Aberrant Expression of BTLA, CD160, SPN, TIM-3, VISTA and TIGIT in Chronic Lymphocytic Leukemia and Psoriasis Patients Compared to Healthy Volunteers
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Isolation
2.2. RNA Isolation and Reverse Transcription Reaction (RT)
2.3. Assessment of BTLA, CD160, SPN, TIM-3, VISTA and TIGIT mRNA Expression Using Real-Time Reverse Transcription–Polymerase Chain Reaction (qRT-PCR)
2.4. Statistical Analyses
3. Results
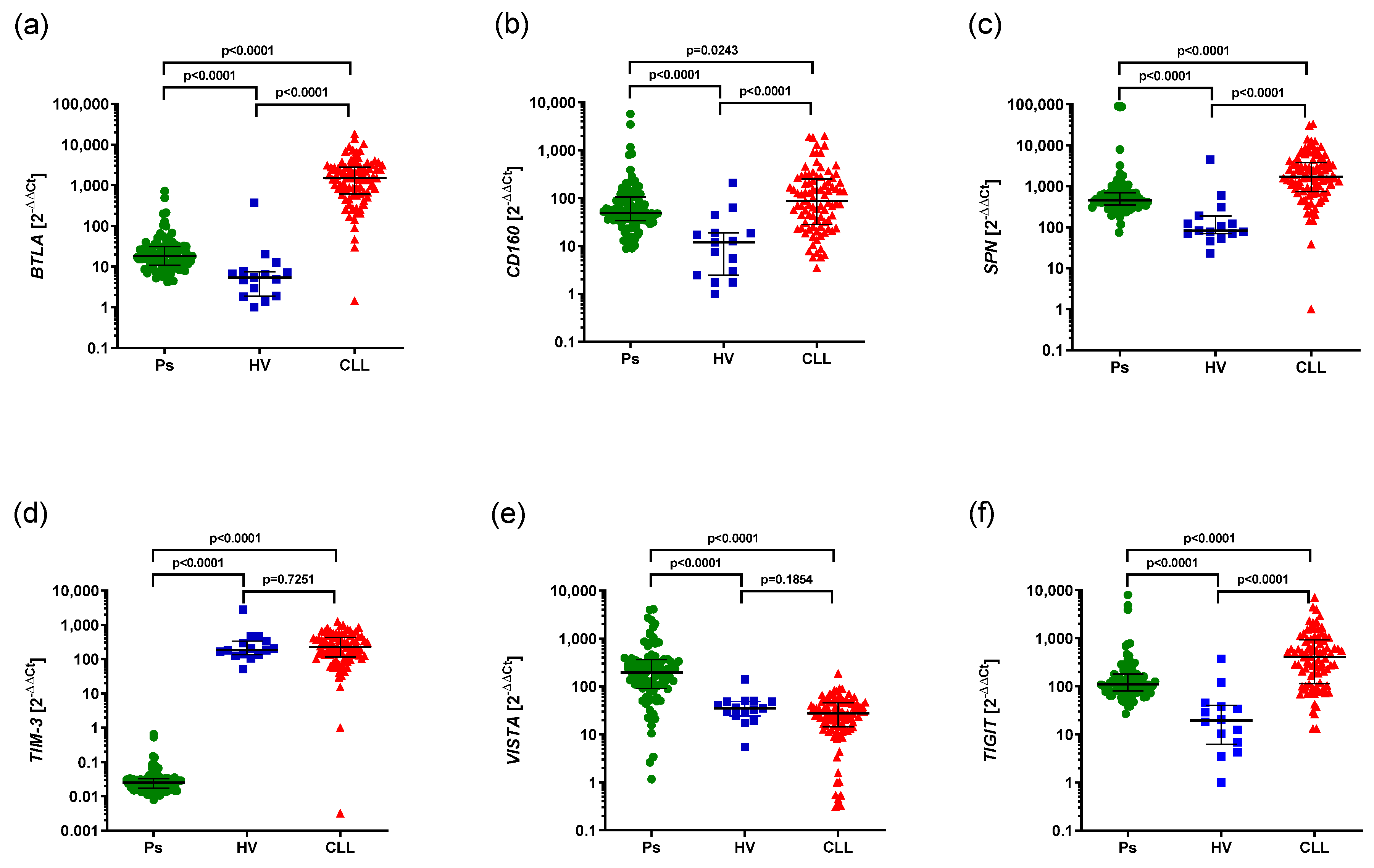
3.1. Aberrant mRNA Expression of BTLA, CD160, SPN, TIM3, VISTA, TIGIT in CLL and Psoriatic Patients Compared to HVs
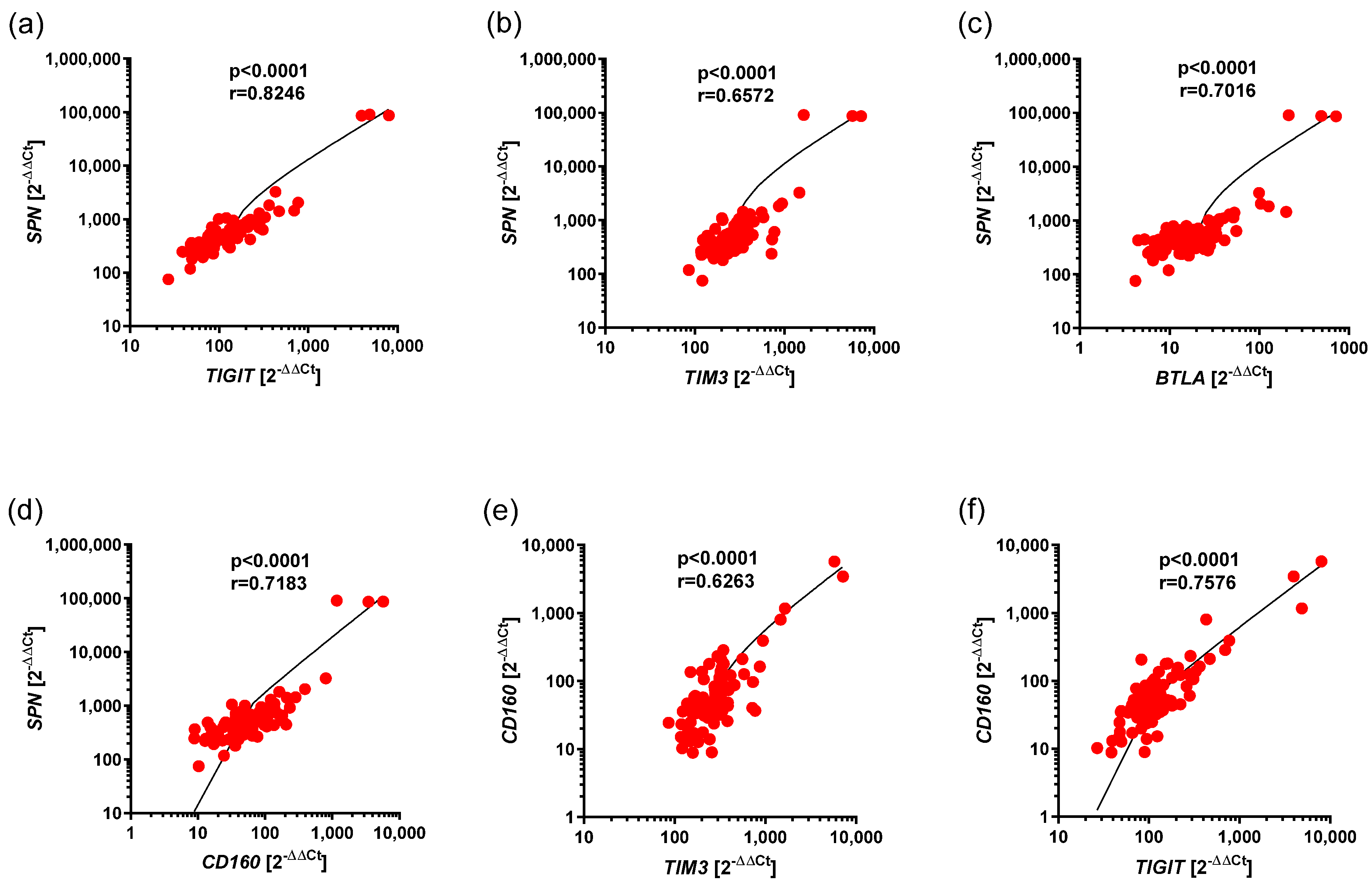
3.2. Correlations Between Expression of BTLA, CD160, SPN, TIM3, VISTA, as Well as TIGIT in CLL
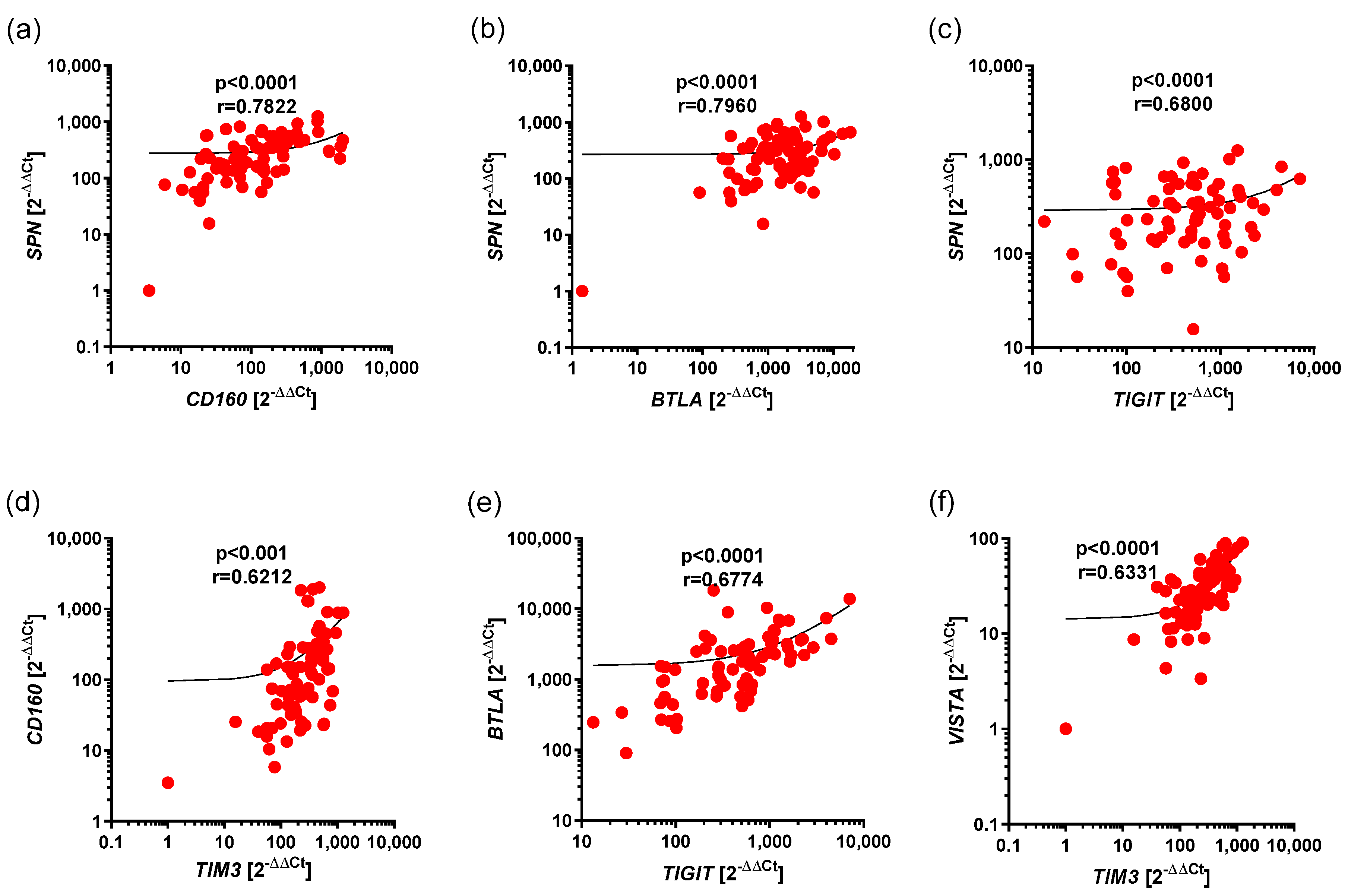
3.3. Correlations Between the Expression of BTLA, CD160, SPN, TIM3, and VISTA, as Well as TIGIT, in Ps
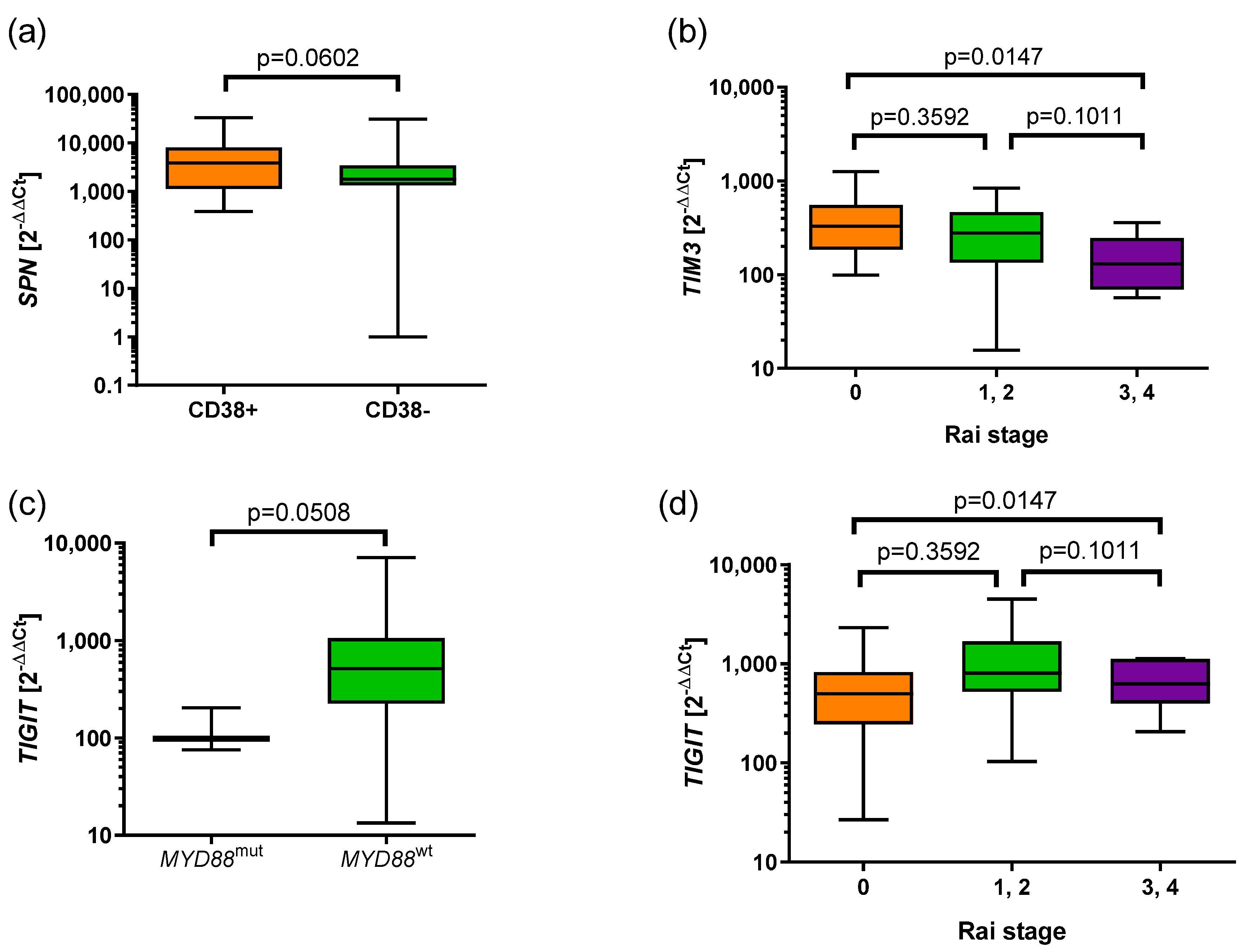
3.4. Associations of the Expression of BTLA, CD160, SPN, TIM3, VISTA, and TIGIT with Prognostic Parameters in CLL
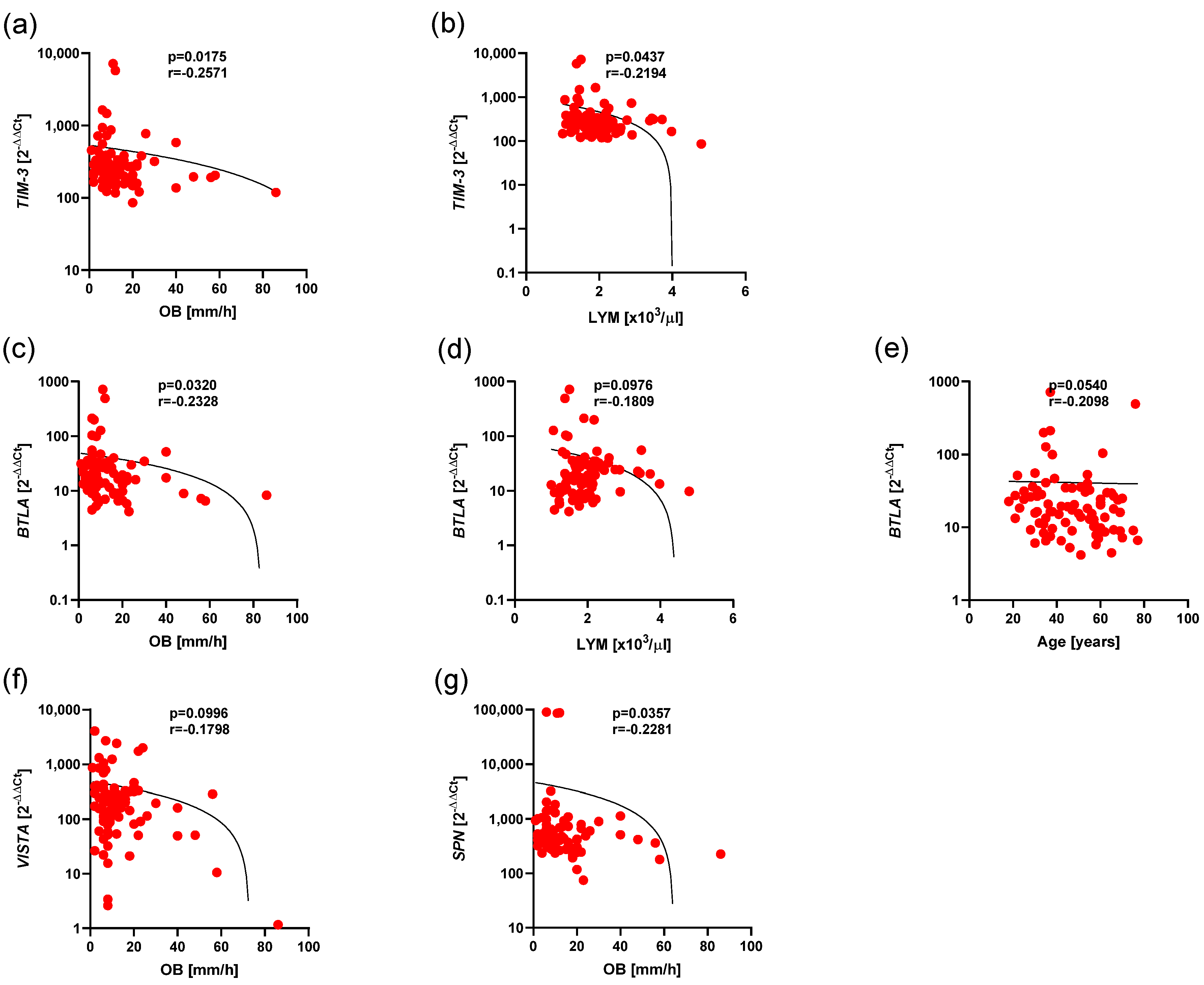
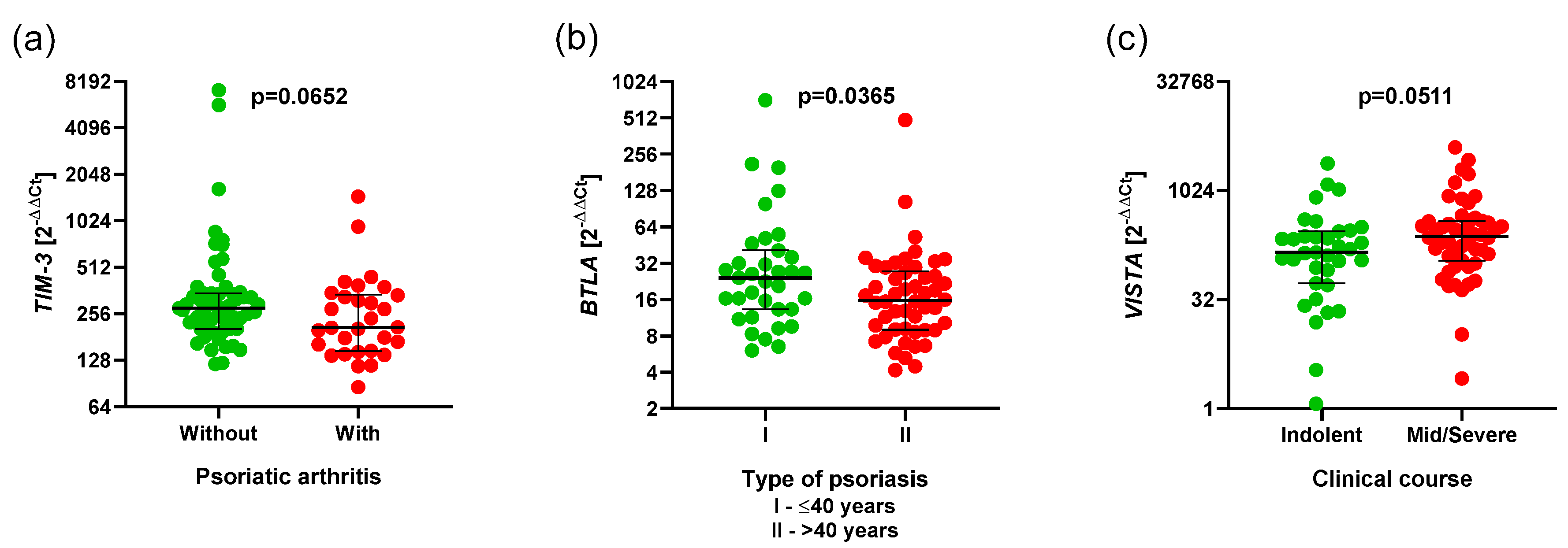
3.5. Associations of the Expression of BTLA, CD160, SPN, TIM3, VISTA, and TIGIT and Clinical Parameters in Ps
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Strand, V.; Balsa, A.; Al-Saleh, J.; Barile-Fabris, L.; Horiuchi, T.; Takeuchi, T.; Lula, S.; Hawes, C.; Kola, B.; Marshall, L. Immunogenicity of Biologics in Chronic Inflammatory Diseases: A Systematic Review. BioDrugs 2017, 31, 299–316. [Google Scholar] [CrossRef] [PubMed]
- Forconi, F.; Moss, P. Perturbation of the Normal Immune System in Patients with CLL. Blood 2015, 126, 573–581. [Google Scholar] [CrossRef]
- Hamblin, A.D.; Hamblin, T.J. The Immunodeficiency of Chronic Lymphocytic Leukaemia. Br. Med. Bull. 2008, 87, 49–62. [Google Scholar] [CrossRef]
- Arruga, F.; Gyau, B.B.; Iannello, A.; Vitale, N.; Vaisitti, T.; Deaglio, S. Immune Response Dysfunction in Chronic Lymphocytic Leukemia: Dissecting Molecular Mechanisms and Microenvironmental Conditions. Int. J. Mol. Sci. 2020, 21, 1825. [Google Scholar] [CrossRef]
- Rendon, A.; Schäkel, K. Psoriasis Pathogenesis and Treatment. Int. J. Mol. Sci. 2019, 20, 1475. [Google Scholar] [CrossRef]
- Cottone, M.; Sapienza, C.; Macaluso, F.S.; Cannizzaro, M. Psoriasis and Inflammatory Bowel Disease. Dig. Dis. 2019, 37, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Strange, A.; Capon, F.; Spencer, C.C.A.; Knight, J.; Weale, M.E.; Allen, M.H.; Barton, A.; Band, G.; Bellenguez, C.; Bergboer, J.G.M.; et al. A Genome-Wide Association Study Identifies New Psoriasis Susceptibility Loci and an Interaction between HLA-C and ERAP1. Nat. Genet. 2010, 42, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Plenge, R. GWASs and the Age of Human as the Model Organism for Autoimmune Genetic Research. Genome Biol. 2010, 11, 212. [Google Scholar] [CrossRef]
- Trafford, A.M.; Parisi, R.; Kontopantelis, E.; Griffiths, C.E.M.; Ashcroft, D.M. Association of Psoriasis with the Risk of Developing or Dying of Cancer: A Systematic Review and Meta-Analysis. JAMA Dermatol. 2019, 155, 1390–1403. [Google Scholar] [CrossRef]
- Loft, N.D.; Vaengebjerg, S.; Skov, L. Cancer Risk in Patients with Psoriasis: Should We Be Paying More Attention? Expert Rev. Clin. Immunol. 2020, 16, 479–492. [Google Scholar] [CrossRef]
- Vaengebjerg, S.; Skov, L.; Egeberg, A.; Loft, N.D. Prevalence, Incidence, and Risk of Cancer in Patients with Psoriasis and Psoriatic Arthritis: A Systematic Review and Meta-Analysis. JAMA Dermatol. 2020, 156, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Htut, T.W.; Han, M.M.; Thein, K.Z. Acalabrutinib-Related Second Primary Malignancies and Nonmelanoma Skin Cancers in Patients with Chronic Lymphocytic Leukaemia (CLL): A Systematic Review and Meta-Analysis of Randomised Controlled Trials (RCTs). EJHaem 2021, 2, 112–117. [Google Scholar] [CrossRef]
- Balda, A.; Wani, I.; Roohi, T.F.; Suman; Krishna, K.L.; Mehdi, S.; Nadiga, A.P.; Makkapati, M.; Baig, M.A.I. Psoriasis and Skin Cancer—Is There a Link? Int. Immunopharmacol. 2023, 121, 110464. [Google Scholar] [CrossRef] [PubMed]
- Zavdy, O.; Coreanu, T.; Bar-On, D.Y.; Ritter, A.; Bachar, G.; Shpitzer, T.; Kurman, N.; Mansour, M.; Ad-El, D.; Rozovski, U.; et al. Cutaneous Squamous Cell Carcinoma in Immunocompromised Patients-A Comparison between Different Immunomodulating Conditions. Cancers 2023, 15, 1764. [Google Scholar] [CrossRef]
- Fried, L.J.; Criscito, M.C.; Stevenson, M.L.; Pomeranz, M.K. Chronic Lymphocytic Leukemia and the Skin: Implications for the Dermatologist. Int. J. Dermatol. 2022, 61, 519–531. [Google Scholar] [CrossRef]
- Hodgson, K.; Ferrer, G.; Montserrat, E.; Moreno, C. Chronic Lymphocytic Leukemia and Autoimmunity: A Systematic Review. Haematologica 2011, 96, 752–761. [Google Scholar] [CrossRef]
- Abecassis, S.; Giustiniani, J.; Meyer, N.; Schiavon, V.; Ortonne, N.; Campillo, J.A.; Bagot, M.; Bensussan, A. Identification of a Novel CD160+CD4+ T-Lymphocyte Subset in the Skin: A Possible Role for CD160 in Skin Inflammation. J. Investig. Dermatol. 2007, 127, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Pallant, A.; Eskenazi, A.; Mattei, M.G.; Fournier, R.E.; Carlsson, S.R.; Fukuda, M.; Frelinger, J.G. Characterization of CDNAs Encoding Human Leukosialin and Localization of the Leukosialin Gene to Chromosome 16. Proc. Natl. Acad. Sci. USA 1989, 86, 1328–1332. [Google Scholar] [CrossRef]
- Shelley, C.S.; Remold-O’Donnell, E.; Davis, A.E.; Bruns, G.A.; Rosen, F.S.; Carroll, M.C.; Whitehead, A.S. Molecular Characterization of Sialophorin (CD43), the Lymphocyte Surface Sialoglycoprotein Defective in Wiskott-Aldrich Syndrome. Proc. Natl. Acad. Sci. USA 1989, 86, 2819–2823. [Google Scholar] [CrossRef]
- Hadadi, L.; Hafezi, M.; Amirzargar, A.A.; Sharifian, R.A.; Abediankenari, S.; Asgarian-Omran, H. Dysregulated Expression of Tim-3 and NKp30 Receptors on NK Cells of Patients with Chronic Lymphocytic Leukemia. Oncol. Res. Treat. 2019, 42, 197–203. [Google Scholar] [CrossRef]
- Li, N.; Xu, W.; Yuan, Y.; Ayithan, N.; Imai, Y.; Wu, X.; Miller, H.; Olson, M.; Feng, Y.; Huang, Y.H.; et al. Immune-Checkpoint Protein VISTA Critically Regulates the IL-23/IL-17 Inflammatory Axis. Sci. Rep. 2017, 7, 1485. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Li, Y.; Tan, J.; Xu, L.; Li, Y. Targeting LAG-3, TIM-3, and TIGIT for Cancer Immunotherapy. J. Hematol. Oncol. 2023, 16, 101. [Google Scholar] [CrossRef] [PubMed]
- Bourque, J.; Hawiger, D. The BTLA–HVEM–CD5 Immunoregulatory Axis–An Instructive Mechanism Governing PTreg Cell Differentiation. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Youssef, R.M.; El-Ramly, A.Z.; Hussien, M.F.; Shoukry, N.M.; Amr, K. Expression of B and T Lymphocyte Attenuator, Retinoid-related Orphan Receptor Gamma-isoform-t and Interleukin 7 in Psoriasis Vulgaris. Australas. J. Dermatol. 2019, 60. [Google Scholar] [CrossRef]
- Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Gonzalez-Rodriguez, A.P.; R Payer, Á.; González-García, E.; López-Soto, A.; Gonzalez, S. BTLA/HVEM Axis Induces NK Cell Immunosuppression and Poor Outcome in Chronic Lymphocytic Leukemia. Cancers 2021, 13, 1766. [Google Scholar] [CrossRef] [PubMed]
- Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Martínez-Pérez, A.; Gonzalez-Rodriguez, A.P.; Payer, Á.R.; González-García, E.; Aguilar-García, C.; González-Rodríguez, S.; López-Soto, A.; García-Torre, A.; et al. BTLA Dysregulation Correlates with Poor Outcome and Diminished T Cell-Mediated Antitumor Responses in Chronic Lymphocytic Leukemia. Cancer Immunol. Immunother. 2023, 72, 2529–2539. [Google Scholar] [CrossRef]
- Vendel, A.C.; Calemine-Fenaux, J.; Izrael-Tomasevic, A.; Chauhan, V.; Arnott, D.; Eaton, D.L. B and T Lymphocyte Attenuator Regulates B Cell Receptor Signaling by Targeting Syk and BLNK. J. Immunol. 2009, 182, 1509–1517. [Google Scholar] [CrossRef]
- Ware, C.F. Targeting Lymphocyte Activation through the Lymphotoxin and LIGHT Pathways. Immunol. Rev. 2008, 223, 186–201. [Google Scholar] [CrossRef]
- Kannan, S.; Kurupati, R.K.; Doyle, S.A.; Freeman, G.J.; Schmader, K.E.; Ertl, H.C.J. BTLA Expression Declines on B Cells of the Aged and Is Associated with Low Responsiveness to the Trivalent Influenza Vaccine. Oncotarget 2015, 6, 19445–19455. [Google Scholar] [CrossRef]
- Karabon, L.; Andrzejczak, A.; Ciszak, L.; Tomkiewicz, A.; Szteblich, A.; Bojarska-Junak, A.; Roliński, J.; Wołowiec, D.; Wróbel, T.; Kosmaczewska, A. BTLA Expression in CLL: Epigenetic Regulation and Impact on CLL B Cell Proliferation and Ability to IL-4 Production. Cells 2021, 10, 3009. [Google Scholar] [CrossRef]
- Pasero, C.; Olive, D. Interfering with Coinhibitory Molecules: BTLA/HVEM as New Targets to Enhance Anti-Tumor Immunity. Immunol. Lett. 2013, 151, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Freeman, G.J. The CD160, BTLA, LIGHT/HVEM Pathway: A Bidirectional Switch Regulating T-cell Activation. Immunol. Rev. 2009, 229, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Muench, G.; Hare, E.; Soroosh, P.; Parmley, S.; Lizzul, P.; Dahl, M. 50585 ANB032, a B and T Cell Lymphocyte Attenuator (BTLA) Checkpoint Receptor Agonist, Modulates Dendritic Cell (DC) Maturation and Function: A Novel Mechanism Addressing Atopic Dermatitis Pathophysiology. J. Am. Acad. Dermatol. 2024, 91, AB17. [Google Scholar] [CrossRef]
- Luu, K.; Randazzo, B.; Hare, E.; Hsu, M.; Haines, C.; Dahl, M.; Sibley, C.; Lizzul, P. 88 ANB032, a Novel BTLA Agonist Monoclonal Antibody, Inhibits T Cell Proliferation, Reduces Inflammatory Cytokines, and down Modulates BTLA Expression on Circulating T and B Cells. J. Investig. Dermatol. 2023, 143, S347. [Google Scholar] [CrossRef]
- Oumeslakht, L.; Aziz, A.; Bensussan, A.; Ben Mkaddem, S. CD160 Receptor in CLL: Current State and Future Avenues. Front. Immunol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- El-Neanaey, W.A.; Swelem, R.S.; Ghallab, O.M.; Mohamed Abu-Shelou, S. Evaluation of CD160 and CD200 Expression as Differentiating Markers between Chronic Lymphocytic Leukemia and Other Mature B-Cell Neoplasms. Int. J. Hematol. Stem cell Res. 2020, 14, 27–37. [Google Scholar] [CrossRef]
- Farren, T.W.; Giustiniani, J.; Liu, F.-T.; Tsitsikas, D.A.; Macey, M.G.; Cavenagh, J.D.; Oakervee, H.E.; Taussig, D.; Newland, A.C.; Calaminici, M.; et al. Differential and Tumor-Specific Expression of CD160 in B-Cell Malignancies. Blood 2011, 118, 2174–2183. [Google Scholar] [CrossRef][Green Version]
- Hobbs, S.J.; Nolz, J.C. Regulation of T Cell Trafficking by Enzymatic Synthesis of O-Glycans. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Ali, A.J.; Abuelela, A.F.; Merzaban, J.S. An Analysis of Trafficking Receptors Shows That CD44 and P-Selectin Glycoprotein Ligand-1 Collectively Control the Migration of Activated Human T-Cells. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Falay, M.; Afacan Öztürk, B.; Güneş, K.; Kalpakçı, Y.; Dağdaş, S.; Ceran, F.; Özet, G. The Role of CD200 and CD43 Expression in Differential Diagnosis between Chronic Lymphocytic Leukemia and Mantle Cell Lymphoma. Turk. J. Hematol. 2018, 35, 94–98. [Google Scholar] [CrossRef]
- Sorigue, M.; Juncà, J.; Sarrate, E.; Grau, J. Expression of CD43 in Chronic Lymphoproliferative Leukemias. Cytom. Part B Clin. Cytom. 2018, 94, 136–142. [Google Scholar] [CrossRef]
- Jung, G.; Eisenmann, J.; Thiébault, S.; Hénon, P. Cell Surface CD43 Determination Improves Diagnostic Precision in Late B-cell Diseases. Br. J. Haematol. 2003, 120, 496–499. [Google Scholar] [CrossRef] [PubMed]
- Mehdizadeh, S.; Bayatipoor, H.; Pashangzadeh, S.; Jafarpour, R.; Shojaei, Z.; Motallebnezhad, M. Immune Checkpoints and Cancer Development: Therapeutic Implications and Future Directions. Pathol. Res. Pract. 2021, 223, 153485. [Google Scholar] [CrossRef]
- Anderson, A.C.; Joller, N.; Kuchroo, V.K. Lag-3, Tim-3, and TIGIT: Co-Inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016, 44, 989–1004. [Google Scholar] [CrossRef]
- Behdad, A.; Schipma, M.J.; Ma, S.; Chen, Y.-H.; Chen, Q.C. Distinct Immune-Response Profile of Richter Transformation Chronic Lymphocytic Leukemia (CLL), Defined by High Expression of PD1, LAG3, TIM3 and IL-10. Leuk. Lymphoma 2022, 63, 3222–3226. [Google Scholar] [CrossRef] [PubMed]
- Taghiloo, S.; Allahmoradi, E.; Ebadi, R.; Tehrani, M.; Hosseini-Khah, Z.; Janbabaei, G.; Shekarriz, R.; Asgarian-Omran, H. Upregulation of Galectin-9 and PD-L1 Immune Checkpoints Molecules in Patients with Chronic Lymphocytic Leukemia. Asian Pac. J. Cancer Prev. 2017, 18, 2269–2274. [Google Scholar] [CrossRef] [PubMed]
- Grzywnowicz, M.; Karczmarczyk, A.; Skorka, K.; Zajac, M.; Zaleska, J.; Chocholska, S.; Tomczak, W.; Giannopoulos, K. Expression of Programmed Death 1 Ligand in Different Compartments of Chronic Lymphocytic Leukemia. Acta Haematol. 2015, 134, 255–262. [Google Scholar] [CrossRef]
- Rezaei, M.; Tan, J.; Zeng, C.; Li, Y.; Ganjalikhani-Hakemi, M. TIM-3 in Leukemia; Immune Response and Beyond. Front. Oncol. 2021, 11, 753677. [Google Scholar] [CrossRef]
- Tao, J.; Li, L.; Fu, R.; Wang, H.; Jiang, H.; Yue, L.; Zhang, W.; Liu, H.; Ruan, E.; Qu, W.; et al. Elevated TIM3+ Hematopoietic Stem Cells in Untreated Myelodysplastic Syndrome Displayed Aberrant Differentiation, Overproliferation and Decreased Apoptosis. Leuk. Res. 2014, 38, 714–721. [Google Scholar] [CrossRef]
- Zeidan, A.M.; Komrokji, R.S.; Brunner, A.M. TIM-3 Pathway Dysregulation and Targeting in Cancer. Expert Rev. Anticancer Ther. 2021, 21, 523–534. [Google Scholar] [CrossRef]
- Bartosińska, J.; Zakrzewska, E.; Król, A.; Raczkiewicz, D.; Purkot, J.; Majdan, M.; Krasowska, D.; Chodorowska, G.; Giannopoulos, K. Differential Expression of Programmed Death 1 (PD-1) on CD4+ and CD8+ T Cells in Rheumatoid Arthritis and Psoriatic Arthritis. Polish Arch. Intern. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jia, B.; Claxton, D.F.; Ehmann, W.C.; Rybka, W.B.; Mineishi, S.; Naik, S.; Khawaja, M.R.; Sivik, J.; Han, J.; et al. VISTA Is Highly Expressed on MDSCs and Mediates an Inhibition of T Cell Response in Patients with AML. Oncoimmunology 2018, 7, e1469594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chen, S.; Luo, T.-T.; Qu, J.-H. Expression and Clinical Significance of Co-Inhibitory Molecules TIGIT/CD155 and PD-1 in Chronic Lymphocytic Leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2025, 33, 54–61. [Google Scholar] [CrossRef]
- Hajiasghar-Sharbaf, R.; Asgarian-Omran, H.; Valadan, R.; Hossein-Nattaj, H.; Shekarriz, R.; Zaboli, E.; Janbabaei, G.; Tehrani, M. CD8+ T-Cells Co-Expressing PD-1 and TIGIT Are Highly Frequent in Chronic Lymphocytic Leukemia. Iran. J. Allergy Asthma Immunol. 2021, 20, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Catakovic, K.; Gassner, F.J.; Ratswohl, C.; Zaborsky, N.; Rebhandl, S.; Schubert, M.; Steiner, M.; Gutjahr, J.C.; Pleyer, L.; Egle, A.; et al. TIGIT Expressing CD4+T Cells Represent a Tumor-Supportive T Cell Subset in Chronic Lymphocytic Leukemia. Oncoimmunology 2017, 7, e1371399. [Google Scholar] [CrossRef]
- Chauvin, J.-M.; Zarour, H.M. TIGIT in Cancer Immunotherapy. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef]

| Characteristic | CLL (n = 74) |
|---|---|
| Sex | |
| Male | 46 |
| Female | 28 |
| Age (years) | |
| Median | 66 |
| Range | 48–84 |
| Rai Stage | |
| 0 | 26 |
| I–II | 34 |
| III–IV | 14 |
| ZAP-70 (cut-off 20%) | |
| Positive | 24 |
| Negative | 34 |
| NA | 16 |
| CD38 (cut-off 30%) | |
| Positive | 22 |
| Negative | 38 |
| NA | 14 |
| IGHV | |
| Mutated | 32 |
| Unmutated | 39 |
| NA | 3 |
| Characteristic | Ps (n = 85) |
|---|---|
| Sex | |
| Male | 71 |
| Female | 14 |
| Age (years) | |
| Median | 47 |
| Range | 18–77 |
| Type | |
| I age ≤ 40 | 35 |
| II age > 40 | 50 |
| Articular Ps | |
| With | 30 |
| Without | 55 |
| Duration | |
| Median | 16 |
| Range | 55 |
| PASI | |
| Median | 12.1 |
| Range | 49.4 |
| Course | |
| Mild | 35 |
| Severe | 50 |
| WBC | |
| Median | 6.53 |
| Range | 3.54–13.42 |
| Neutrophils | |
| Median | 3.61 |
| Range | 1.4–10.96 |
| Lymphocytes | |
| Median | 1.83 |
| Range | 1–4.79 |
| CRP | |
| Median | 1.6 |
| Range | 0.8–57.3 |
| OB | |
| Median | 9 |
| Range | 1–86 |
| Pairs of Genes | r | Statistical Significance (p) |
|---|---|---|
| TIGIT and TIM3 | 0.2522 | 0.068 |
| TIM3 and BTLA | 0.4003 | <0.001 |
| CD160 and VISTA | 0.3861 | <0.001 |
| CD160 and BTLA | 0.5504 | <0.0001 |
| CD160 and SPN | 0.5821 | <0.0001 |
| Pairs of Genes | r | Statistical Significance (p) |
|---|---|---|
| TIGIT and TIM3 | 0.5951 | <0.0001 |
| TIM3 and BTLA | 0.5312 | <0.0001 |
| TIGIT and BTLA | 0.6012 | <0.0001 |
| BTLA and CD160 | 0.4773 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skórka, K.; Wdowiak-Filip, A.; Stasiak, G.; Bartosińska, J.; Krasowska, D.; Giannopoulos, K. Aberrant Expression of BTLA, CD160, SPN, TIM-3, VISTA and TIGIT in Chronic Lymphocytic Leukemia and Psoriasis Patients Compared to Healthy Volunteers. Cancers 2025, 17, 2116. https://doi.org/10.3390/cancers17132116
Skórka K, Wdowiak-Filip A, Stasiak G, Bartosińska J, Krasowska D, Giannopoulos K. Aberrant Expression of BTLA, CD160, SPN, TIM-3, VISTA and TIGIT in Chronic Lymphocytic Leukemia and Psoriasis Patients Compared to Healthy Volunteers. Cancers. 2025; 17(13):2116. https://doi.org/10.3390/cancers17132116
Chicago/Turabian StyleSkórka, Katarzyna, Anita Wdowiak-Filip, Grażyna Stasiak, Joanna Bartosińska, Dorota Krasowska, and Krzysztof Giannopoulos. 2025. "Aberrant Expression of BTLA, CD160, SPN, TIM-3, VISTA and TIGIT in Chronic Lymphocytic Leukemia and Psoriasis Patients Compared to Healthy Volunteers" Cancers 17, no. 13: 2116. https://doi.org/10.3390/cancers17132116
APA StyleSkórka, K., Wdowiak-Filip, A., Stasiak, G., Bartosińska, J., Krasowska, D., & Giannopoulos, K. (2025). Aberrant Expression of BTLA, CD160, SPN, TIM-3, VISTA and TIGIT in Chronic Lymphocytic Leukemia and Psoriasis Patients Compared to Healthy Volunteers. Cancers, 17(13), 2116. https://doi.org/10.3390/cancers17132116









