Clinical Application of Next-Generation Sequencing for Molecular Classification in the Management of Endometrial Cancer: An Observational Cohort Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Selections
2.2. Next-Generation Sequencing (NGS) Assay
2.3. Immunohistochemistry
2.4. Statistical Analysis
3. Results
3.1. Clinicopathological Characteristics
3.2. Molecular Subtyping
3.3. POLE
3.4. MMRd/MSI
3.5. p53abn/TP53mut
3.6. NSMP
3.7. Concordance Between IHC, NGS, and PCR Results
3.8. Identification of Additional Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tung, H.J.; Wu, R.C.; Lin, C.Y.; Lai, C.H. Rare Subtype of Endometrial Cancer: Undifferentiated/Dedifferentiated Endometrial Carcinoma, from Genetic Aspects to Clinical Practice. Int. J. Mol. Sci. 2022, 30, 23. [Google Scholar] [CrossRef] [PubMed]
- Levine, D.; The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Plotkin, A.; Kuzeljevic, B.; De Villa, V.; Thompson, E.F.; Gilks, C.B.; Clarke, B.A.; Köbel, M.; McAlpine, J.N. Interlaboratory Concordance of ProMisE Molecular Classification of Endometrial Carcinoma Based on Endometrial Biopsy Specimens. Int. J. Gynecol. Pathol. 2020, 39, 537–545. [Google Scholar] [CrossRef]
- Stelloo, E.; Bosse, T.; Nout, R.A.; MacKay, H.J.; Church, D.N.; Nijman, H.W.; Bosse, T.; Smit, V.T.; Colman, J.M.; van Wezel, T.; et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod. Pathol. 2015, 28, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jurgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Bulten, J.; Creutzberg, C.L.; de Kroon, C.D. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial Cancer-Combined Analysis of the PORTEC Cohorts. Clin. Cancer Res. 2016, 22, 4215–4224. [Google Scholar] [CrossRef] [PubMed]
- Kommoss, S.; McConechy, M.K.; Kommoss, F.; Leung, S.; Bunz, A.; Magrill, J.; Huntsman, D.G.; Gilks, C.B.; Talhouk, A.; Köbel, M.; et al. Final validation of the ProMisE molecular classifier for endometrial carcinoma in a large population-based case series. Ann. Oncol. 2018, 29, 1180–1188. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Blake Gilks, C.; Huntsman, D.; Köbel, M.; et al. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Blake Gilks, C.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of next-generation sequencing technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef]
- Huvila, J.; Orte, K.; Vainio, P.; Mettala, T.; Joutsiniemi, T.; Hietanen, S. Molecular subtype diagnosis of endometrial carcinoma: Comparison of the next-generation sequencing panel and proactive molecular risk classifier for endometrial cancer classifier. Hum. Pathol. 2021, 111, 98–109. [Google Scholar] [CrossRef]
- Berek, J.S.; Matias-Guiu, X.; Creutzberg, C.; Fotopoulou, C.; Gaffney, D.; Kehoe, S.; Lax, S.; Ray-Coquard, I.; Mirza, M.; Morice, P.; et al. FIGO staging of endometrial cancer: 2023. Int. J. Gynaecol. Obstet. 2023, 162, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Yang, X.; Guo, L.; Liu, B.; Lin, J.; Liang, H.; Wei, L.; Zhao, Z.; Zhang, H.; Wang, J.; et al. MSI sensor-pro: Fast, accurate, and matched-normal-sample-free detection of microsatellite instability. Genom. Proteom. Bioinform. 2020, 18, 65–71. [Google Scholar] [CrossRef]
- Köbel, M.; Ronnett, B.M.; Singh, N.; Soslow, R.A.; Gilks, C.B.; McCluggage, W.G.; Kommoss, S.; Leung, S.; Huntsman, D.; Talhouk, A.; et al. Interpretation of P53 immunohistochemistry in endometrial carcinomas: Toward increased reproducibility. Int. J. Gynecol. Pathol. 2019, 38, S123–S131. [Google Scholar] [CrossRef]
- León-Castillo, A.; Gilvazquez, E.; Nout, R.; Smith, V.; McAlpine, J.N.; Church, D.N.; McConechy, M.K.; Kommoss, S.; Talhouk, A.; Singh, N.; et al. Clinicopathological and molecular characterisation of ‘multiple-classifier’ endometrial carcinomas. J. Pathol. 2020, 250, 312–322. [Google Scholar] [CrossRef]
- León-Castillo, A.; Britton, H.; McConechy, M.K.; McAlpine, J.N.; Nout, R.; Kommoss, S.; Grevenkamp, F.; Hoang, L.; Sisler, J.; Leung, S.; et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J. Pathol. 2020, 250, 323–335. [Google Scholar] [CrossRef]
- Huvila, J.; Thompson, E.F.; Vanden Broek, J.; Lum, A.; Senz, J.; Leung, S.; Kommoss, S.; McConechy, M.K.; Gilks, C.B.; Köbel, M.; et al. Subclonal P53 Immunostaining in the Diagnosis of Endometrial Carcinoma Molecular Subtype. Histopathology 2023, 83, 880–890. [Google Scholar] [CrossRef] [PubMed]
- Catasus, L.; Gallardo, A.; Cuatrecasas, M.; Prat, J. PIK3CA mutations in the kinase domain (exon 20) of uterine endometrial adenocarcinomas are associated with adverse prognostic parameters. Mod. Pathol. 2008, 21, 131–139. [Google Scholar] [CrossRef]
- Schultheis, A.M.; Martelotto, L.G.; De Filippo, M.R.; Piscuglio, S.; Ng, C.K.; Hussein, Y.R.; Reis-Filho, J.S.; Soslow, R.A.; Weigelt, B. TP53 mutational spectrum in endometrioid and serous endometrial cancers. Int. J. Gynecol. Pathol. 2016, 35, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Stokoe, D.; Taketani, Y.; McCormick, F. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005, 65, 10669–10673. [Google Scholar] [CrossRef]
- Konstantinova, D.; Kaneva, R.; Dimitrov, R.; Savov, A.; Ivanov, S.; Dyankova, T.; Shivarov, V.; Jordanova, D.; Angelova, S.; Toncheva, D.; et al. Rare mutations in the PIK3CA gene contribute to aggressive endometrial cancer. DNA Cell Biol. 2010, 29, 65–70. [Google Scholar] [CrossRef]
- Toss, A.; Piacentini, F.; Cortesi, L.; Artuso, L.; Bernardis, I.; Parenti, S.; Gambini, D.; Moscetti, L.; Cattaneo, L.; Dominici, S.; et al. Genomic alterations at the basis of treatment resistance in metastatic breast cancer: Clinical applications. Oncotarget 2018, 9, 31606–31619. [Google Scholar] [CrossRef]
- AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Konopka, B.; Janiec-Jankowska, A.; Kwiatkowska, E.; Najmoła, U.; Bidziński, M.; Olszewski, W.; Czapiewski, P.; Stawinski, J.; Kotarski, J.; Kowalewska, M.; et al. PIK3CA mutations and amplification in endometrioid endometrial carcinomas: Relation to other genetic defects and clinicopathologic status of the tumors. Hum. Pathol. 2011, 42, 1710–1719. [Google Scholar] [CrossRef]
- Rudd, M.L.; Price, J.C.; Fogoros, S.; Godwin, A.K.; Sgroi, D.C.; Merino, M.J.; Chen, X.; Wang, Y.; Romero, L.; Broaddus, R.R.; et al. A unique spectrum of somatic PIK3CA (p110alpha) mutations within primary endometrial carcinomas. Clin. Cancer Res. 2011, 17, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Backes, F.J.; Walker, C.J.; Goodfellow, P.J.; Hade, E.M.; Agarwal, G.; Mutch, D.; Suarez, A.A.; Cohn, D.E.; Fowler, J.M.; Copeland, L.J.; et al. Estrogen receptor-alpha as a predictive biomarker in endometrioid endometrial cancer. Gynecol. Oncol. 2016, 141, 312–317. [Google Scholar] [CrossRef]
- Gibson, W.J.; Hoivik, E.A.; Halle, M.K.; Taylor-Weiner, A.; Cherniack, A.D.; Berg, A.; Holst, F.; Werner, H.M.; Bjorge, L.; Anglesio, M.S.; et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat. Genet. 2016, 48, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, Z.; Vahrenkamp, J.M.; Berrett, K.C.; Arnesen, S.; Gertz, J.; Hall, J.M.; Nevins, J.R.; Adams, J.; Malovannaya, A.; Aakre, C.A.; et al. Estrogen-independent molecular actions of mutant estrogen receptor 1 in endometrial cancer. Genome Res. 2019, 29, 1429–1441. [Google Scholar] [CrossRef]
- Velasco, A.; Bussaglia, E.; Pallares, J.; Dolcet, X.; Llobet, D.; Encinas, M.; Matias-Guiu, X.; Prat, J.; Montironi, R.; Furlanetto, B.; et al. PIK3CA gene mutations in endometrial carcinoma: Correlation with PTEN and K-RAS alterations. Hum. Pathol. 2006, 37, 1465–1472. [Google Scholar] [CrossRef]
- Rao, Q.; Liao, J.; Li, Y.; Zhang, X.; Xu, G.; Zhu, C.; Chen, Y.; Wang, H.; Huang, J.; Wang, X.; et al. Application of NGS molecular classification in the diagnosis of endometrial carcinoma: A supplement to traditional pathological diagnosis. Cancer Med. 2023, 12, 5409–5419. [Google Scholar] [CrossRef]
- León-Castillo, A.; de Boer, S.M.; Powell, M.E.; Mileshkin, L.R.; Mackay, H.J.; Leary, A.; Nijman, H.W.; Edmondson, R.J.; Altman, A.D.; Sisodia, R.C.; et al. Molecular Classification of the PORTEC-3 Trial for High-Risk Endometrial Cancer: Impact on Prognosis and Benefit From Adjuvant Therapy. J. Clin. Oncol. 2020, 38, 3388–3397. [Google Scholar] [CrossRef]
- Thompson, E.F.; Huvila, J.; Jamieson, A.; Leung, S.; Lum, A.; Offman, S.; McConechy, M.K.; Kommoss, S.; Brucker, S.Y.; Taran, F.A.; et al. Variability in endometrial carcinoma pathology practice: Opportunities for improvement with molecular classification. Mod. Pathol. 2022, 35, 1974–1982. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, A.; Thompson, E.F.; Huvila, J.; Gilks, C.B.; McAlpine, J.N. p53abn endometrial cancer: Understanding the most aggressive endometrial cancers in the era of molecular classification. Int. J. Gynecol. Cancer 2021, 31, 907–913. [Google Scholar] [CrossRef]
- Bosse, T.; Nout, R.A.; McAlpine, J.N.; McConechy, M.K.; Britton, H.; Hussein, Y.R.; Kommoss, S.; Brucker, S.Y.; Leitao, M.M.; Soslow, R.A.; et al. Molecular Classification of Grade 3 Endometrioid Endometrial Cancers Identifies Distinct Prognostic Subgroups. Am. J. Surg. Pathol. 2018, 42, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, A.; Lisa, M.; Barroilhet, L.M.; McAlpine, J.N. Molecular classification in endometrial cancer: Opportunities for precision oncology in a changing landscape. Cancer 2022, 128, 2853–2857. [Google Scholar] [CrossRef] [PubMed]
- Zannoni, G.F.; Bragantini, E.; Castiglione, F.; Fassan, M.; Troncone, G.; Inzani, F.; Santoro, A.; Fraggetta, F.; Angelico, G.; Capodanno, A.; et al. Current Prognostic and Predictive Biomarkers for Endometrial Cancer in Clinical Practice: Recommendations/Proposal from the Italian Study Group. Front. Oncol. 2022, 12, 805613. [Google Scholar] [CrossRef]
- De Vitis, L.A.; Schivardi, G.; Caruso, G.; Fumagalli, C.; Vacirca, D.; Achilarre, M.T.; Cossu Rocca, P.; Alessandrini, L.; Tozzi, L.; Scaglione, G.; et al. Clinicopathological characteristics of multiple-classifier endometrial cancers: A cohort study and systematic review. Int. J. Gynecol. Cancer 2024, 34, 229–238. [Google Scholar] [CrossRef]
- Bartley, A.N.; Mills, A.M.; Konnick, E.Q.; Overman, M.J.; Ventura, C.B.; Souter, L.H.; Lindor, N.M.; Pearlman, R.; Azad, N.S.; Church, J.M.; et al. Mismatch Repair and Microsatellite Instability Testing for Immune Checkpoint Inhibitor Therapy: Guideline from the College of American Pathologists in Collaboration With the Association for Molecular Pathology and Fight Colorectal Cancer. Arch. Pathol. Lab. Med. 2022, 146, 1194–1210. [Google Scholar] [CrossRef]
- Bateman, A.C. DNA mismatch repair proteins: Scientific update and practical guide. J. Clin. Pathol. 2021, 74, 264–268. [Google Scholar] [CrossRef]
- Dedeurwaerdere, F.; Claes, K.B.M.; Van Dorpe, J.; Rottiers, I.; Van Der Meulen, J.; Breyne, J.; Van Maerken, T.; Devriendt, K.; Segers, K.; Smeets, D.; et al. Comparison of microsatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Sci. Rep. 2021, 11, 12880. [Google Scholar] [CrossRef]
- NCCN. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Uterine Neoplasms Version 1.2024. Plymouth Meeting: NCCN. Available online: https://www.nccn.org/professionals/physician_gls/pdf/uterine.pdf (accessed on 3 March 2025).
- Li, C.; Song, W.; Xu, Y.; Guo, T.; Zhou, X.; Liu, F.; Chen, Z.; Yang, J.; Zhang, H.; Wang, L.; et al. A one-stop approach to diagnosing hereditary colorectal cancer in the Chinese population. J. Gastroenterol. Hepatol. 2023, 38, 1980–1987. [Google Scholar] [CrossRef]
- Guo, Q.; Tang, S.; Ju, X.; Feng, Z.; Zhang, Z.; Peng, D.; Li, M.; Zhou, F.; Wei, Y.; Wu, H.; et al. Identification of molecular subtypes for endometrial carcinoma using a 46-gene next-generation sequencing panel: A retrospective study on a consecutive color. ESMO Open 2024, 9, 103710. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shi, C.; Eisenberg, R.; Vnencak-Jones, C.L. Differences in Microsatellite Instability Profiles between Endometrioid and Colorectal Cancers: A Potential Cause for False-Negative Results? J. Mol. Diagn. 2017, 19, 57–64. [Google Scholar] [CrossRef]
- Amemiya, K.; Hirotsu, Y.; Nagakubo, Y.; Watanabe, S.; Amemiya, S.; Mochizuki, H.; Oikawa, R.; Nakagomi, T.; Oyama, T.; Omata, M. Simple IHC reveals complex MMR alternations than PCR assays: Validation by LCM and next-generation sequencing. Cancer Med. 2022, 11, 4479–4490. [Google Scholar] [CrossRef]
- Goodfellow, P.J.; Buttin, B.M.; Herzog, T.J.; Rader, J.S.; Gibb, R.K.; Swisher, E.; Look, K.Y.; Shih, I.M.; Mutoh, M.; Kolasa, I.; et al. Prevalence of defective DNA mismatch repair and MSH6 mutation in an unselected series of endometrial cancers. Proc. Natl. Acad. Sci. USA 2003, 100, 5908–5913. [Google Scholar] [CrossRef] [PubMed]
- Vermij, L.; Léon-Castillo, A.; Singh, N.; Bosse, T.; Kommoss, S.; Creutzberg, C.L.; Smit, V.T.; Powell, M.E.; Mackay, H.J.; Mileshkin, L.R.; et al. p53 immunohistochemistry in endometrial cancer: Clinical and molecular correlates in the PORTEC-3 trial. Mod. Pathol. 2022, 35, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Piskorz, A.M.; Bosse, T.; Jimenez-Linan, M.; Rous, B.; Brenton, J.D.; Gilks, C.B.; Kommoss, S.; Lax, S.F.; Djordjevic, B.; et al. p53 immunohistochemistry is an accurate surrogate for TP53 mutational analysis in endometrial carcinoma biopsies. J. Pathol. 2020, 250, 336–345. [Google Scholar] [CrossRef]
- Di Donato, V.; Giannini, A.; Bogani, G.; D’Oria, O.; Ghezzi, F.; Benedetti Panici, P.; Muzii, L.; Mancini, E.; Montico, M.; Marchetti, C. Recent Advances in Endometrial Cancer Management. J. Clin. Med. 2023, 12, 2241. [Google Scholar] [CrossRef]
- Golia, D.; Cuccu, I.; Santangelo, G.; Muzii, L.; Giannini, A.; Bogani, G.; D’Oria, O.; De Angelis, E.; Sgamba, L.; Panici, P.B.; et al. Novel Insights into Molecular Mechanisms of Endometrial Diseases. Biomolecules 2023, 13, 499. [Google Scholar] [CrossRef]
- Cuccu, I.; D’Oria, O.; Sgamba, L.; De Angelis, E.; D’Augè, T.G.; Turetta, C.; Di Dio, C.; Scudo, M.; Bogani, G.; Di Donato, V.; et al. Role of genomic and molecular biology in the modulation of the treatment of Endometrial Cancer: Narrative Review and Perspectives. Healthcare 2023, 11, 571. [Google Scholar] [CrossRef]
- Bogani, G.; Chiappa, V.; Lopez, S.; Salvatore, C.; Interlenghi, M.; D’Oria, O.; De Angelis, E.; Sgamba, L.; Muzii, L.; Giannini, A.; et al. Radiomics and Molecular Classification in Endometrial Cancer (The ROME Study): A Step Forward to a Simplified Precision Medicine. Healthcare 2022, 10, 2464. [Google Scholar] [CrossRef]
- McIntyre, J.B.; Nelson, G.S.; Ghatage, P.; Morris, D.; Duggan, M.A.; Lee, C.H.; Köbel, M.; Kalloger, S.E.; Gilks, C.B.; Kommoss, S.; et al. PIK3CA missense mutation is associated with unfavorable outcome in grade 3 endometrioid carcinoma but not in serous endometrial carcinoma. Gynecol. Oncol. 2014, 132, 188–193. [Google Scholar] [CrossRef]
- Dong, Y.; Yang, X.; Wong, O.; Zhang, X.; Liang, Y.; Zhang, Y.; Li, X.; Qiu, S.; He, F.; Wu, X.; et al. PIK3CA mutations in endometrial carcinomas in Chinese women: Phosphatidylinositol 3′-kinase pathway alterations might be associated with favorable prognosis. Hum. Pathol. 2012, 43, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Sideris, M.; Emin, E.I.; Abdullah, Z.; Hanrahan, J.; Stefatou, K.M.; Sevas, V.; Mitra, A.; Payne, F.; Kitchener, H.C.; Crosbie, E.J. The Role of KRAS in Endometrial Cancer: A Mini-Review. Anticancer Res. 2019, 39, 533–539. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-fam-trastuzumab-deruxtecan-nxki-unresectable-or-metastatic-her2 (accessed on 27 May 2024).
- Aro, K.; Loukovaara, M.; Bützow, R.; Pasanen, A. HER2 amplification and HER2 low expression in endometrial carcinoma: Prevalence across molecular, histological and clinicopathological risk groups. BJC Rep. 2025, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.S.; Devereaux, K.A.; Jin, C.; Shumansky, K.; Singhi, A.D.; Lee, M.; Rabban, J.T.; Chen, L.M.; Soslow, R.A.; Garcia-Closas, M.; et al. Histopathologic features and molecular genetic landscape of HER2-amplified endometrial carcinomas. Mod. Pathol. 2022, 35, 962–971. [Google Scholar] [CrossRef] [PubMed]
- Drouyer, A.; Beaussire, L.; Jorda, P.; Leheurteur, M.; Guillemet, C.; Berghian, A.; Lejeune, S.; Rousseau, F.; Vielh, P.; Sabourin, J.C.; et al. Clinical relevance of circulating ESR1 mutations during endocrine therapy for advanced hormone-dependent endometrial carcinoma. BMC Cancer 2023, 23, 1061. [Google Scholar] [CrossRef]
- Walsh, C.S.; Hacker, K.E.; Secord, A.A.; De Lair, D.F.; McCourt, C.K.; Urban, R.R.; Berchuck, A.; Lankes, H.A.; Backes, F.J.; Leath, C.A.; et al. Molecular testing for endometrial cancer: An SGO clinical practice statement. Gynecol. Oncol. 2023, 168, 48–55. [Google Scholar] [CrossRef]
- Abstract ESMO 2024. ESR1 Mutation in Untreated Endometrial Cancer: Prevalence, Characteristics and Prognostic Implications from the UTOLA Trial. Available online: https://www.esmoopen.com/article/S2059-7029(24)01310-3/pdf (accessed on 21 February 2025).
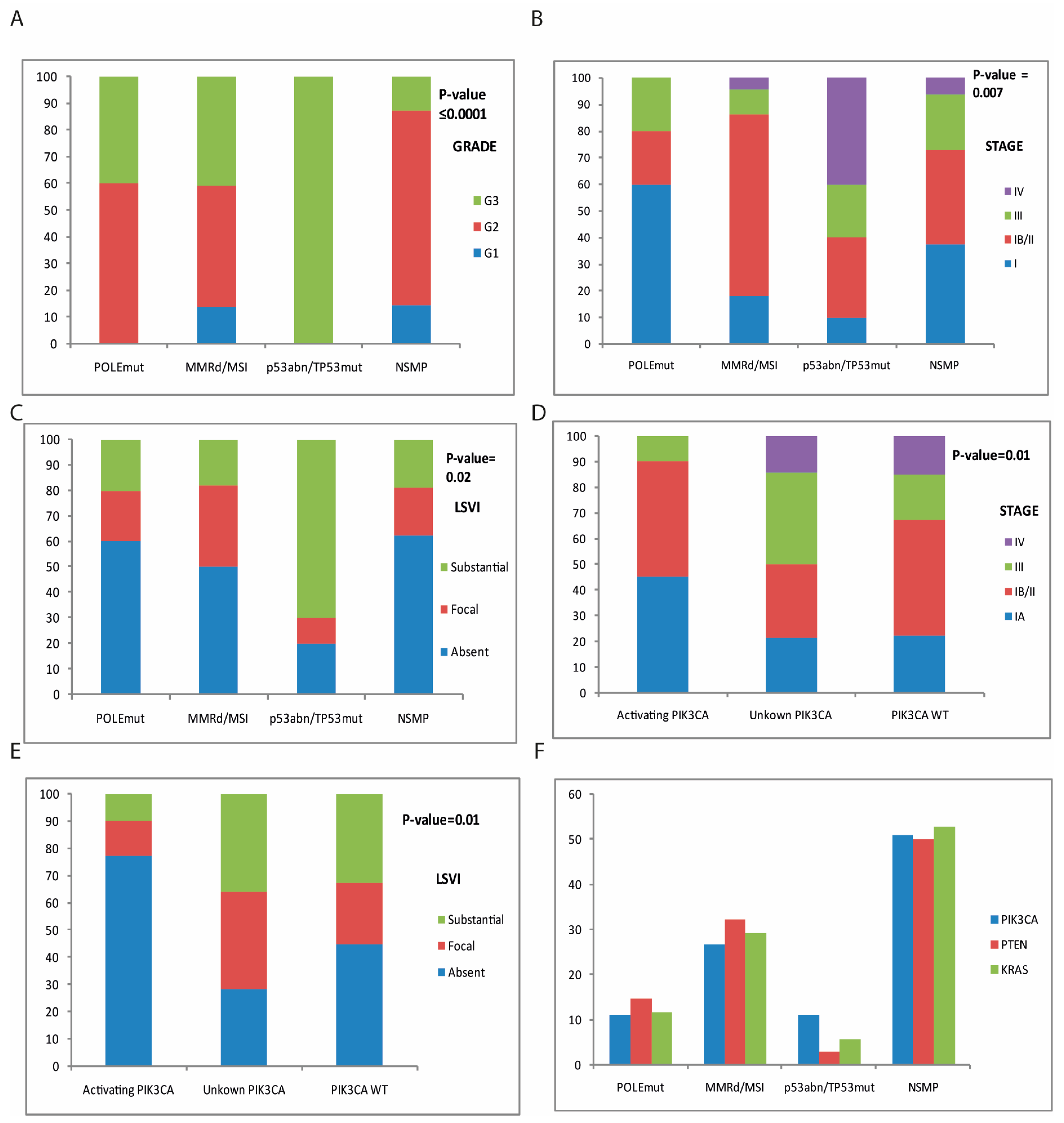
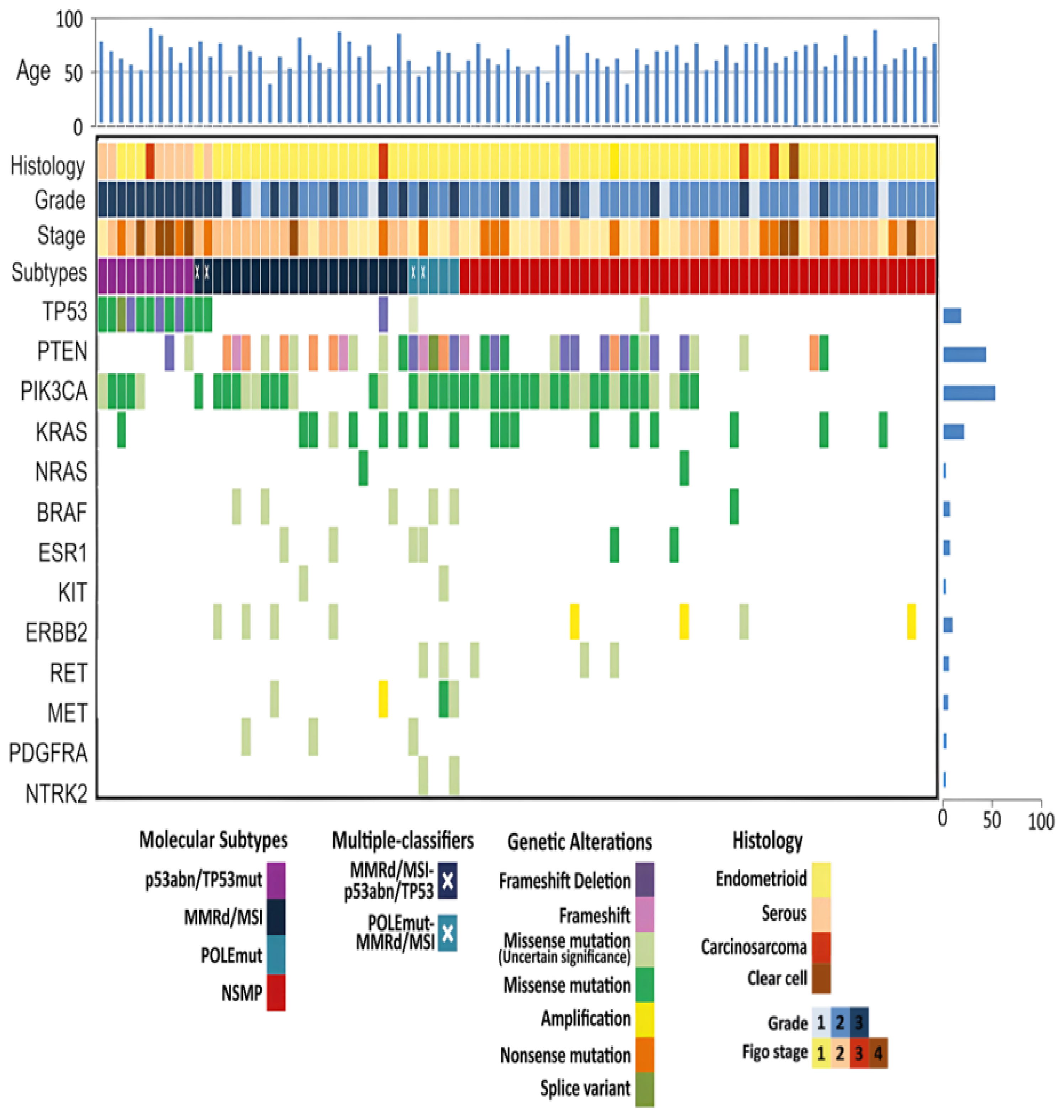
| Total Cases n = 85 | POLEmut n = 5 (5.9%) | MMRd/MSI n = 22 (25.8%) | p53abn/TP53 n = 10 (11.8%) | NSMP n = 48 (56.5%) | Univariate Analysis p-Value | Multivariate Analysis p-Value | ||
|---|---|---|---|---|---|---|---|---|
| Age, years | 64 ± 11 | 56 ± 10 | 63 ± 12 | 70 ± 12 | 64 ± 11 | 0.13 | ||
| Histotype | 0.01 | |||||||
| Endometrioid | 72 (84.7%) | 5 (100%) | 20 (91%) | 3 (30%) | 44 (92%) | <0.0001 | ||
| Serous | 8 (9.4%) | 0 | 1 (4.54%) | 6 (60%) | 1 (2%) | |||
| Carcinosarcoma | 4 (4.7%) | 0 | 1 (4.54%) | 1 (10%) | 2 (4%) | |||
| Clear Cell | 1 (1.2%) | 0 | 0 | 0 | 1 (2%) | |||
| Grade | <0.0001 | 0.01 | ||||||
| G1 | 10 (11.8%) | 0 | 3 (13.5%) | 0 | 7 (14.5%) | |||
| G2 | 48 (56.5%) | 3 (60%) | 10 (45.5%) | 0 | 35 (73%) | |||
| G3 | 27 (31.7%) | 2 (40%) | 9 (41%) | 10 (100%) | 6 (12.5%) | |||
| FIGO stage | 0.007 | 0.03 | ||||||
| IA | 26 (30.6%) | 3 (60%) | 4 (18.4%) | 1 (10%) | 18 (37.5%) | |||
| IB/II | 36 (42.3%) | 1 (20%) | 15 (68%) | 3 (30%) | 17 (35.5%) | |||
| III | 15 (17.7%) | 1 (20%) | 2 (9%) | 2 (20%) | 10 (20.8%) | |||
| IV | 8 (9.4%) | 0 | 1 (4.6%) | 4 (40%) | 3 (6.2%) | |||
| Lymph node status | 0.94 | |||||||
| Negative | 77 (90.6%) | 5 (100%) | 21 (95.5%) | 9 (90%) | 42 (87.5%) | |||
| Positive | 8 (9.5%) | 0 | 1 (4.6%) | 1 (10%) | 6 (12.5%) | |||
| LVSI | 0.02 | 0.05 | ||||||
| Absent | 46 (54.1%) | 3 (60%) | 11 (50%) | 2 (20%) | 30 (62.6%) | |||
| Focal | 18 (21.2%) | 1 (20%) | 7 (31.8%) | 1 (10%) | 9 (18.7%) | |||
| Substantial | 21 (24.7%) | 1 (20%) | 4 (18.1%) | 7 (70%) | 9 (18.7%) | |||
| Adjuvant Treatment | Survival | |||||||
| None | 50 | Recurrence | 0 | |||||
| Vaginal brachytherapy (VBRT) | 2 | Died of disease | 0 | |||||
| External beam radiation therapy + vaginal brachytherapy (EBRT + VBRT) | 19 | |||||||
| Chemo-radiotherapy + vaginal brachytherapy (CTRT + VBRT) | 12 | |||||||
| PIK3CA Activating Mutations (n = 31) | PIK3CA Unknown Mutations (n = 14) | PIK3CA Wild Type (n = 40) | Univariate Analysis p-Value | Multivariate Analysis p-Value | ||
|---|---|---|---|---|---|---|
| Age, years | 61.8 ± 13 | 62.5 ± 11 | 67 ± 10 | 0.147 | ||
| Histotype | 0.11 | |||||
| Endometrioid | 30 (96.7%) | 12 (85.7%) | 30 (75%) | |||
| Serous | 1 (3.2%) | 2 (14.3%) | 5 (12.5%) | |||
| Carcinosarcoma | 0 | 0 | 4 (10%) | |||
| Clear cell | 0 | 0 | 1 (2.5%) | |||
| Grade | 0.31 | |||||
| G1 | 4 (13%) | 2 (14.3%) | 4 (10%) | |||
| G2 | 18 (58%) | 5 (35.7%) | 27 (67.5%) | |||
| G3 | 9 (29%) | 7 (50%) | 9 (22.5%) | |||
| FIGO stage | 0.05 | 0.17 | ||||
| IA | 14 (45.2%) | 3 (14.3%) | 9 (25%) | |||
| IB/II | 14 (45.2%) | 4 (35.7%) | 18 (45%) | |||
| III | 3 (9.6%) | 5 (35.7%) | 7 (17.5%) | |||
| IV | 0 | 2 (14.3%) | 6 (12.5%) | |||
| Lymph node status | 0.36 | |||||
| Negative | 30 (96.8%) | 12 (85.7%) | 35 (87.5%) | |||
| Positive | 1 (3.2%) | 2 (14.3%) | 5 (12.5%) | |||
| LVSI | 0.01 | 0.05 | ||||
| Absent | 24 (77%) | 4 (28.6%) | 18 (45%) | |||
| Focal | 4 (13%) | 5 (35.7%) | 9 (22.5%) | |||
| Substantial | 3 (9.7%) | 5 (35.7%) | 13 (32.5%) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paratore, S.; Russo, A.; Blanco, G.; Lanzafame, K.; Giurato, E.; Bartoloni, G.; D’Asta, M.; Sapienza, M.; Solarino, V.; Vinci, V.; et al. Clinical Application of Next-Generation Sequencing for Molecular Classification in the Management of Endometrial Cancer: An Observational Cohort Study. Cancers 2025, 17, 1806. https://doi.org/10.3390/cancers17111806
Paratore S, Russo A, Blanco G, Lanzafame K, Giurato E, Bartoloni G, D’Asta M, Sapienza M, Solarino V, Vinci V, et al. Clinical Application of Next-Generation Sequencing for Molecular Classification in the Management of Endometrial Cancer: An Observational Cohort Study. Cancers. 2025; 17(11):1806. https://doi.org/10.3390/cancers17111806
Chicago/Turabian StyleParatore, Sabrina, Angela Russo, Giusi Blanco, Katia Lanzafame, Eliana Giurato, Giovanni Bartoloni, Marco D’Asta, Mirella Sapienza, Valeria Solarino, Valentina Vinci, and et al. 2025. "Clinical Application of Next-Generation Sequencing for Molecular Classification in the Management of Endometrial Cancer: An Observational Cohort Study" Cancers 17, no. 11: 1806. https://doi.org/10.3390/cancers17111806
APA StyleParatore, S., Russo, A., Blanco, G., Lanzafame, K., Giurato, E., Bartoloni, G., D’Asta, M., Sapienza, M., Solarino, V., Vinci, V., Bonanno, G. M., Ettore, G., & Bordonaro, R. (2025). Clinical Application of Next-Generation Sequencing for Molecular Classification in the Management of Endometrial Cancer: An Observational Cohort Study. Cancers, 17(11), 1806. https://doi.org/10.3390/cancers17111806





