Harnessing Artificial Intelligence in Pediatric Oncology Diagnosis and Treatment: A Review
Simple Summary
Abstract
1. Introduction
2. AI-Based Application for Pediatric Diagnosis and Treatment
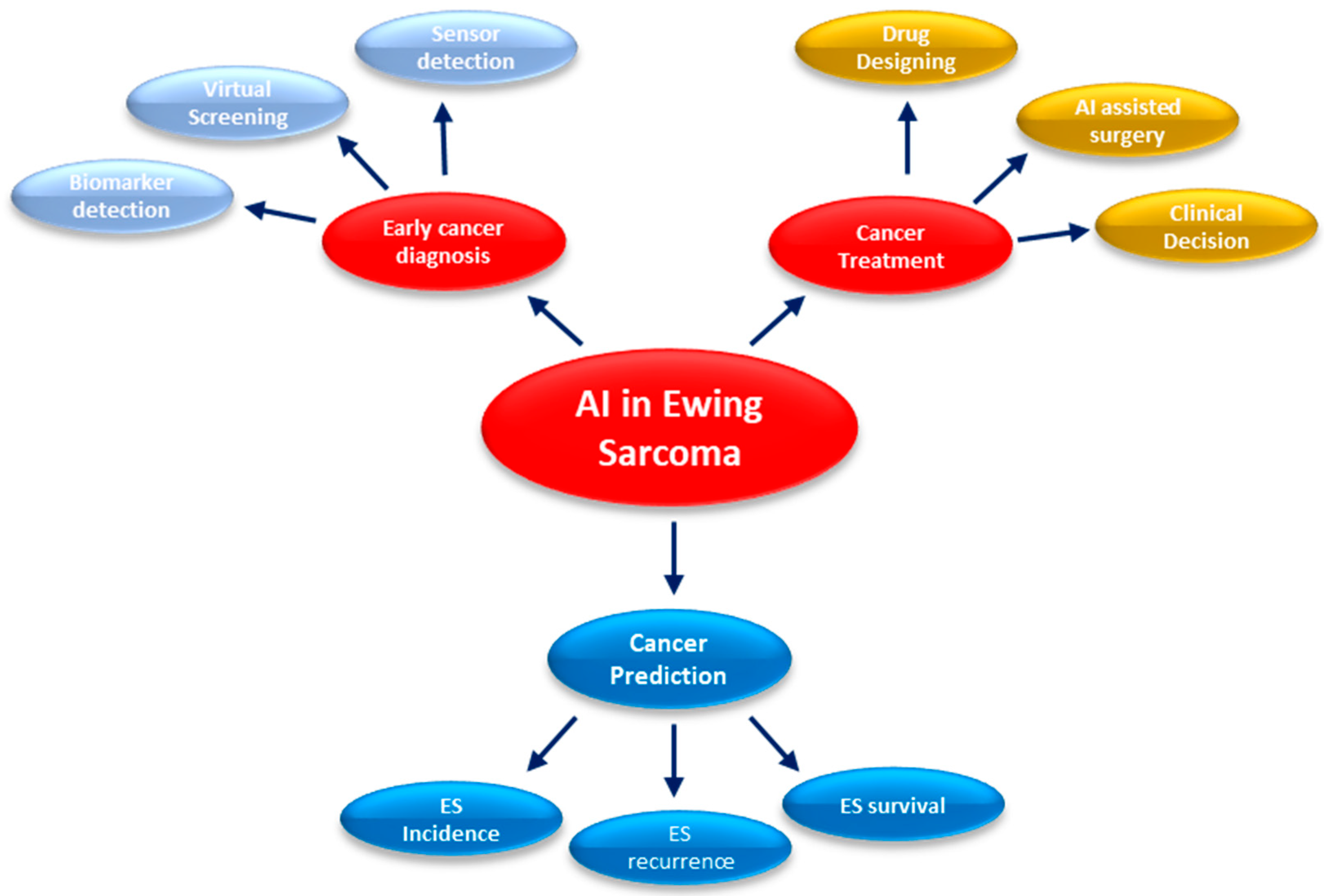
2.1. Early Detection and Diagnosis for ES
2.1.1. Image Analysis
2.1.2. Tumor Detection and Segmentation
2.1.3. Quantitative Analysis of Tumor Characteristics
2.1.4. Risk Stratification and Prognostic Assessment
2.1.5. Radiomics and Texture Analysis
2.1.6. Clinical Decision Support Systems (CDSS)
2.2. AI Image Analysis Tools and Algorithms in Pediatrics Cancers
2.2.1. Convolutional Neural Networks (CNNs)
2.2.2. U-Net
2.2.3. DeepLab
3. AI Approaches in Drug Discovery and Development
3.1. Target Identification for Pediatric Cancers
3.2. AI in Anticancer Drug Repurposing
3.3. AI in Anti-Drug Repositioning Based on Drug–Target Interaction
3.4. AI in Anti-Drug Repositioning Considering Drug–Disease Interactions
3.5. Virtual Screening and Molecular Docking
3.6. AI’s Role in Evaluating Drugs for Cancer Prevention
3.6.1. AI and Structure-Based Virtual Screening (SBVS)
3.6.2. AI and Ligand-Based Virtual Screening (LBVS)
3.6.3. AI and Fragment-Based Virtual Screening (FBVS)
3.7. AI-Based Target Identification for Anticancer Drugs
3.8. Determining the Druggability of Cancer Drug Targets Using AI
3.9. Modelling Applications in Drug Discovery
3.9.1. Variational Auto-Encoder (VAE) Model
3.9.2. Recurrent Neural Network (RNN) Model
3.9.3. Generative Adversarial Network (GAN)
3.10. Practical Application of AI to Treat Pediatric Cancer
4. Recent Advancements in Pediatric Oncology Diagnosis and Treatment
5. Analysis of Genomic and Molecular Data
5.1. Pediatrics Malignancies and Genome Landscape
5.2. AI Resources for Genome Landscape Research
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Agarwal, S. Pediatric Cancers: Insights and Novel Therapeutic Approaches. Cancers 2023, Volume 15, 3537. [Google Scholar]
- Al Lamki, Z. Improving cancer care for children in the developing world: Challenges and strategies. Curr. Pediatr. Rev. 2017, 13, 13–23. [Google Scholar] [CrossRef]
- Grovas, A.; Fremgen, A.; Rauck, A.; Ruymann, F.B.; Hutchinson, C.L.; Winchester, D.P.; Menck, H.R. The National Cancer Data Base report on patterns of childhood cancers in the United States. Cancer: Interdiscip. Int. J. Am. Cancer Soc. 1997, 80, 2321–2332. [Google Scholar] [CrossRef]
- Arya, L. Childhood cancer-challenges and opportunities. Indian. J. Pediatr. 2003, 70, 159–162. [Google Scholar] [CrossRef]
- Carroll, W.L.; Bhojwani, D.; Min, D.-J.; Raetz, E.; Relling, M.; Davies, S.; Downing, J.R.; Willman, C.L.; Reed, J.C. Pediatric acute lymphoblastic leukemia. ASH Educ. Program. Book. 2003, 2003, 102–131. [Google Scholar] [CrossRef][Green Version]
- Board, P.P.T.E. Childhood Brain and Spinal Cord Tumors Treatment Overview (PDQ®). In PDQ Cancer Information Summaries [Internet]; National Cancer Institute (US): Bethesda, MD, USA, 2019. [Google Scholar]
- Ness, K.K.; Gurney, J.G. Adverse late effects of childhood cancer and its treatment on health and performance. Annu. Rev. Public. Health 2007, 28, 279–302. [Google Scholar] [CrossRef]
- Landier, W.; Bhatia, S. Cancer survivorship: A pediatric perspective. Oncologist 2008, 13, 1181–1192. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.; Li, Y. Artificial intelligence applications in pediatric oncology diagnosis. Explor. Target. Anti-Tumor Ther. 2023, 4, 157. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, X.; Cao, X.; Huang, C.; Liu, E.; Qian, S.; Liu, X.; Wu, Y.; Dong, F.; Qiu, C.-W. Artificial intelligence: A powerful paradigm for scientific research. Innovation 2021, 2, 100179. [Google Scholar] [CrossRef]
- Chen, Z.H.; Lin, L.; Wu, C.F.; Li, C.F.; Xu, R.H.; Sun, Y. Artificial intelligence for assisting cancer diagnosis and treatment in the era of precision medicine. Cancer Commun. 2021, 41, 1100–1115. [Google Scholar] [CrossRef]
- Schork, N.J. Artificial intelligence and personalized medicine. In Precision Medicine in Cancer Therapy; Springer Nature: Berlin/Heidelberg, Germany, 2019; pp. 265–283. [Google Scholar]
- Esposito, D.; Esposito, F. Introducing Machine Learning; Microsoft Press: Redmond, WA, USA, 2020. [Google Scholar]
- Cui, S.; Tseng, H.H.; Pakela, J.; Ten Haken, R.K.; El Naqa, I. Introduction to machine and deep learning for medical physicists. Med. Phys. 2020, 47, e127–e147. [Google Scholar] [CrossRef]
- Baduge, S.K.; Thilakarathna, S.; Perera, J.S.; Arashpour, M.; Sharafi, P.; Teodosio, B.; Shringi, A.; Mendis, P. Artificial intelligence and smart vision for building and construction 4.0: Machine and deep learning methods and applications. Autom. Constr. 2022, 141, 104440. [Google Scholar] [CrossRef]
- Shandilya, S.K.; Datta, A.; Kartik, Y.; Nagar, A. Role of artificial intelligence and machine learning. In Digital Resilience: Navigating Disruption and Safeguarding Data Privacy; Springer: Berlin/Heidelberg, Germany, 2024; pp. 313–399. [Google Scholar]
- Kourou, K.; Exarchos, K.P.; Papaloukas, C.; Sakaloglou, P.; Exarchos, T.; Fotiadis, D.I. Applied machine learning in cancer research: A systematic review for patient diagnosis, classification and prognosis. Comput. Struct. Biotechnol. J. 2021, 19, 5546–5555. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Javed, Z.; Sadia, H.; Qureshi, I.A.; Irshad, A.; Ahmed, R.; Malik, K.; Raza, S.; Abbas, A.; Pezzani, R. Clinical applications of artificial intelligence and machine learning in cancer diagnosis: Looking into the future. Cancer Cell Int. 2021, 21, 270. [Google Scholar] [CrossRef]
- Nedungadi, P.; Iyer, A.; Gutjahr, G.; Bhaskar, J.; Pillai, A.B. Data-driven methods for advancing precision oncology. Curr. Pharmacol. Rep. 2018, 4, 145–156. [Google Scholar] [CrossRef]
- Alum, E.U. AI-driven biomarker discovery: Enhancing precision in cancer diagnosis and prognosis. Discov. Oncol. 2025, 16, 1–12. [Google Scholar] [CrossRef]
- Mirtaheri, S.L.; Shahbazian, R. Machine Learning: Theory to Applications; CRC Press: Boca Raton, FL, USA, 2022. [Google Scholar]
- Deshmukh, M.T.; Wankhede, P.; Chakole, N.; Kale, P.D.; Jadhav, M.R.; Kulkarni, M.B.; Bhaiyya, M. Towards Intelligent Food Safety: Machine Learning Approaches for Aflatoxin Detection and Risk Prediction. Trends Food Sci. Technol. 2025, 161, 105055. [Google Scholar] [CrossRef]
- Gangas, P.; Graf, N.; Wen, S.; Cattaneo, C.; Bicchieri, M.; Lennox-Chhugani, N. Artificial intelligence in paediatric cancer: Insights from innovation experts in the UNICA4EU project. EJC Paediatr. Oncol. 2025, 5, 100208. [Google Scholar] [CrossRef]
- Siddiq, M. ML-based Medical Image Analysis for Anomaly Detection in CT Scans, X-rays, and MRIs. Devot. J. Community Serv. 2020, 2, 53–64. [Google Scholar] [CrossRef]
- Kaur, C.; Garg, U. Artificial intelligence techniques for cancer detection in medical image processing: A review. Mater. Today Proc. 2021, 81, 806–809. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Kumar, M.J.; Kumar, D.G.R.; Reddy, R.V.K. Review on image segmentation techniques. Int. J. Sci. Res. Eng. Technol. 2014, 3, 993–997. [Google Scholar]
- Cai, W.-L.; Hong, G.-B. Quantitative image analysis for evaluation of tumor response in clinical oncology. Chronic Dis. Transl. Med. 2018, 4, 18–28. [Google Scholar] [CrossRef]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA: A Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef]
- Cheung, H.; Rubin, D. Challenges and opportunities for artificial intelligence in oncological imaging. Clin. Radiol. 2021, 76, 728–736. [Google Scholar] [CrossRef]
- Consalvo, S.; Hinterwimmer, F.; Neumann, J.; Steinborn, M.; Salzmann, M.; Seidl, F.; Lenze, U.; Knebel, C.; Rueckert, D.; Burgkart, R.H. Two-phase deep learning algorithm for detection and differentiation of ewing sarcoma and acute osteomyelitis in paediatric radiographs. Anticancer. Res. 2022, 42, 4371–4380. [Google Scholar] [CrossRef]
- Rogers, W.; Thulasi Seetha, S.; Refaee, T.A.; Lieverse, R.I.; Granzier, R.W.; Ibrahim, A.; Keek, S.A.; Sanduleanu, S.; Primakov, S.P.; Beuque, M.P. Radiomics: From qualitative to quantitative imaging. Br. J. Radiol. 2020, 93, 20190948. [Google Scholar] [CrossRef]
- Gitto, S.; Corino, V.D.; Annovazzi, A.; Milazzo Machado, E.; Bologna, M.; Marzorati, L.; Albano, D.; Messina, C.; Serpi, F.; Anelli, V. 3D vs. 2D MRI radiomics in skeletal Ewing sarcoma: Feature reproducibility and preliminary machine learning analysis on neoadjuvant chemotherapy response prediction. Front. Oncol. 2022, 12, 1016123. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Callegaro, D.; Miceli, R.; Bonvalot, S.; Ferguson, P.; Strauss, D.C.; Levy, A.; Griffin, A.; Hayes, A.J.; Stacchiotti, S.; Le Pechoux, C. Development and external validation of two nomograms to predict overall survival and occurrence of distant metastases in adults after surgical resection of localised soft-tissue sarcomas of the extremities: A retrospective analysis. Lancet Oncol. 2016, 17, 671–680. [Google Scholar] [CrossRef]
- Navarro, F.; Dapper, H.; Asadpour, R.; Knebel, C.; Spraker, M.B.; Schwarze, V.; Schaub, S.K.; Mayr, N.A.; Specht, K.; Woodruff, H.C. Development and external validation of deep-learning-based tumor grading models in soft-tissue sarcoma patients using MR imaging. Cancers 2021, 13, 2866. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, C.; Zhou, S. Artificial intelligence-based risk stratification, accurate diagnosis and treatment prediction in gynecologic oncology. Semin. Cancer Biol. 2023, 96, 82–99. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; De Jong, E.E.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Mori, Y.; Ren, H.; Mori, N.; Watanuki, M.; Hitachi, S.; Watanabe, M.; Mugikura, S.; Takase, K. Magnetic Resonance Imaging Texture Analysis Based on Intraosseous and Extraosseous Lesions to Predict Prognosis in Patients with Osteosarcoma. Diagnostics 2024, 14, 2562. [Google Scholar] [CrossRef]
- Chiesa, A.M.; Spinnato, P.; Miceli, M.; Facchini, G. Radiologic assessment of osteosarcoma lung metastases: State of the art and recent advances. Cells 2021, 10, 553. [Google Scholar] [CrossRef]
- Osheroff, J.A.; Teich, J.; Levick, D.; Saldana, L.; Velasco, F.; Sittig, D.; Rogers, K.; Jenders, R. Improving Outcomes with Clinical Decision Support: An Implementer’s Guide; Himss Publishing: Chicago, IL, USA, 2012. [Google Scholar]
- Sutton, R.T.; Pincock, D.; Baumgart, D.C.; Sadowski, D.C.; Fedorak, R.N.; Kroeker, K.I. An overview of clinical decision support systems: Benefits, risks, and strategies for success. NPJ Digit. Med. 2020, 3, 17. [Google Scholar] [CrossRef]
- Malin, J.L. Envisioning Watson as a rapid-learning system for oncology. J. Oncol. Pract. 2013, 9, 155. [Google Scholar] [CrossRef]
- Sloane, E.B.; Silva, R.J. Artificial intelligence in medical devices and clinical decision support systems. In Clinical Engineering Handbook; Elsevier: Amsterdam, The Netherlands, 2020; pp. 556–568. [Google Scholar]
- Tian, Y. Artificial intelligence image recognition method based on convolutional neural network algorithm. Ieee Access 2020, 8, 125731–125744. [Google Scholar] [CrossRef]
- Singha, A.; Thakur, R.S.; Patel, T. Deep learning applications in medical image analysis. In Biomedical Data Mining for Information Retrieval: Methodologies, Techniques and Applications; Wiley: Hoboken, NJ, USA, 2021; pp. 293–350. [Google Scholar]
- Yin, P.; Wang, W.; Wang, S.; Liu, T.; Sun, C.; Liu, X.; Chen, L.; Hong, N. The potential for different computed tomography-based machine learning networks to automatically segment and differentiate pelvic and sacral osteosarcoma from Ewing’s sarcoma. Quant. Imaging Med. Surg. 2023, 13, 3174. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W., Frangi, A., Eds.; Lecture Notes in Computer Science; Proceedings, Part III 18; Springer: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar]
- Yuan, Y.; Cheng, Y. Medical image segmentation with UNet-based multi-scale context fusion. Sci. Rep. 2024, 14, 15687. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Liu, C.; Chen, L.-C.; Schroff, F.; Adam, H.; Hua, W.; Yuille, A.L.; Fei-Fei, L. Auto-deeplab: Hierarchical neural architecture search for semantic image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 82–92. [Google Scholar]
- Mittal, M.; Arora, M.; Pandey, T.; Goyal, L.M. Image segmentation using deep learning techniques in medical images. In Advancement of Machine Intelligence in Interactive Medical Image Analysis; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 41–63. [Google Scholar]
- Yurtkulu, S.C.; Şahin, Y.H.; Unal, G. Semantic segmentation with extended DeepLabv3 architecture. In Proceedings of the 2019 27th Signal Processing and Communications Applications Conference (SIU), Sivas, Turkey, 24–26 April 2019; pp. 1–4. [Google Scholar]
- Pun, F.W.; Ozerov, I.V.; Zhavoronkov, A. AI-powered therapeutic target discovery. Trends Pharmacol. Sci. 2023, 44, 561–572. [Google Scholar] [CrossRef]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artificial intelligence in cancer research and precision medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef]
- Challa, A.P.; Zaleski, N.M.; Jerome, R.N.; Lavieri, R.R.; Shirey-Rice, J.K.; Barnado, A.; Lindsell, C.J.; Aronoff, D.M.; Crofford, L.J.; Harris, R.C. Human and machine intelligence together drive drug repurposing in rare diseases. Front. Genet. 2021, 12, 707836. [Google Scholar] [CrossRef]
- Alves, V.M.; Korn, D.; Pervitsky, V.; Thieme, A.; Capuzzi, S.J.; Baker, N.; Chirkova, R.; Ekins, S.; Muratov, E.N.; Hickey, A. Knowledge-based approaches to drug discovery for rare diseases. Drug Discov. Today 2022, 27, 490–502. [Google Scholar] [CrossRef]
- Tanoli, Z.; Vähä-Koskela, M.; Aittokallio, T. Artificial intelligence, machine learning, and drug repurposing in cancer. Expert. Opin. Drug Discov. 2021, 16, 977–989. [Google Scholar] [CrossRef]
- Islam, M.T.; Newaz, A.A.H.; Paul, R.; Hassan Melon, M.M.; Hussen, M. Ai-Driven Drug Repurposing: Uncovering Hidden Potentials Of Established Medications For Rare Disease Treatment. Libr. Prog. -Libr. Sci. Inf. Technol. Comput. 2024, 44, 21949–21965. [Google Scholar] [CrossRef]
- Rahman, M.M.; Islam, M.R.; Rahman, F.; Rahaman, M.S.; Khan, M.S.; Abrar, S.; Ray, T.K.; Uddin, M.B.; Kali, M.S.K.; Dua, K. Emerging promise of computational techniques in anti-cancer research: At a glance. Bioengineering 2022, 9, 335. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Issa, N.T.; Stathias, V.; Schürer, S.; Dakshanamurthy, S. Machine and deep learning approaches for cancer drug repurposing. Semin. Cancer Biol. 2021, 68, 132–142. [Google Scholar] [CrossRef]
- Yaseen, B.T.; Kurnaz, S. Drug–target interaction prediction using artificial intelligence. Appl. Nanosci. 2023, 13, 3335–3345. [Google Scholar] [CrossRef]
- Parisi, D.; Adasme, M.F.; Sveshnikova, A.; Bolz, S.N.; Moreau, Y.; Schroeder, M. Drug repositioning or target repositioning: A structural perspective of drug-target-indication relationship for available repurposed drugs. Comput. Struct. Biotechnol. J. 2020, 18, 1043–1055. [Google Scholar] [CrossRef]
- Cheng, F.; Lu, W.; Liu, C.; Fang, J.; Hou, Y.; Handy, D.E.; Wang, R.; Zhao, Y.; Yang, Y.; Huang, J. A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat. Commun. 2019, 10, 3476. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Y.; Chen, S.; Wang, J. DeepDRK: A deep learning framework for drug repurposing through kernel-based multi-omics integration. Brief. Bioinform. 2021, 22, bbab048. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cui, H.; Zhang, T.; Cao, Y.; Xuan, P. Learning multi-scale heterogenous network topologies and various pairwise attributes for drug–disease association prediction. Brief. Bioinform. 2022, 23, bbac009. [Google Scholar] [CrossRef]
- Wang, L.; Song, Y.; Wang, H.; Zhang, X.; Wang, M.; He, J.; Li, S.; Zhang, L.; Li, K.; Cao, L. Advances of Artificial Intelligence in Anti-Cancer Drug Design: A Review of the Past Decade. Pharmaceuticals 2023, 16, 253. [Google Scholar] [CrossRef] [PubMed]
- Jarada, T.N.; Rokne, J.G.; Alhajj, R. SNF-NN: Computational method to predict drug-disease interactions using similarity network fusion and neural networks. BMC Bioinform. 2021, 22, 1–20. [Google Scholar] [CrossRef]
- Luo, H.; Wang, J.; Li, M.; Luo, J.; Peng, X.; Wu, F.-X.; Pan, Y. Drug repositioning based on comprehensive similarity measures and bi-random walk algorithm. Bioinformatics 2016, 32, 2664–2671. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, S.; Lu, J.; Ngom, A. An Integrative Heterogeneous Graph Neural Network–Based Method for Multi-Labeled Drug Repurposing. Front. Pharmacol. 2022, 13, 908549. [Google Scholar] [CrossRef]
- Doshi, S.; Chepuri, S.P. A computational approach to drug repurposing using graph neural networks. Comput. Biol. Med. 2022, 150, 105992. [Google Scholar] [CrossRef]
- Oliveira, T.A.d.; Silva, M.P.d.; Maia, E.H.B.; Silva, A.M.d.; Taranto, A.G. Virtual Screening Algorithms in Drug Discovery: A Review Focused on Machine and Deep Learning Methods. Drugs Drug Candidates 2023, 2, 311–334. [Google Scholar] [CrossRef]
- Li, Q.; Ma, Z.; Qin, S.; Zhao, W.-J. Virtual screening-based drug development for the treatment of nervous system diseases. Curr. Neuropharmacol. 2023, 21, 2447–2464. [Google Scholar] [CrossRef] [PubMed]
- Muhammed, M.T.; Aki-Yalcin, E. Molecular docking: Principles, advances, and its applications in drug discovery. Lett. Drug Des. Discov. 2024, 21, 480–495. [Google Scholar] [CrossRef]
- Hassan, M.; Ashraf, Z.; Abbas, Q.; Raza, H.; Seo, S.-Y. Exploration of novel human tyrosinase inhibitors by molecular modeling, docking and simulation studies. Interdiscip. Sci. Comput. Life Sci. 2018, 10, 68–80. [Google Scholar] [CrossRef]
- Hassan, M.; Yasir, M.; Shahzadi, S.; Kloczkowski, A. Exploration of Potential Ewing Sarcoma Drugs from FDA-Approved Pharmaceuticals through Computational Drug Repositioning, Pharmacogenomics, Molecular Docking, and MD Simulation Studies. ACS Omega 2022, 7, 19243–19260. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Park, J.; Han, E.-T.; Park, W.S.; Han, J.-H.; Kwon, Y.-S.; Lee, H.-J.; Hassan, M.; Kloczkowski, A.; Chun, W. Exploration of Flavonoids as Lead Compounds against Ewing Sarcoma through Molecular Docking, Pharmacogenomics Analysis, and Molecular Dynamics Simulations. Molecules 2023, 28, 414. [Google Scholar] [CrossRef]
- Nandy, A.; Duan, C.; Taylor, M.G.; Liu, F.; Steeves, A.H.; Kulik, H.J. Computational discovery of transition-metal complexes: From high-throughput screening to machine learning. Chem. Rev. 2021, 121, 9927–10000. [Google Scholar] [CrossRef]
- Bleicher, K.H.; Böhm, H.-J.; Müller, K.; Alanine, A.I. Hit and lead generation: Beyond high-throughput screening. Nat. Rev. Drug Discov. 2003, 2, 369–378. [Google Scholar] [CrossRef] [PubMed]
- del Carmen Quintal Bojórquez, N.; Campos, M.R. Traditional and Novel Computer-Aided Drug Design (CADD) Approaches in the Anticancer Drug Discovery Process. Curr. Cancer Drug Targets 2023, 23, 333–345. [Google Scholar]
- Lavecchia, A.; Di Giovanni, C. Virtual screening strategies in drug discovery: A critical review. Curr. Med. Chem. 2013, 20, 2839–2860. [Google Scholar] [CrossRef]
- Yasuo, N.; Sekijima, M. Improved method of structure-based virtual screening via interaction-energy-based learning. J. Chem. Inf. Model. 2019, 59, 1050–1061. [Google Scholar] [CrossRef]
- Kumar, N.; Acharya, V. Advances in machine intelligence-driven virtual screening approaches for big-data. Med. Res. Rev. 2023, 44, 939–974. [Google Scholar] [CrossRef] [PubMed]
- Suay-García, B.; Bueso-Bordils, J.I.; Falcó, A.; Antón-Fos, G.M.; Alemán-López, P.A. Virtual combinatorial chemistry and pharmacological screening: A short guide to drug design. Int. J. Mol. Sci. 2022, 23, 1620. [Google Scholar] [CrossRef]
- Berrhail, F.; Belhadef, H.; Haddad, M. Deep Convolutional Neural Network to improve the performances of screening process in LBVS. Expert. Syst. Appl. 2022, 203, 117287. [Google Scholar] [CrossRef]
- Kalaszi, A.; Szisz, D.; Imre, G.; Polgar, T. Screen3D: A novel fully flexible high-throughput shape-similarity search method. J. Chem. Inf. Model. 2014, 54, 1036–1049. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Raza, H.; Abbasi, M.A.; Moustafa, A.A.; Seo, S.-Y. The exploration of novel Alzheimer’s therapeutic agents from the pool of FDA approved medicines using drug repositioning, enzyme inhibition and kinetic mechanism approaches. Biomed. Pharmacother. 2019, 109, 2513–2526. [Google Scholar] [CrossRef]
- Hassan, M.; Abbas, Q.; Ashraf, Z.; Moustafa, A.A.; Seo, S.-Y. Pharmacoinformatics exploration of polyphenol oxidases leading to novel inhibitors by virtual screening and molecular dynamic simulation study. Comput. Biol. Chem. 2017, 68, 131–142. [Google Scholar] [CrossRef]
- Niazi, S.K.; Mariam, Z. Recent advances in machine-learning-based chemoinformatics: A comprehensive review. Int. J. Mol. Sci. 2023, 24, 11488. [Google Scholar] [CrossRef]
- Konaklieva, M.I.; Plotkin, B.J. Fragment-Based Lead Discovery Strategies in Antimicrobial Drug Discovery. Antibiotics 2023, 12, 315. [Google Scholar] [CrossRef]
- Singh, M.; Tam, B.; Akabayov, B. NMR-fragment based virtual screening: A brief overview. Molecules 2018, 23, 233. [Google Scholar] [CrossRef]
- Kawatkar, S.; Wang, H.; Czerminski, R.; Joseph-McCarthy, D. Virtual fragment screening: An exploration of various docking and scoring protocols for fragments using Glide. J. Comput.-Aided Mol. Des. 2009, 23, 527–539. [Google Scholar] [CrossRef]
- You, Y.; Lai, X.; Pan, Y.; Zheng, H.; Vera, J.; Liu, S.; Deng, S.; Zhang, L. Artificial intelligence in cancer target identification and drug discovery. Signal Transduct. Target. Ther. 2022, 7, 156. [Google Scholar] [CrossRef]
- D’Souza, S.; Prema, K.; Balaji, S. Machine learning models for drug–target interactions: Current knowledge and future directions. Drug Discov. Today 2020, 25, 748–756. [Google Scholar] [CrossRef]
- Peng, J.; Wang, Y.; Guan, J.; Li, J.; Han, R.; Hao, J.; Wei, Z.; Shang, X. An end-to-end heterogeneous graph representation learning-based framework for drug–target interaction prediction. Brief. Bioinform. 2021, 22, bbaa430. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.; Zhang, Y.; Wen, Y.; Zhang, Z.; He, S.; Bo, X. DTI-HETA: Prediction of drug–target interactions based on GCN and GAT on heterogeneous graph. Brief. Bioinform. 2022, 23, bbac109. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhong, W.; Zhao, L.; Chen, C.Y.-C. ML-DTI: Mutual learning mechanism for interpretable drug–target interaction prediction. J. Phys. Chem. Lett. 2021, 12, 4247–4261. [Google Scholar] [CrossRef]
- Raies, A.; Tulodziecka, E.; Stainer, J.; Middleton, L.; Dhindsa, R.S.; Hill, P.; Engkvist, O.; Harper, A.R.; Petrovski, S.; Vitsios, D. DrugnomeAI is an ensemble machine-learning framework for predicting druggability of candidate drug targets. Commun. Biol. 2022, 5, 1291. [Google Scholar] [CrossRef]
- Guo, J.; Liu, H.; Zheng, J. SynLethDB: Synthetic lethality database toward discovery of selective and sensitive anticancer drug targets. Nucleic Acids Res. 2016, 44, D1011–D1017. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xu, F.; Li, Y.; Wang, J.; Zhang, K.; Liu, Y.; Wu, M.; Zheng, J. KG4SL: Knowledge graph neural network for synthetic lethality prediction in human cancers. Bioinformatics 2021, 37, i418–i425. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Born, J.; Manica, M. Trends in deep learning for property-driven drug design. Curr. Med. Chem. 2021, 28, 7862–7886. [Google Scholar] [CrossRef]
- Samanta, B.; De, A.; Jana, G.; Gómez, V.; Chattaraj, P.; Ganguly, N.; Gomez-Rodriguez, M. Nevae: A deep generative model for molecular graphs. J. Mach. Learn. Res. 2020, 21, 1–33. [Google Scholar] [CrossRef]
- Grisoni, F.; Moret, M.; Lingwood, R.; Schneider, G. Bidirectional molecule generation with recurrent neural networks. J. Chem. Inf. Model. 2020, 60, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014, 27, 2672–2680. [Google Scholar]
- Maziarka, Ł.; Pocha, A.; Kaczmarczyk, J.; Rataj, K.; Danel, T.; Warchoł, M. Mol-CycleGAN: A generative model for molecular optimization. J. Cheminformatics 2020, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, M.; Santos, B.P.; Pereira, T.C.; Sofia, R.; Monteiro, N.R.; Simões, C.J.; Brito, R.M.; Ribeiro, B.; Oliveira, J.L.; Arrais, J.P. Designing optimized drug candidates with Generative Adversarial Network. J. Cheminformatics 2022, 14, 40. [Google Scholar] [CrossRef]
- Salih, A.M.; Menegaz, G.; Pillay, T.; Boyle, E.M. Explainable Artificial Intelligence in Paediatric: Challenges for the Future. Health Sci. Rep. 2024, 7, e70271. [Google Scholar] [CrossRef]
- Tippur, A. AI-Powered Precision Oncology: Computational Insights Redefining Therapeutic Landscapes. DHR Proc. 2023, 3, 1–10. [Google Scholar] [CrossRef]
- Jiang, X.; Hu, Z.; Wang, S.; Zhang, Y. Deep learning for medical image-based cancer diagnosis. Cancers 2023, 15, 3608. [Google Scholar] [CrossRef]
- Yeasmin, M.N.; Al Amin, M.; Joti, T.J.; Aung, Z.; Azim, M.A. Advances of AI in image-based computer-aided diagnosis: A review. Array 2024, 23, 100357. [Google Scholar] [CrossRef]
- Mann, M.; Kumar, C.; Zeng, W.-F.; Strauss, M.T. Artificial intelligence for proteomics and biomarker discovery. Cell Syst. 2021, 12, 759–770. [Google Scholar] [CrossRef]
- Kumar, A. AI-driven precision oncology: Predictive biomarker discovery and personalized treatment optimization using genomic data. Int. J. Adv. Res. Publ. Rev. 2024, 1, 21–38. [Google Scholar] [CrossRef]
- Huynh, E.; Hosny, A.; Guthier, C.; Bitterman, D.S.; Petit, S.F.; Haas-Kogan, D.A.; Kann, B.; Aerts, H.J.; Mak, R.H. Artificial intelligence in radiation oncology. Nat. Rev. Clin. Oncol. 2020, 17, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Sarker, I.H. Machine learning: Algorithms, real-world applications and research directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef] [PubMed]
- Gurumurthy, G.; Gurumurthy, J.; Gurumurthy, S. Machine learning in paediatric haematological malignancies: A systematic review of prognosis, toxicity and treatment response models. Pediatr. Res. 2025, 97, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.; Chokkara, S.; Shen, T.; Major, A.; Volchenboum, S.L.; Mayampurath, A.; Applebaum, M.A. Applications of artificial intelligence in pediatric oncology: A systematic review. JCO Clin. Cancer Inform. 2021, 5, 1208–1219. [Google Scholar] [CrossRef]
- Elsayed, B.; Elhadary, M.; Elshoeibi, R.M.; Elshoeibi, A.M.; Badr, A.; Metwally, O.; ElSherif, R.A.; Salem, M.E.; Khadadah, F.; Alshurafa, A. Deep learning enhances acute lymphoblastic leukemia diagnosis and classification using bone marrow images. Front. Oncol. 2023, 13, 1330977. [Google Scholar] [CrossRef]
- Huang, J.; Shlobin, N.A.; Lam, S.K.; DeCuypere, M. Artificial intelligence applications in pediatric brain tumor imaging: A systematic review. World Neurosurg. 2022, 157, 99–105. [Google Scholar] [CrossRef]
- Pacchiano, F.; Tortora, M.; Doneda, C.; Izzo, G.; Arrigoni, F.; Ugga, L.; Cuocolo, R.; Parazzini, C.; Righini, A.; Brunetti, A. Radiomics and artificial intelligence applications in pediatric brain tumors. World J. Pediatr. 2024, 20, 747–763. [Google Scholar] [CrossRef]
- Fu, S.W.; Tang, C.; Tan, X.; Srivastava, S. Liquid biopsy for early cancer detection: Technological revolutions and clinical dilemma. Expert. Rev. Mol. Diagn. 2024, 24, 937–955. [Google Scholar] [CrossRef]
- Eloranta, S.; Boman, M. Predictive models for clinical decision making: Deep dives in practical machine learning. J. Intern. Med. 2022, 292, 278–295. [Google Scholar] [CrossRef]
- Joshi, H. Natural Language Processing of Electronic Health Records for Predicting Alzheimer’s Disease. In Deep Generative Models for Integrative Analysis of Alzheimer’s Biomarkers; IGI Global: Hershey, PA, USA, 2025; pp. 141–174. [Google Scholar]
- Sweet-Cordero, E.A.; Biegel, J.A. The genomic landscape of pediatric cancers: Implications for diagnosis and treatment. Science 2019, 363, 1170–1175. [Google Scholar] [CrossRef] [PubMed]
- Umeda, M.; Ma, J.; Westover, T.; Ni, Y.; Song, G.; Maciaszek, J.L.; Rusch, M.; Rahbarinia, D.; Foy, S.; Huang, B.J.; et al. A new genomic framework to categorize pediatric acute myeloid leukemia. Nat Genet 2024, 56, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Wen, J.; Meggendorfer, M.; Choi, J.K.; Shi, L.; Pounds, S.B.; Carmichael, C.L.; Masih, K.E.; Morris, S.M.; Lindsley, R.C. Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat. Genet. 2019, 51, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Bertrums, E.J.; Smith, J.L.; Harmon, L.; Ries, R.E.; Wang, Y.-C.J.; Alonzo, T.A.; Menssen, A.J.; Chisholm, K.M.; Leonti, A.R.; Tarlock, K. Comprehensive molecular and clinical characterization of NUP98 fusions in pediatric acute myeloid leukemia. Haematologica 2023, 108, 2044. [Google Scholar] [CrossRef]
- Dlamini, Z.; Francies, F.Z.; Hull, R.; Marima, R. Artificial intelligence (AI) and big data in cancer and precision oncology. Comput. Struct. Biotechnol. J. 2020, 18, 2300–2311. [Google Scholar] [CrossRef]
- Li, Q.; Ren, Z.; Cao, K.; Li, M.M.; Wang, K.; Zhou, Y. CancerVar: An artificial intelligence–empowered platform for clinical interpretation of somatic mutations in cancer. Sci. Adv. 2022, 8, eabj1624. [Google Scholar] [CrossRef]
- da Rocha, J.L.D.; Lai, J.; Pandey, P.; Myat, P.S.M.; Loschinskey, Z.; Bag, A.K.; Sitaram, R. Artificial intelligence for neuroimaging in pediatric cancer. Cancers 2025, 17, 622. [Google Scholar]
- Biswas, N.; Chakrabarti, S. Artificial intelligence (AI)-based systems biology approaches in multi-omics data analysis of cancer. Front. Oncol. 2020, 10, 588221. [Google Scholar] [CrossRef]
- Wei, L.; Niraula, D.; Gates, E.D.; Fu, J.; Luo, Y.; Nyflot, M.J.; Bowen, S.R.; El Naqa, I.M.; Cui, S. Artificial intelligence (AI) and machine learning (ML) in precision oncology: A review on enhancing discoverability through multiomics integration. Br. J. Radiol. 2023, 96, 20230211. [Google Scholar] [CrossRef]
- Kaliappan, S.; Maranan, R.; Ali, H.M. AI-enabled predictive modeling and management of rare pediatric cancers. In Proceedings of the 2023 IEEE International Conference on ICT in Business Industry & Government (ICTBIG), Indore, India, 8–9 December 2023; pp. 1–6. [Google Scholar]
- Ramgopal, S.; Sanchez-Pinto, L.N.; Horvat, C.M.; Carroll, M.S.; Luo, Y.; Florin, T.A. Artificial intelligence-based clinical decision support in pediatrics. Pediatr. Res. 2023, 93, 334–341. [Google Scholar] [CrossRef]
- Mohsin, F.M.; Nesa, M.; Mostafa, S.M.; Das, A.; Mostafa, Z.; Noor, R.; Nisa, Z.U. The Role of AI in Predicting Cancer Recurrence and Patient Survival Rates. Am. J. Multidiscip. Res. Dev. (AJMRD) 2025, 7, 73–79. [Google Scholar]
- Shah, A.C.; Badawy, S.M. Telemedicine in pediatrics: Systematic review of randomized controlled trials. JMIR Pediatr. Parent. 2021, 4, e22696. [Google Scholar] [CrossRef]
- Jeddi, Z.; Bohr, A. Remote patient monitoring using artificial intelligence. In Artificial Intelligence in Healthcare; Elsevier: Amsterdam, The Netherlands, 2020; pp. 203–234. [Google Scholar]
- Ganatra, H.A. Machine Learning in Pediatric Healthcare: Current Trends, Challenges, and Future Directions. J. Clin. Med. 2025, 14, 807. [Google Scholar] [CrossRef]
- Elsayid, N.N.; Adam, E.I.A.; Mahmoud, S.M.Y.; Saadeldeen, H.; Nauman, M.; Ahmed, T.A.A.; Yousif, B.A.H.; Taha, A.I.A.; Adam Sr, E.I.A.; Yousif Sr, B.A.H. The Role of Machine Learning Approaches in Pediatric Oncology: A Systematic Review. Cureus 2025, 17, e77524. [Google Scholar] [CrossRef]
- Di Biasi, L. The Use of Artificial Intelligence for the Melanoma Binary Classification Problem on Images (MIBCP). 2023. Available online: http://elea.unisa.it/handle/10556/7303 (accessed on 27 May 2025).
| AI Method | Medical Field | Task | Tumor |
|---|---|---|---|
| CNN/GAN | Pathology | Detecting ALL and AML using a deep learner classifier using microscopic blood images | ALL and AML |
| CNN and GAN | Pathology/Genomics | Constructing a hybrid model using a genetic algorithm and a residual CNN to predict ALL using microscopy images | ALL |
| SVM | Pathology | Building a model to classify acute leukemias using flow cytometry | Acute promyelocytic leukemia |
| ANN/FFNN/SVM | Pathology | Proposing an ML-based model for ALL categorization using microscopic blood images | ALL |
| CNN | Pathology | Building an aggregated DL model for leukemic B-lymphoblast classification | Leukemic B-lymphoblast |
| CNN | Pathology | Using bone marrow cell microscopy images for the classification of AML, ALL, and CML | AML, ALL, and CML |
| RF | Others-mRNA sequencing | Developing transcriptome-wide biomarkers for ALL subtyping | ALL |
| ANN | Others-DNA methylation | Identifying reliable cancer-associated methylation signals in gene regions from leukemia patients | Leukemia |
| Nearest shrunken centroids | Others-DNA methylation | Investigating the utility of CpG methylation status to differentiate blood from patients with ALL and AML from normal blood | ALL and AML |
| AI Models | Cancer Type | Data Type | Application | Performance Metrics | Key Features |
|---|---|---|---|---|---|
| Deep Learning CNN | Acute Lymphoblastic Leukemia (ALL), AML, CML | Bone marrow cell microscopy images | Classification of leukemias | Accuracy: 90–99%; Some models >98% | Outperforms expert hematologists; hybrid models improve with genetic algorithms |
| Transcriptomic & Methylation-Based ML | ALL Subtyping | mRNA sequencing, DNA methylation profiles | Subtyping and diagnosis | Accuracy: 93.8–100%; AUC up to 99.98 | Uses multi-omics; provides reliable leukemia subtype discrimination |
| ML Classification (SVM, RF) | Acute Promyelocytic Leukemia | Flow cytometry | Leukemia classification | ACC: 94.2%; AUC: 99.5 | Effective in flow cytometry data |
| DL-CNN | Pediatric Brain Tumors (Medulloblastoma, Gliomas, Ependymomas) | MRI sequences, Histological images | Tumor subtype classification and diagnosis | Accuracy: 75–95.5%; AUC: 0.81–0.99 | Improved sensitivity and specificity; supports real-time and intraoperative diagnostics |
| Temporal Deep Learning Model | Pediatric Gliomas | Sequential brain MRI scans | Predict recurrence risk | Accuracy: 75–89% | Uses temporal learning on multiple longitudinal scans, outperforming single-timepoint methods |
| DL-Based PET/MR Image Augmentation | Pediatric Lymphoma | Ultralow-dose PET/MR images | Image quality enhancement, dose reduction | Radiation dose reduction >90% | Augments image quality to reduce radiation exposure in imaging |
| ML Radiomics + CT | Osteosarcoma | CT scan of primary tumor | Predict lung metastases | Accuracy: 73% | Early metastatic risk prediction; needs further validation |
| DL-CNN + Multimodal MRI | Osteosarcoma | MRI images (T1, STIR, postcontrast) | Chemotherapy response evaluation | Accuracy: >90% | Differentiates necrotic vs. viable tumor areas |
| ML Model Using FDG PET | Osteosarcoma | Baseline FDG PET imaging | Predict neoadjuvant chemotherapy response | AUC: Up to 0.863 | Texture feature analysis improved response prediction |
| CNN Classifier | Ewing Sarcoma | Radiographs | Lesion detection and differentiation | Accuracy: ~90–94% | Differentiates Ewing sarcoma and osteomyelitis effectively |
| DL-CNN Classifier | Wilms Tumor | Triphasic CT images | Tumor differentiation and staging | Sensitivity: 78.1%; Accuracy: ~79% | Outperforms human experts for non-Wilms tumor detection |
| ML Classifiers (SVM, RF etc.) | Soft-Tissue Sarcomas | Radiological images, histopathology slides | Malignant vs. benign differentiation | Accuracy: 80.8–90.5%; AUC: 0.88–0.96 | Applied on histopathology and imaging, effective in pediatric soft-tissue masses classification |
| CNN-Based Dermatology AI | Infantile hemangiomas | Clinical and dermoscopic photos | Disease diagnosis | Accuracy: 91.7% | Non-invasive clinical image-based AI diagnosis |
| ML & Proteomics | Pediatric Brain Tumors | CSF proteomic profiles | Tumor subtype classification | AUC: 0.97–1 | Classifies brain tumors accurately with proteomics and ML algorithms |
| Raman Spectroscopy + ML | Intraoperative Pediatric Brain Tumors | Raman spectroscopy data | Real-time tumor vs. normal tissue differentiation | AUC: 0.91–0.94 | Enables safe tumor resections intraoperatively |
| AI Approaches | Explanation | Ref. |
|---|---|---|
| Variant identification | Identify and classify the somatic and other genomic alterations from sequencing data. | [130] |
| Pattern recognition and discovery | Discover hidden relationships and patterns in large datasets to identify new driver genes, pathways, and possible treatment targets. | [9,131] |
| Integration of multi-omics data | Integrate information from genomics, transcriptomics, proteomics, and other omics data sources for treatment prediction. | [132,133] |
| Predictive modeling | Make informed clinical decisions and enhance the care of individual patients by using their genetic profiles to predict treatment success, relapse risk, and patient outcomes. | [134] |
| Clinical Trial Design and Patient Stratification | Optimize clinical trial design and predict treatment responses based on patient characteristics and molecular profiles. | [135] |
| Prognostic and Predictive Analytics | Survival prediction and recurrence risk for the patients | [136] |
| Telemedicine and Remote Monitoring | Remote consultations and continuous monitoring of patients | [137,138] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, M.; Shahzadi, S.; Kloczkowski, A. Harnessing Artificial Intelligence in Pediatric Oncology Diagnosis and Treatment: A Review. Cancers 2025, 17, 1828. https://doi.org/10.3390/cancers17111828
Hassan M, Shahzadi S, Kloczkowski A. Harnessing Artificial Intelligence in Pediatric Oncology Diagnosis and Treatment: A Review. Cancers. 2025; 17(11):1828. https://doi.org/10.3390/cancers17111828
Chicago/Turabian StyleHassan, Mubashir, Saba Shahzadi, and Andrzej Kloczkowski. 2025. "Harnessing Artificial Intelligence in Pediatric Oncology Diagnosis and Treatment: A Review" Cancers 17, no. 11: 1828. https://doi.org/10.3390/cancers17111828
APA StyleHassan, M., Shahzadi, S., & Kloczkowski, A. (2025). Harnessing Artificial Intelligence in Pediatric Oncology Diagnosis and Treatment: A Review. Cancers, 17(11), 1828. https://doi.org/10.3390/cancers17111828







