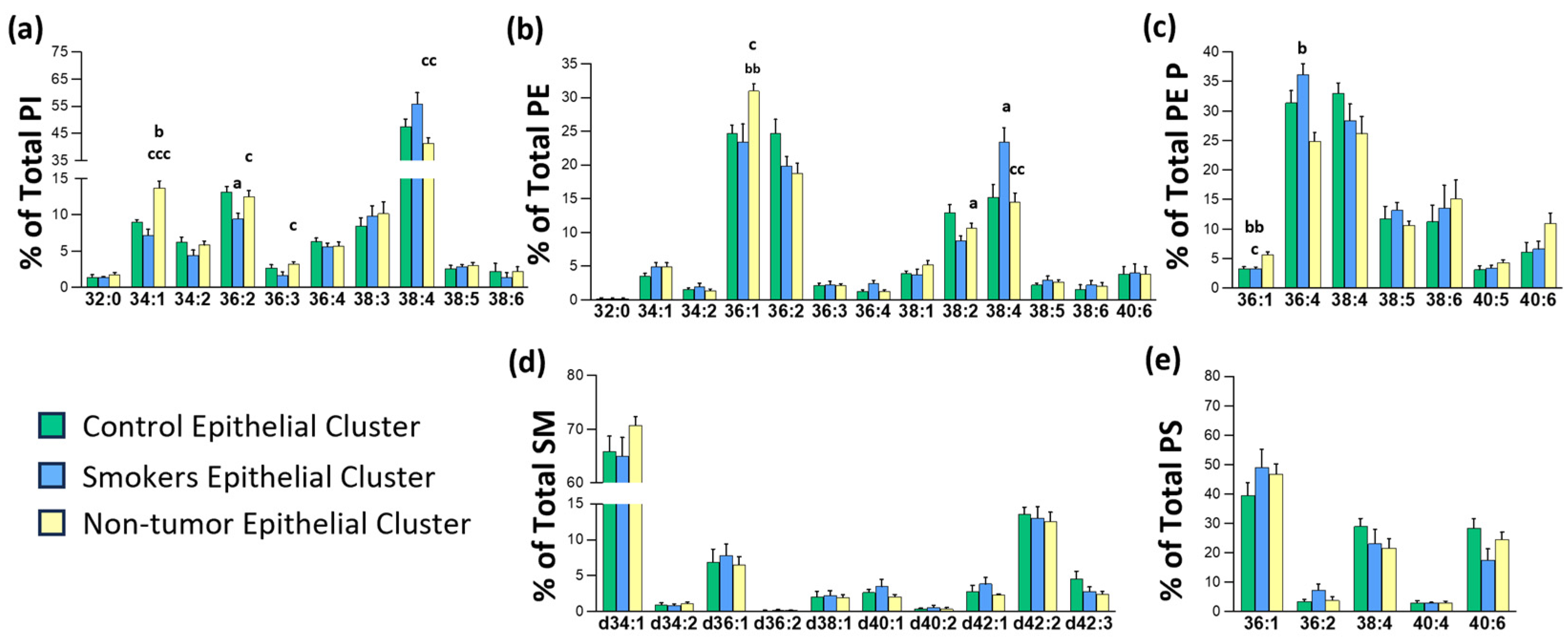
3.2. Identification of a Tissue-Type Dependent Response to Lung Malignization at the Lipid Profile Level
3.3. Impact of Chronic Inflammation on Healthy Lung Tissue
3.4. Changes in Lipid Composition in Non-Malignant Tissues
3.5. Lipid Metabolism Gene Expression Regulation in Non-Small Cell Lung Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | arachidonic acid |
| COPD | chronic obstructive pulmonary disease |
| DHA | docosahexaenoic acid |
| MALDI | matrix-assisted laser desorption/ionization |
| MSI | mass spectrometry imaging |
| PE | phosphatidylethanolamine |
| PE P | PE plasmalogens |
| PI | phosphatidylinositol |
| PS | phosphatidylserine |
| SM | sphingomyelin |
References
- Allam, M.; Cai, S.; Coskun, A.F. Multiplex bioimaging of single-cell spatial profiles for precision cancer diagnostics and therapeutics. npj Precis. Oncol. 2020, 4, 11. [Google Scholar] [CrossRef]
- Ivanova, P.T.; Milnse, S.B.; Myers, D.S.; Brown, H.A. Lipidomics: A mass spectrometry based systems level analysis of cellular lipids. Curr. Opin. Chem. Biol. 2009, 13, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Philipsen, M.H.; Phan, N.T.N.N.; Fletcher, J.S.; Malmberg, P.; Ewing, A.G. Mass Spectrometry Imaging Shows Cocaine and Methylphenidate Have Opposite Effects on Major Lipids in Drosophila Brain. ACS Chem. Neurosci. 2018, 9, 1462–1468. [Google Scholar] [CrossRef] [PubMed]
- Barceló-Coblijn, G.; Fernández, J.A. Mass spectrometry coupled to imaging techniques: The better the view the greater the challenge. Front. Physiol. 2015, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Hillenkamp, F.; Karas, M.; Beavis, R.C.; Chait, B.T. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry of Biopolymers. Anal. Chem. 1991, 63, 1193A–1203A. [Google Scholar] [CrossRef]
- Tanaka, K.; Waki, H.; Ido, Y.; Akita, S.; Yoshida, Y.; Yoshida, T.; Matsuo, T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988, 2, 151–153. [Google Scholar] [CrossRef]
- Maimó-Barceló, A.; Pérez-Romero, K.; Rodríguez, R.M.; Huergo, C.; Calvo, I.; Fernández, J.A.; Barceló-Coblijn, G. To image or not to image: Use of imaging mass spectrometry in biomedical lipidomics. Prog. Lipid Res. 2025, 97, 101319. [Google Scholar] [CrossRef]
- Bestard-Escalas, J.; Garate, J.; Maimó-Barceló, A.; Fernández, R.; Lopez, D.H.; Lage, S.; Reigada, R.; Khorrami, S.; Ginard, D.; Reyes, J.; et al. Lipid fingerprint image accurately conveys human colon cell pathophysiologic state: A solid candidate as biomarker. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2016, 1861, 1942–1950. [Google Scholar] [CrossRef]
- Lopez, D.H.; Bestard-Escalas, J.; Garate, J.; Maimó-Barceló, A.; Fernández, R.; Reigada, R.; Khorrami, S.; Ginard, D.; Okazaki, T.; Fernández, J.A.; et al. Tissue-selective alteration of ethanolamine plasmalogen metabolism in dedifferentiated colon mucosa. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2018, 1863, 928–938. [Google Scholar] [CrossRef]
- Maimó-Barceló, A.; Martín-Saiz, L.; Barceló-Nicolau, M.; Salivo, S.; Pérez-Romero, K.; Rodriguez, R.M.; Martín, J.; Martínez, M.A.; García, M.; Amengual, I.; et al. Lipid signature associated with chronic colon inflammation reveals a dysregulation in colonocyte differentiation process. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2024, 1869, 159528. [Google Scholar] [CrossRef]
- Marien, E.; Meister, M.; Muley, T.; del Pulgar, T.G.; Derua, R.; Spraggins, J.M.; Van de Plas, R.; Vanderhoydonc, F.; Machiels, J.; Binda, M.M.; et al. Phospholipid profiling identifies acyl chain elongation as a ubiquitous trait and potential target for the treatment of lung squamous cell carcinoma. Oncotarget 2016, 7, 12582–12597. [Google Scholar] [CrossRef]
- Lee, G.K.; Lee, H.S.; Park, Y.S.; Lee, J.H.; Lee, S.C.; Lee, J.H.; Lee, S.J.; Shanta, S.R.; Park, H.M.; Kim, H.R.; et al. Lipid MALDI profile classifies non-small cell lung cancers according to the histologic type. Lung Cancer 2012, 76, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Quaderi, S.A.; Hurst, J.R. The unmet global burden of COPD. Glob. Health Epidemiol. Genom. 2018, 3, e4. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Congleton, J.; Muers, M.F. The incidence of airflow obstruction in bronchial carcinoma, its relation to breathlessness, and response to bronchodilator therapy. Respir. Med. 1995, 89, 291–296. [Google Scholar] [CrossRef]
- Loganathan, R.S.; Stover, D.E.; Shi, W.; Venkatraman, E. Prevalence of COPD in women compared to men around the time of diagnosis of primary lung cancer. Chest 2006, 129, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Garate, J.; Martín-Saiz, L.; Galetich, I.; Fernández, J.A. Matrix Sublimation Device for MALDI Mass Spectrometry Imaging. Anal. Chem. 2019, 91, 803–807. [Google Scholar] [CrossRef]
- Astigarraga, E.; Barreda-Gómez, G.; Lombardero, L.; Fresnedo, O.; Castaño, F.; Giralt, M.T.; Ochoa, B.; Rodríguez-Puertas, R.; Fernández, J.A. Profiling and Imaging of Lipids on Brain and Liver Tissue by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Using 2-Mercaptobenzothiazole as a Matrix. Anal. Chem. 2008, 80, 9105–9114. [Google Scholar] [CrossRef]
- Garate, J.; Fernández, R.; Lage, S.; Bestard-Escalas, J.; Lopez, D.H.; Reigada, R.; Khorrami, S.; Ginard, D.; Reyes, J.; Amengual, I.; et al. Imaging mass spectrometry increased resolution using 2-mercaptobenzothiazole and 2,5-diaminonaphtalene matrices: Application to lipid distribution in human colon. Anal. Bioanal. Chem. 2015, 407, 4697–4708. [Google Scholar] [CrossRef]
- Xiong, X.; Xu, W.; Eberlin, L.S.; Wiseman, J.M.; Fang, X.; Jiang, Y.; Huang, Z.; Zhang, Y.; Cooks, R.G.; Ouyang, Z. Data Processing for 3D Mass Spectrometry Imaging. J. Am. Soc. Mass Spectrom. 2012, 23, 1147–1156. [Google Scholar] [CrossRef][Green Version]
- Fernández, R.; Garate, J.; Tolentino-Cortez, T.; Herraiz, A.; Lombardero, L.; Ducrocq, F.; Rodríguez-Puertas, R.; Trifilieff, P.; Astigarraga, E.; Barreda-Gómez, G.; et al. Microarray and Mass Spectrometry-Based Methodology for Lipid Profiling of Tissues and Cell Cultures. Anal. Chem. 2019, 91, 15967–15973. [Google Scholar] [CrossRef] [PubMed]
- Dombrowsky, H.; Clark, G.T.; Rau, G.A.; Bernhard, W.; Postle, A.D. Molecular Species Compositions of Lung and Pancreas Phospholipids in the cftrtm1HGU/tm1HGU Cystic Fibrosis Mouse. Pediatr. Res. 2003, 53, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Karnati, S.; Garikapati, V.; Liebisch, G.; Van Veldhoven, P.P.; Spengler, B.; Schmitz, G.; Baumgart-Vogt, E. Quantitative lipidomic analysis of mouse lung during postnatal development by electrospray ionization tandem mass spectrometry. PLoS ONE 2018, 13, e0203464. [Google Scholar] [CrossRef]
- Zemski Berry, K.A.; Murphy, R.C.; Kosmider, B.; Mason, R.J. Lipidomic characterization and localization of phospholipids in the human lung. J. Lipid Res. 2017, 58, 926–933. [Google Scholar] [CrossRef]
- Russo, P.S.T.; Ferreira, G.R.; Cardozo, L.E.; Bürger, M.C.; Arias-Carrasco, R.; Maruyama, S.R.; Hirata, T.D.C.; Lima, D.S.; Passos, F.M.; Fukutani, K.F.; et al. CEMiTool: A Bioconductor package for performing comprehensive modular co-expression analyses. BMC Bioinform. 2018, 19, 56. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Deprez, M.; Zaragosi, L.-E.; Truchi, M.; Becavin, C.; Ruiz García, S.; Arguel, M.-J.; Plaisant, M.; Magnone, V.; Lebrigand, K.; Abelanet, S.; et al. A Single-Cell Atlas of the Human Healthy Airways. Am. J. Respir. Crit. Care Med. 2020, 202, 1636–1645. [Google Scholar] [CrossRef]
- Marien, E.; Meister, M.; Muley, T.; Fieuws, S.; Bordel, S.; Derua, R.; Spraggins, J.; Van de Plas, R.; Dehairs, J.; Wouters, J.; et al. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int. J. Cancer 2015, 137, 1539–1548. [Google Scholar] [CrossRef]
- Gouw, A.M.; Eberlin, L.S.; Margulis, K.; Sullivan, D.K.; Toal, G.G.; Tong, L.; Zare, R.N.; Felsher, D.W. Oncogene KRAS activates fatty acid synthase, resulting in specific ERK and lipid signatures associated with lung adenocarcinoma. Proc. Natl. Acad. Sci. USA 2017, 114, 4300–4305. [Google Scholar] [CrossRef]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Ko, T.; Hatsuse, S.; Nomura, S.; Zhang, B.; Dai, Z.; Inoue, S.; Kubota, M.; Sawami, K.; Yamada, T.; et al. Spatiotemporal transcriptome analysis reveals critical roles for mechano-sensing genes at the border zone in remodeling after myocardial infarction. Nat. Cardiovasc. Res. 2022, 1, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Kundishora, A.J.; Allington, G.; McGee, S.; Mekbib, K.Y.; Gainullin, V.; Timberlake, A.T.; Nelson-Williams, C.; Kiziltug, E.; Smith, H.; Ocken, J.; et al. Multiomic analyses implicate a neurodevelopmental program in the pathogenesis of cerebral arachnoid cysts. Nat. Med. 2023, 29, 667–678. [Google Scholar] [CrossRef]
- Leader, A.M.; Grout, J.A.; Maier, B.B.; Nabet, B.Y.; Park, M.D.; Tabachnikova, A.; Chang, C.; Walker, L.; Lansky, A.; Le Berichel, J.; et al. Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell 2021, 39, 1594–1609.e12. [Google Scholar] [CrossRef] [PubMed]
- Salcher, S.; Sturm, G.; Horvath, L.; Untergasser, G.; Kuempers, C.; Fotakis, G.; Panizzolo, E.; Martowicz, A.; Trebo, M.; Pall, G.; et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell 2022, 40, 1503–1520.e8. [Google Scholar] [CrossRef]
- Schiller, H.B.; Montoro, D.T.; Simon, L.M.; Rawlins, E.L.; Meyer, K.B.; Strunz, M.; Vieira Braga, F.A.; Timens, W.; Koppelman, G.H.; Budinger, G.R.S.; et al. The Human Lung Cell Atlas: A High-Resolution Reference Map of the Human Lung in Health and Disease. Am. J. Respir. Cell Mol. Biol. 2019, 61, 31–41. [Google Scholar] [CrossRef]
- Bestard-Escalas, J.; Maimó-Barceló, A.; Pérez-Romero, K.; Lopez, D.H.; Barceló-Coblijn, G. Ins and Outs of Interpreting Lipidomic Results. J. Mol. Biol. 2019, 431, 5039–5062. [Google Scholar] [CrossRef]
- Veloso, A.; Fernández, R.; Astigarraga, E.; Barreda-Gómez, G.; Manuel, I.; Giralt, M.T.; Ferrer, I.; Ochoa, B.; Rodríguez-Puertas, R.; Fernández, J.A. Distribution of lipids in human brain. Anal. Bioanal. Chem. 2011, 401, 89–101. [Google Scholar] [CrossRef]
- Mendis, L.H.S.; Grey, A.C.; Faull, R.L.M.; Curtis, M.A. Hippocampal lipid differences in Alzheimer’s disease: A human brain study using matrix-assisted laser desorption/ionization-imaging mass spectrometry. Brain Behav. 2016, 6, e00517. [Google Scholar] [CrossRef]
- Hart, P.J.; Francese, S.; Claude, E.; Woodroofe, M.N.; Clench, M.R. MALDI-MS imaging of lipids in ex vivo human skin. Anal. Bioanal. Chem. 2011, 401, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Margulis, K.; Chiou, A.S.; Aasi, S.Z.; Tibshirani, R.J.; Tang, J.Y.; Zare, R.N. Distinguishing malignant from benign microscopic skin lesions using desorption electrospray ionization mass spectrometry imaging. Proc. Natl. Acad. Sci. USA 2018, 115, 6347–6352. [Google Scholar] [CrossRef]
- Garate, J.; Lage, S.; Fernández, R.; Velasco, V.; Abad, B.; Asumendi, A.; Gardeazabal, J.; Arroyo-Berdugo, Y.; Rodríguez, M.Á.; Artola, J.L.; et al. Imaging Mass Spectrometry–Based Lipidomic Approach to Classification of Architectural Features in Nevi. J. Investig. Dermatol. 2019, 139, 2055–2058.e7. [Google Scholar] [CrossRef]
- Martín-Saiz, L.; Mosteiro, L.; Solano-Iturri, J.D.; Rueda, Y.; Martín-Allende, J.; Imaz, I.; Olano, I.; Ochoa, B.; Fresnedo, O.; Fernández, J.A.; et al. High-Resolution Human Kidney Molecular Histology by Imaging Mass Spectrometry of Lipids. Anal. Chem. 2021, 93, 9364–9372. [Google Scholar] [CrossRef]
- Ide, Y.; Waki, M.; Hayasaka, T.; Nishio, T.; Morita, Y.; Tanaka, H.; Sasaki, T.; Koizumi, K.; Matsunuma, R.; Hosokawa, Y.; et al. Human Breast Cancer Tissues Contain Abundant Phosphatidylcholine(36:1) with High Stearoyl-CoA Desaturase-1 Expression. PLoS ONE 2013, 8, e61204. [Google Scholar] [CrossRef]
- Tata, A.; Woolman, M.; Ventura, M.; Bernards, N.; Ganguly, M.; Gribble, A.; Shrestha, B.; Bluemke, E.; Ginsberg, H.J.; Vitkin, A.; et al. Rapid Detection of Necrosis in Breast Cancer with Desorption Electrospray Ionization Mass Spectrometry. Sci. Rep. 2016, 6, 35374. [Google Scholar] [CrossRef] [PubMed]
- Berry, K.A.Z.; Li, B.; Reynolds, S.D.; Barkley, R.M.; Gijón, M.A.; Hankin, J.A.; Henson, P.M.; Murphy, R.C. MALDI imaging MS of phospholipids in the mouse lung. J. Lipid Res. 2011, 52, 1551–1560. [Google Scholar] [CrossRef]
- Wang, G.; Qiu, M.; Xing, X.; Zhou, J.; Yao, H.; Li, M.; Yin, R.; Hou, Y.; Li, Y.; Pan, S.; et al. Lung cancer scRNA-seq and lipidomics reveal aberrant lipid metabolism for early-stage diagnosis. Sci. Transl. Med. 2022, 14, eabk2756. [Google Scholar] [CrossRef]
- Fernández, L.P.; Gómez de Cedrón, M.; Ramírez de Molina, A. Alterations of Lipid Metabolism in Cancer: Implications in Prognosis and Treatment. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Hao, Y.; Li, D.; Xu, Y.; Ouyang, J.; Wang, Y.; Zhang, Y.; Li, B.; Xie, L.; Qin, G. Investigation of lipid metabolism dysregulation and the effects on immune microenvironments in pan-cancer using multiple omics data. BMC Bioinform. 2019, 20, 195. [Google Scholar] [CrossRef]
- Durham, A.L.; Adcock, I.M. The relationship between COPD and lung cancer. Lung Cancer 2015, 90, 121–127. [Google Scholar] [CrossRef]
- Woolhouse, I.S.; Bayley, D.L.; Stockley, R.A. Sputum chemotactic activity in chronic obstructive pulmonary disease: Effect of alpha(1)-antitrypsin deficiency and the role of leukotriene B(4) and interleukin 8. Thorax 2002, 57, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Montuschi, P.; Kharitonov, S.A.; Ciabattoni, G.; Barnes, P.J. Exhaled leukotrienes and prostaglandins in COPD. Thorax 2003, 58, 585–588. [Google Scholar] [CrossRef]
- Lukowski, J.K.; Olson, H.; Velickovic, M.; Wang, J.; Kyle, J.E.; Kim, Y.-M.; Williams, S.M.; Zhu, Y.; Huyck, H.L.; McGraw, M.D.; et al. An optimized approach and inflation media for obtaining complimentary mass spectrometry-based omics data from human lung tissue. Front. Mol. Biosci. 2022, 9, 1022775. [Google Scholar] [CrossRef]
- Ellis, S.R.; Hall, E.; Panchal, M.; Flinders, B.; Madsen, J.; Koster, G.; Heeren, R.M.A.; Clark, H.W.; Postle, A.D. Mass spectrometry imaging of phosphatidylcholine metabolism in lungs administered with therapeutic surfactants and isotopic tracers. J. Lipid Res. 2021, 62, 100023. [Google Scholar] [CrossRef] [PubMed]
- Thiele, C.; Wunderling, K.; Leyendecker, P. Multiplexed and single cell tracing of lipid metabolism. Nat. Methods 2019, 16, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Yagnik, G.; Liu, Z.; Rothschild, K.J.; Lim, M.J. Highly Multiplexed Immunohistochemical MALDI-MS Imaging of Biomarkers in Tissues. J. Am. Soc. Mass Spectrom. 2021, 32, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–862. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calvo, I.; Maimó-Barceló, A.; Garate, J.; Bestard-Escalas, J.; Scrimini, S.; Sauleda, J.; Cosío, B.G.; Fernández, J.A.; Barceló-Coblijn, G. Challenges and Advantages of Using Spatially Resolved Lipidomics to Assess the Pathological State of Human Lung Tissue. Cancers 2025, 17, 2160. https://doi.org/10.3390/cancers17132160
Calvo I, Maimó-Barceló A, Garate J, Bestard-Escalas J, Scrimini S, Sauleda J, Cosío BG, Fernández JA, Barceló-Coblijn G. Challenges and Advantages of Using Spatially Resolved Lipidomics to Assess the Pathological State of Human Lung Tissue. Cancers. 2025; 17(13):2160. https://doi.org/10.3390/cancers17132160
Chicago/Turabian StyleCalvo, Ibai, Albert Maimó-Barceló, Jone Garate, Joan Bestard-Escalas, Sergio Scrimini, Jaume Sauleda, Borja G. Cosío, José Andrés Fernández, and Gwendolyn Barceló-Coblijn. 2025. "Challenges and Advantages of Using Spatially Resolved Lipidomics to Assess the Pathological State of Human Lung Tissue" Cancers 17, no. 13: 2160. https://doi.org/10.3390/cancers17132160
APA StyleCalvo, I., Maimó-Barceló, A., Garate, J., Bestard-Escalas, J., Scrimini, S., Sauleda, J., Cosío, B. G., Fernández, J. A., & Barceló-Coblijn, G. (2025). Challenges and Advantages of Using Spatially Resolved Lipidomics to Assess the Pathological State of Human Lung Tissue. Cancers, 17(13), 2160. https://doi.org/10.3390/cancers17132160







