Circulating Tumor DNA Profiling in Liver Transplant for Hepatocellular Carcinoma, Cholangiocarcinoma, and Colorectal Liver Metastases: A Programmatic Proof of Concept
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hagness, M.; Foss, A.; Line, P.-D.; Scholz, T.; Jørgensen, P.F.; Fosby, B.; Boberg, K.M.; Mathisen, O.; Gladhaug, I.P.; Egge, T.S.; et al. Liver Transplantation for Nonresectable Liver Metastases from Colorectal Cancer. Ann. Surg. 2013, 257, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Dueland, S.; Guren, T.K.; Hagness, M.; Glimelius, B.; Line, P.-D.; Pfeiffer, P.; Foss, A.; Tveit, K.M. Chemotherapy or Liver Transplantation for Nonresectable Liver Metastases from Colorectal Cancer? Ann. Surg. 2015, 261, 956–960. [Google Scholar] [CrossRef]
- Dueland, S.; Syversveen, T.; Solheim, J.M.; Solberg, S.; Grut, H.; Bjørnbeth, B.A.; Hagness, M.; Line, P.-D. Survival Following Liver Transplantation for Patients with Nonresectable Liver-Only Colorectal Metastases. Ann. Surg. 2020, 271, 212–218. [Google Scholar] [CrossRef]
- Tabrizian, P.; Holzner, M.L.; Mehta, N.; Halazun, K.; Agopian, V.G.; Yao, F.; Busuttil, R.W.; Roberts, J.; Emond, J.C.; Samstein, B.; et al. Ten-Year Outcomes of Liver Transplant and Downstaging for Hepatocellular Carcinoma. JAMA Surg. 2022, 157, 779–788. [Google Scholar] [CrossRef]
- Twohig, P.; Peeraphatdit, T.B.; Mukherjee, S. Current Status of Liver Transplantation for Cholangiocarcinoma. World J. Gastrointest. Surg. 2022, 14, 1–11. [Google Scholar] [CrossRef]
- Gorji, L.; Brown, Z.J.; Limkemann, A.; Schenk, A.D.; Pawlik, T.M. Liver Transplant as a Treatment of Primary and Secondary Liver Neoplasms. JAMA Surg. 2024, 159, 211–218. [Google Scholar] [CrossRef]
- Agarwal, P.D.; Lucey, M.R. Management of Hepatocellular Carcinoma Recurrence after Liver Transplantation. Ann. Hepatol. 2022, 27, 100654. [Google Scholar] [CrossRef]
- Wu, T.C.; Smith, C.P.; Li, J.S.; Burton, J.; Jackson, N.J.; Tao, R.; Ludmir, E.B.; Raldow, A.C. A Systematic Review and Meta-Analysis of Pathologic Complete Response Rates for Patients with Cholangiocarcinoma Treated on Liver Transplant Protocols. J. Surg. Oncol. 2023, 129, 574–583. [Google Scholar] [CrossRef]
- Solheim, J.M.; Dueland, S.; Line, P.-D.; Hagness, M. Transplantation for Nonresectable Colorectal Liver Metastases: Long-Term Follow-Up of the First Prospective Pilot Study. Ann. Surg. 2023, 278, 239. [Google Scholar] [CrossRef] [PubMed]
- Berenguer, M.; Burra, P.; Ghobrial, M.; Hibi, T.; Metselaar, H.; Sapisochin, G.; Bhoori, S.; Kwan Man, N.; Mas, V.; Ohira, M.; et al. Posttransplant Management of Recipients Undergoing Liver Transplantation for Hepatocellular Carcinoma. Working Group Report From the ILTS Transplant Oncology Consensus Conference. Transplantation 2020, 104, 1143. [Google Scholar] [CrossRef] [PubMed]
- Hanif, H.; Ali, M.J.; Susheela, A.T.; Khan, I.W.; Luna-Cuadros, M.A.; Khan, M.M.; Lau, D.T.-Y. Update on the Applications and Limitations of Alpha-Fetoprotein for Hepatocellular Carcinoma. World J. Gastroenterol. 2022, 28, 216–229. [Google Scholar] [CrossRef]
- Lin, M.-S.; Huang, J.-X.; Yu, H. Elevated Serum Level of Carbohydrate Antigen 19-9 in Benign Biliary Stricture Diseases Can Reduce Its Value as a Tumor Marker. Int. J. Clin. Exp. Med. 2014, 7, 744–750. [Google Scholar] [PubMed]
- Roles of Serum and Biliary CEA, CA19-9, VEGFR3, and TAC in Differentiating between Malignant and Benign Biliary Obstructions. Available online: http://turkjgastroenterol.org/en/roles-of-serum-and-biliary-cea-ca19-9-vegfr3-and-tac-in-differentiating-between-malignant-and-benign-biliary-obstructions-134337 (accessed on 25 January 2024).
- Sekiguchi, M.; Matsuda, T. Limited Usefulness of Serum Carcinoembryonic Antigen and Carbohydrate Antigen 19-9 Levels for Gastrointestinal and Whole-Body Cancer Screening. Sci. Rep. 2020, 10, 18202. [Google Scholar] [CrossRef] [PubMed]
- Liquid Biopsy at the Frontier of Detection, Prognosis and Progression Monitoring in Colorectal Cancer—PMC. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8951719/ (accessed on 25 January 2024).
- Bent, A.; Raghavan, S.; Dasari, A.; Kopetz, S. The Future of ctDNA-Defined Minimal Residual Disease: Personalizing Adjuvant Therapy in Colorectal Cancer. Clin. Color. Cancer 2022, 21, 89–95. [Google Scholar] [CrossRef]
- Kopystecka, A.; Patryn, R.; Leśniewska, M.; Budzyńska, J.; Kozioł, I. The Use of ctDNA in the Diagnosis and Monitoring of Hepatocellular Carcinoma—Literature Review. Int. J. Mol. Sci. 2023, 24, 9342. [Google Scholar] [CrossRef]
- Cabel, L.; Proudhon, C.; Buecher, B.; Pierga, J.-Y.; Bidard, F.-C. Circulating Tumor DNA Detection in Hepatocellular Carcinoma. Ann. Oncol. 2018, 29, 1094–1096. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Y.; Wu, L.; Li, J.; Ji, J.; Yu, Q.; Dai, W.; Feng, J.; Wu, J.; Guo, C. Current Status of ctDNA in Precision Oncology for Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 140. [Google Scholar] [CrossRef]
- Levitsky, J.; Kandpal, M.; Guo, K.; Kleiboeker, S.; Sinha, R.; Abecassis, M. Donor-Derived Cell-Free DNA Levels Predict Graft Injury in Liver Transplant Recipients. Am. J. Transplant. 2022, 22, 532–540. [Google Scholar] [CrossRef]
- Wehrle, C.J.; Raj, R.; Aykun, N.; Orabi, D.; Estfan, B.; Kamath, S.; Krishnamurthi, S.; Fujiki, M.; Hashimoto, K.; Quintini, C.; et al. Liquid Biopsy by ctDNA in Liver Transplantation for Colorectal Cancer Liver Metastasis. J. Gastrointest. Surg. Off. J. Soc. Surg. Aliment. Tract 2023, 27, 1498–1509. [Google Scholar] [CrossRef]
- Halazun, K.J.; Najjar, M.; Abdelmessih, R.M.; Samstein, B.; Griesemer, A.D.; Guarrera, J.V.; Kato, T.; Verna, E.C.; Emond, J.C.; Brown, R.S. Recurrence After Liver Transplantation for Hepatocellular Carcinoma. Ann. Surg. 2017, 265, 557–564. [Google Scholar] [CrossRef]
- Huang, A.; Guo, D.-Z.; Zhang, X.; Sun, Y.; Zhang, S.-Y.; Zhang, X.; Fu, X.-T.; Wang, Y.-P.; Yang, G.-H.; Sun, Q.-M.; et al. Serial Circulating Tumor DNA Profiling Predicts Tumor Recurrence after Liver Transplantation for Liver Cancer. Hepatol. Int. 2023, 18, 254–264. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N. Engl. J. Med. 2022, 386, 2261–2272. [Google Scholar] [CrossRef]
- Powles, T.; Assaf, Z.J.; Davarpanah, N.; Banchereau, R.; Szabados, B.E.; Yuen, K.C.; Grivas, P.; Hussain, M.; Oudard, S.; Gschwend, J.E.; et al. ctDNA Guiding Adjuvant Immunotherapy in Urothelial Carcinoma. Nature 2021, 595, 432–437. [Google Scholar] [CrossRef]
- Liu, W.; Jin, K.-M.; Zhang, M.-H.; Bao, Q.; Liu, M.; Xu, D.; Wang, K.; Xing, B.-C. Recurrence Prediction by Circulating Tumor DNA in the Patient with Colorectal Liver Metastases After Hepatectomy: A Prospective Biomarker Study. Ann. Surg. Oncol. 2023, 30, 4916–4926. [Google Scholar] [CrossRef]
- Nishioka, Y.; Chun, Y.S.; Overman, M.J.; Cao, H.S.T.; Tzeng, C.-W.D.; Mason, M.C.; Kopetz, S.W.; Bauer, T.W.; Vauthey, J.-N.; Newhook, T.E.; et al. Effect of Co-Mutation of RAS and TP53 on Postoperative ctDNA Detection and Early Recurrence after Hepatectomy for Colorectal Liver Metastases. J. Am. Coll. Surg. 2022, 234, 474. [Google Scholar] [CrossRef]
- Kotani, D.; Oki, E.; Nakamura, Y.; Yukami, H.; Mishima, S.; Bando, H.; Shirasu, H.; Yamazaki, K.; Watanabe, J.; Kotaka, M.; et al. Molecular Residual Disease and Efficacy of Adjuvant Chemotherapy in Patients with Colorectal Cancer. Nat. Med. 2023, 29, 127–134. [Google Scholar] [CrossRef]
- Yoo, C.; Laliotis, G.; Jeong, H.; Jeong, J.H.; Kim, K.-P.; Lee, S.; Ryoo, B.-Y.; Sharma, S.; Dutta, P.; Malhotra, M.; et al. Utility of Circulating Tumor DNA (ctDNA) as a Predictive Biomarker for Disease Monitoring in Patients (Pts) with Cholangiocarcinoma (CCA) before and during Adjuvant Chemotherapy (ACT): Sub-Analysis of the Randomized Phase 2 STAMP Trial. J. Clin. Oncol. 2023, 41 (Suppl. S16), 4123. [Google Scholar] [CrossRef]
- Wang, J.; Huang, A.; Wang, Y.-P.; Yin, Y.; Fu, P.-Y.; Zhang, X.; Zhou, J. Circulating Tumor DNA Correlates with Microvascular Invasion and Predicts Tumor Recurrence of Hepatocellular Carcinoma. Ann. Transl. Med. 2020, 8, 237. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Zeng, X.; Tang, J.; Liang, Z.; Qi, X.; Huang, L.; Pang, F. Circulating Tumor DNA Is a Potential Prognostic Risk Factor of Recurrence in Patients with Hepatocellular Carcinoma Treated by Liver Transplantation. J. Clin. Oncol. 2022, 40 (Suppl. S16), e16196. [Google Scholar] [CrossRef]
- Chan, H.T.; Nagayama, S.; Otaki, M.; Chin, Y.M.; Fukunaga, Y.; Ueno, M.; Nakamura, Y.; Low, S.-K. Tumor-Informed or Tumor-Agnostic Circulating Tumor DNA as a Biomarker for Risk of Recurrence in Resected Colorectal Cancer Patients. Front. Oncol. 2023, 12, 1055968. [Google Scholar] [CrossRef] [PubMed]
- Ambrozkiewicz, F.; Trailin, A.; Červenková, L.; Vaclavikova, R.; Hanicinec, V.; Allah, M.A.O.; Palek, R.; Třeška, V.; Daum, O.; Tonar, Z.; et al. CTNNB1 Mutations, TERT Polymorphism and CD8+ Cell Densities in Resected Hepatocellular Carcinoma Are Associated with Longer Time to Recurrence. BMC Cancer 2022, 22, 884. [Google Scholar] [CrossRef]
- Tavolari, S.; Brandi, G. Mutational Landscape of Cholangiocarcinoma According to Different Etiologies: A Review. Cells 2023, 12, 1216. [Google Scholar] [CrossRef]
- Zhang, L.; Shay, J.W. Multiple Roles of APC and Its Therapeutic Implications in Colorectal Cancer. JNCI J. Natl. Cancer Inst. 2017, 109, djw332. [Google Scholar] [CrossRef]
- Lamlum, H.; Ilyas, M.; Rowan, A.; Clark, S.; Johnson, V.; Bell, J.; Frayling, I.; Efstathiou, J.; Pack, K.; Payne, S.; et al. The Type of Somatic Mutation at APC in Familial Adenomatous Polyposis Is Determined by the Site of the Germline Mutation: A New Facet to Knudson’s “two-Hit” Hypothesis. Nat. Med. 1999, 5, 1071–1075. [Google Scholar] [CrossRef]
- Wang, R.; Li, J.; Zhou, X.; Mao, Y.; Wang, W.; Gao, S.; Wang, W.; Gao, Y.; Chen, K.; Yu, S.; et al. Single-Cell Genomic and Transcriptomic Landscapes of Primary and Metastatic Colorectal Cancer Tumors. Genome Med. 2022, 14, 93. [Google Scholar] [CrossRef]
- Michel, M.; Kaps, L.; Maderer, A.; Galle, P.R.; Moehler, M. The Role of P53 Dysfunction in Colorectal Cancer and Its Implication for Therapy. Cancers 2021, 13, 2296. [Google Scholar] [CrossRef]
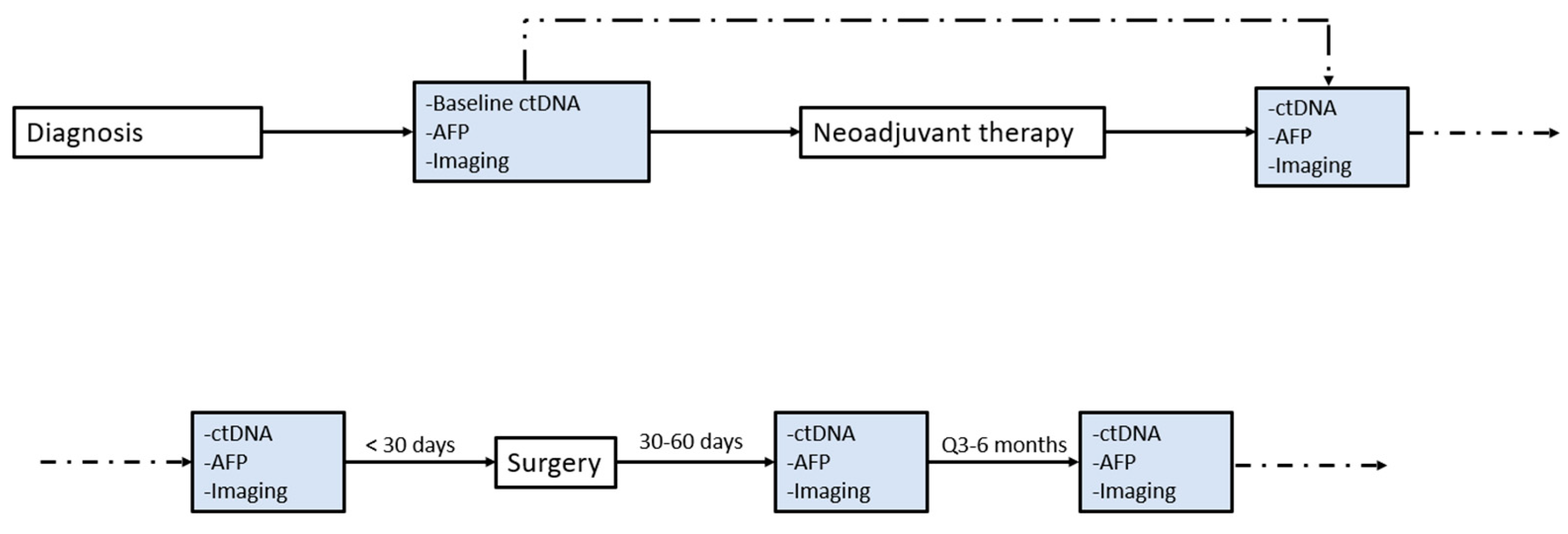
| ALL N = 21 | HCC N = 9 | HCC/CCA N = 1 | CCA N = 3 | CRLM N = 8 | |
|---|---|---|---|---|---|
| Male Sex, N (%) | 16 (76%) | 8 (89%) | 0 | 2 (67%) | 6 (75%) |
| Race, N (%) White Black Other/Unknown | 18 (86%) 2 (10%) 1 (5%) | 8 (89%) 0 1 (11%) | 1 (100%) 0 0 | 2 (50%) 1 (25%) 0 | 7 (88%) 1 (13%) 0 |
| Age at Transplant Surgery, Median (IQR) | 55 (50–68) | 70 (46–73) | 60 | 51 (25–55) | 54 (49–60) |
| Cirrhosis, N (%) Non-Malignancy Cirrhosis Factors A1AT ETOH HBV HCV NASH PSC Biliary Atresia Chemotherapy-Induced PBC | 19 (90%) 1 (5%) 2 (10%) 3 (14%) 1 (5%) 6 (29%) 2 (10%) 2 (10%) 1 (5%) 1 (5%) | 9 (100%) 0 1 (11%) 2 (22%) 1 (11%) 5 (56%) 0 2 (22%) 0 0 | 1 (100%) 1 (100%) 0 1 (100%) 0 0 0 0 0 0 | 3 (75%) 0 1 (25%) 0 0 0 1 (25%) 0 0 0 | 6 (86%) 0 0 0 0 1 (14%) 1 (14%) 0 1 (14%) 1 (14%) |
| MELD Score, Median (IQR) | 15 (11–24) | 22 (14–25) | 24 | 12 (10–29) | 11 (7–19) |
| Pre-Treatment Tumor Marker Level, Mean (SD) AFP (ng/mL) CA19-9 (U/mL) CEA (ng/mL) | 8 (6, 14) 23 (12, 168) 31 (1, 64) | 8 (6, 13) | 39 216 | 22 (8, 24) | 31 (1, 64) |
| Pre-Transplant Number of Lesions, N (%) 1 2–3 Innumerable | 11 (52%) 6 (29%) 2 (10%) | 7 (78%) 1 (11%) 0 | 0 1 (100%) 0 | 4 (100%) 0 0 | 0 4 (50%) 2 (25%) |
| Pre-Transplant Size of Biggest Lesion (cm), Median (IQR) | 4 (2–6) | 1 (1–4) | 3 | 1 | 6.7 (3–8) |
| Pre-Transplant Treatment, N (%) Systemic Chemotherapy Radiotherapy SBRT Prior Surgery Ablation Chemoembolization Radioembolization Immunotherapy | 18 (86%) 10 (48%) 6 (29%%) 4 (19%) 3 (14%) 4 (19%) 6 (29%) 5 (24%) 1 (5%) | 7 (78%) 0 0 0 0 1 (11%) 2 (22%) 4 (44%) 0 | 0 0 0 0 0 0 0 0 0 | 3 (100%) 2 (67%) 2 (67%) 2 (67%) 0 0 0 0 0 | 8 (100%) 8 (100%) 4 (50%) 2 (25%) 3 (38%) 3 (38%) 4 (50%) 1 (13%) 1 (13%) |
| Post-Transplant Tumor Marker Level, Median (IQR) AFP (ng/mL) CA19-9 (U/mL) CEA (ng/mL) | 3 (3–7) 13 (6–20) 2 (1–2) | 3 (3–6.8) | 4.8 | 13 (6–20) | 1.7 (1–2) |
| Recurrence, N (%) | 6 (29%) | 3 (33%) | 0 | 1 (25%) | 2 (25%) |
| Patient Status, N (%) Alive Dead | 17 (81%) 4 (19%) | 6 (67%) 3 (33%) | 1 (100%) 0 | 2 (67%) 1 (33%) | 8 (100%) 0 |
| Cancer-Related Deaths, N (%) | 2 (10%) | 2 (22%) | 0 | 0 | 0 |
| Recurrence Survival (Days), Median (IQR) Overall Survival (Days), Median (IQR) | 13 (7–28) 14 (8–39) | 12 (5–31) 14 (6–34) | 16.7 16.7 | 8 (6–15) 8 (6–32) | 14 (10–40) 25 (10–60) |
| Pt | Age | Sex | Cancer Type | Cirrhosis Factors | MELD at Tx | Tx Type | Liver Transplant Technique | Aberrant Liver Vasculature | Type of Arterial Anastomosis | Type of Venous Anastomosis | Biliary Anastomosis | Real Warm Ischemia Time (min) | LT Duration (min) | RBCs, FFP (Units) | Reperfusion Order | Post-Reperfusion Syndrome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 39 | M | HCC | Biliary atresia, PBC | 12 | LDLT | Piggyback | - | Standard | Interposition | HJ | 38 | 776 | 0, 0 | Vein first | No |
| 2 | 70 | M | HCC | NASH | 22 | DCD | Piggyback | - | Standard | End-to-end | Duct-to-duct | 43 | 541 | 6, 3 | Vein first | No |
| 3 | 52 | M | HCC | HCV | 23 | DCD | Conventional | Replaced RHA | Standard | End-to-end | Duct-to-duct | - | - | - | Vein first | Yes |
| 4 | 75 | M | HCC | NASH | 25 | DBD | Piggyback | - | Standard | End-to-end | Duct-to-duct | 46 | 505 | 2, 0 | Vein first | No |
| 5 | 66 | M | HCC | NASH, ETOH, HBV | 9 | LDLT | Piggyback | Accessory LHA | Standard | End-to-end | Duct-to-duct | 39 | 720 | 0, 0 | Vein first | No |
| 6 | 70 | M | HCC | HBV | 25 | DBD | Piggyback | Accessory RHA | Standard | End-to-end | Duct-to-duct | 46 | 430 | 0, 0 | Vein first | No |
| 7 | 72 | M | HCC | NASH | 11 | LDLT | Piggyback | - | Standard | End-to-end | Duct-to-duct | 42 | 557 | 7, 5 | Vein first | No |
| 8 | 73 | M | HCC | NASH | 12 | DCD | Piggyback | - | Standard | End-to-end | Duct-to-duct | 58 | 410 | 4, 1 | Both | No |
| 9 | 32 | F | HCC | Biliary atresia | 40 | Split, DBD | Conventional | - | Standard | End-to-end | HJ | 45 | 657 | 18, 13 | Vein first | No |
| 10 | 60 | F | HCC/CCA | HBV, A1AT | 24 | DCD | Piggyback | - | Standard | End-to-end | Duct-to-duct | 48 | 385 | 8, 1 | Both | Yes |
| 11 | 25 | M | CCA | PSC | 12 | DCD | Conventional | - | Standard | End-to-end | HJ | 42 | 490 | 3, 0 | Vein first | Yes |
| 12 | 51 | F | CCA | ETOH | 29 | DBD | Piggyback | Replaced LHA | Standard | Conduit | HJ | 27 | 683 | 17, 11 | Vein first | Yes |
| 13 | 55 | M | CCA | - | 10 | DBD | Conventional | Replaced RHA | Infra-renal | Conduit | HJ | 40 | 452 | 1, 0 | Vein first | No |
| 14 | 50 | M | CRLM | - | 6 | LDLT | Piggyback | - | Standard | End-to-end | Duct-to-duct | 39 | 584 | 0, 0 | Vein first | No |
| 15 | 53 | M | CRLM | - | 6 | DBD | Conventional | - | Standard | End-to-end | HJ | 49 | 427 | 0, 0 | Vein first | Yes |
| 16 | 61 | M | CRLM | - | 13 | LDLT | Conventional | - | Standard | End-to-end | Duct-to-duct | 32 | 869 | 4, 0 | Vein first | No |
| 17 | 64 | M | CRLM | - | 11 | LDLT | Piggyback | - | Standard | End-to-end | Duct-to-duct | 43 | 594 | 7, 8 | Vein first | No |
| 18 | 54 | M | CRLM | - | 14 | LDLT | Piggyback | - | Standard | Interposition | Duct-to-duct | 67 | 992 | 5, 0 | Vein first | No |
| 19 | 49 | F | CRLM | PBC | 23 | LDLT | Piggyback | - | Standard | End-to-end | Duct-to-duct | 27 | 685 | 2, 0 | Vein first | No |
| 20 | 49 | M | CRLM | - | 21 | DBD | Piggyback | - | Standard | End-to-end | HJ | 39 | 708 | 20, 12 | Vein first | No |
| 21 | 56 | F | CRLM | NASH | 8 | LDLT | Piggyback | - | Standard | Interposition | Duct-to-duct | 45 | 700 | 4, 0 | Vein first | Yes |
| Patient | Induction IS | Initial IS | IS 12 month | Biliary Complications | Biliary Intervention | Arterial Complications, Intervention | Acute Rejection Grade | Treatment of Acute Rejection | Chronic Rejection |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Basiliximab | GC + Tacrolimus | - | Leak | PTHC | - | - | - | - |
| 2 | - | GC + MMF + Tacrolimus | - | - | - | - | - | - | - |
| 3 | Basiliximab | GC + Tacrolimus | Tacrolimus + Sirolimus | - | - | - | - | - | - |
| 4 | Basiliximab | GC + MMF + Tacrolimus | Cyclosporine + Everolimus | - | - | - | - | - | - |
| 5 | Basiliximab | GC + MMF + Tacrolimus | Tacrolimus + MMF | Leak | ERCP | - | Mild | IV steroids | - |
| 6 | Basiliximab | GC + MMF + Tacrolimus | Tacrolimus + MMF + Sirolimus | - | - | - | - | - | - |
| 7 | Basiliximab | GC + MMF + Tacrolimus | - | - | - | - | - | - | - |
| 8 | - | GC + MMF + Tacrolimus | Tacrolimus + Everolimus | Stricture | ERCP | - | - | - | - |
| 9 | - | GC + MMF + Tacrolimus | - | - | - | - | - | - | - |
| 10 | Basiliximab | GC + MMF + Tacrolimus | Tacrolimus + Everolimus | - | - | - | - | - | - |
| 11 | - | GC + MMF + Tacrolimus | - | Leak, ischemic cholangiopathy | HJ reconstruction, PTHC, re-transplant | HA stenosis and pseudoaneurysm, stent placement | - | - | - |
| 12 | Basiliximab | GC + MMF + Tacrolimus | Tacrolimus + GC + MMF | - | - | - | - | - | - |
| 13 | - | GC + MMF + Tacrolimus | - | - | - | - | - | - | - |
| 14 | Basiliximab | GC + MMF + Tacrolimus | - | - | - | - | - | - | - |
| 15 | - | GC + MMF + Tacrolimus | Tacrolimus + Everolimus | Leak | Re-operation | - | - | - | - |
| 16 | Basiliximab | GC + MMF + Tacrolimus | - | Stricture | ERCP | - | Mild | IV steroids | - |
| 17 | Basiliximab | GC + MMF + Tacrolimus | Tacrolimus + Everolimus | Leak | ERCP + PTC | - | - | - | - |
| 18 | Basiliximab | GC + MMF + Tacrolimus | Tacrolimus + Everolimus | Stricture | ERCP | - | - | - | - |
| 19 | - | GC + MMF + Tacrolimus | - | - | - | - | - | - | - |
| 20 | - | GC + MMF + Tacrolimus | - | - | - | - | - | - | - |
| 21 | - | GC + MMF + Tacrolimus | - | - | - | - | - | - | - |
| Pt | Cancer Type | Date Liver Cancer dx | Dx Date Tumor Marker Level | Dx Tumor Marker Level | Date Pre-Transplant Marker | Pre-Transplant Tumor Marker | Date of Pre-Transplant ctDNA | Pre- ctDNA Results | Date of Transplant | Date Post-Transplant Marker | Post-Transplant Tumor Marker | Date of Post-Transplant ctDNA | Post-Transplant ctDNA Results | ctDNA Timing | Date of Recurrence | Date of Tumor Marker Level with Recurrence | Recurrence Tumor Marker Level |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HCC | 7/2022 | 8/2/2023 | AFP: 15 | 8/15/23 | AFP: 24.1 | 8/15/23 | + (CDx) | 8/21/23 | 10/26/23 | AFP: <3.0 | ||||||
| 2 | HCC | 2/16/2023 | 4/17/2023 | AFP: <3 | 4/17/2023 | AFP: <3 | 4/19/2023 | + (CDx) | 6/9/2023 | 12/5/23 | AFP: <3.0 | 6/20/23 | + | Both | |||
| 3 | HCC | 8/10/2010 | 8/18/2010 | AFP: 8.7 | 9/24/2010 | AFP: 4.6 | 10/12/2010 | 10/21/2010 | AFP:7.1 | 12/19/2019 | + | Post- | 12/16/2019 | 12/17/2019 | AFP: 4398.6 | ||
| 4 | HCC | 12/18/2020 | 12/18/2020 | AFP: 6.2 | 3/7/2022 | AFP: 9.3 | 4/9/2022 | 6/21/2022 | AFP: <3.0 | 4/11/2023 | + | Post- | |||||
| 5 | HCC | 7/11/2022 | 1/27/2022 | AFP: 11 | 1/27/2022 | AFP: 11 | 7/11/2022 | 7/28/2022 | AFP: <3.0 | 8/10/2022 | + | Post- | |||||
| 6 | HCC | 2/18/2019 | 2/18/2019 | AFP: 7.6 | 2/3/2020 | AFP: 4.9 | 5/1/2020 | 10/23/2020 | AFP: <3 | 11/12/2021 | + | Post- | 10/23/2020 | 6/2/21 | AFP: <3.0 | ||
| 7 | HCC | 3/15/2022 | 3/15/2022 | AFP: 7.1 | 09/08/2022 | AFP: 9.5 | 9/18/2022 | 10/10/2022 | AFP: 10.5 | 11/14/2022 | + | Post- | 10/4/2023 | 10/4/23 | AFP: <3.0 | ||
| 8 | HCC | 11/8/2019 | 8/2/2019 | AFP: 5.6 | 2/20/2020 | AFP: 6.3 | 12/18/2019 | - | 4/12/2020 | 12/16/2020 | AFP: <3 | Pre- | |||||
| 9 | HCC | 7/19/2022 | 7/19/2022 | AFP: 14 | 12/8/2022 | AFP: 8.1 | 8/15/2022 | + | 12/30/2022 | 11/18/2022 | AFP: 6 | 9/1/2023 | - | Both | |||
| 10 | HCC/CCA | 5/4/2021 | 5/4/2021; 5/20/2021 | AFP: 38.8, CA19-9: 216 | 7/5/2022 | AFP: 36.6, CA 19-9: 834 | 6/6/2022 | + | 7/7/2022 | 7/22/2022 | AFP: 4.8 | Pre- | |||||
| 11 | CCA | 7/14/2021 | 6/9/2021 | CA19-9: 8 | 1/12/2023 | CA 19-9: 46 | 7/20/2022, 9/2/22 | +, + | 2/7/2023, 07/13/23 | 9/28/2023 | - | Both | |||||
| 12 | CCA | 7/3/2020 | 6/19/2020 | CA19-9: 22 | 1/10/2023 | CA 19-9: 15 | 3/1/2023 | 7/31/2023 | CA19-9: 6.3 | 6/14/2022 | + | Post- | 11/3/2021 | 10/25/21; 5/25/21 | CA 19-9: 146; AFP: <3 | ||
| 13 | CCA | 11/25/2022 | 1/21/2021 | CA19-9: 24 | 8/4/2020 | CA 19-9: 45 | 12/13/2022 | - | 8/6/2020 | 12/1/2020 | CA19-9: 20 | 8/1/2023 | - | Both | |||
| 14 | CRLM | 6/2017 | 12/10/2018 | CEA: 2.4 | 9/21/22 | CEA: 1 | 10/27/22, 9/25/23 | -, - | 10/11/23 | 12/11/23 | CEA: 1.6 | 11/1/23 | + | Both | |||
| 15 | CRLM | 2/20/2020 | 3/3/2020 | CEA: 6854 | 8/9/2022 | CEA: 4.9 | 5/19/2022 | + | 9/14/2022 | 1/10/2023 | CEA: 1.8 | 11/15/2022 | - (GR) | Both | |||
| 16 | CRLM | 10/5/2017 | 9/15/2017 | CEA: 60.1 | 1/6/2020 | CEA: 10.4 | 11/11/2019 | + | 1/12/2020 | 2/6/2020 | CEA: 1.2 | 1/12/2022, 7/15/22, 1/16/23 | -, -, - (GR) | Both | |||
| 17 | CRLM | 2019 | 8/26/2021 | CEA: 1 | 10/28/2022 | CEA: 2.7 | 11/1/2022 | + | 11/1/2022 | 12/15/2022 | CEA: 0.9 | 12/8/2022, 6/7/23 | +, + | Both | |||
| 18 | CRLM | 11/12/2011 | 8/23/2011 | CEA: 1.6 | 9/10/2020 | CEA: 1.6 | 6/25/2019 | + | 9/13/2020 | 1/9/2023 | CEA: 1.7 | 11/8/2021, 5/5/22 | - (GR) | Both | |||
| 19 | CRLM | 4/1/2016 | 4/14/2016 | CEA: 64.4 | 11/27/2017 | CEA: 3.7 | 4/22/2018 | 5/24/2018 | CEA: 3.8 | 1/23/2020, 4/19/22 | +, + | Post- | |||||
| 20 | CRLM | 6/16/2012 | N/A | N/A | 5/27/2018 | CEA: 2.9 | 5/27/2018 | 8/30/2018 | CEA: 2 | 5/27/2022, 2/23/22, 7/28/23 | -, -, - (GR) | Post- | 9/19/2019 | 9/19/2019 | CEA: 1.8 | ||
| 21 | CRLM | 11/9/2020 | 11/2/2020 | CEA: 30.7 | 8/9/2022 | 8/6/2022, 11/10/22 | +, - | 2/6/2023 | 7/3/23 | CEA: 16.5 | 10/31/23 | + | Both | 9/25/23 | 7/17/23; 9/25/23 | CEA: 17.2; CEA: 17.9 |
| Pt | Cancer Type | Liver Cancer dx | Pre-Transplant Treatment | Chemotherapy Details | Radiation Therapy Details | Surgery Details | Pathologic Response | Date of Transplant | Date of Recurrence | Recurrence, Number of Tumors, Sites | Largest Tumor Size (cm) | Treatment of Recurrence | Recurrence Treatment Details | Death, Cause |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HCC | 07/2022 | TARE | 9/2022 | PR | 8/15/23 | ||||||||
| 2 | HCC | 2/16/2023 | - | 6/9/2023 | No | |||||||||
| 3 | HCC | 8/10/2010 | Microwave ablation | 10/12/2010 | 12/16/2019 | Intrahepatic; multifocal | 11.5 | Chemotherapy | 02/2/2020–11/6/2020: levatinib; switched to cobozanrinib after progression until 11/6/2020 | 12/17/2020; HCC | ||||
| 4 | HCC | 12/18/2020 | TACE, TARE | 03/2/12, 5/11/21, 8/18/21 | 1/14/22 | PR | 4/9/2022 | No | ||||||
| 5 | HCC | 7/11/2022 | - | 7/11/2022 | No | |||||||||
| 6 | HCC | 2/18/2019 | TARE | 09/19/2019, 11/19/2019 | SD | 5/1/2020 | 10/23/2020 | Extrahepatic; multifocal—lung, adrenal fossa, retrocaval lymph nodes | 1.3 | Chemotherapy, radiation | 9/7/21: radiation; 12/3/21–2/3/22: levatinib | 5/13/2022: HCC | ||
| 7 | HCC | 3/15/2022 | TARE | 5/18/22 | SD | 9/18/2022 | 10/4/2023 | Extrahepatic; multifocal—lung | 1.5 | Chemotherapy | 11/22/23: levatinib | |||
| 8 | HCC | 11/8/2019 | TACE | 12/2/2019 | PR | 4/12/2020 | No | 10/8/2023: metastatic melanoma | ||||||
| 9 | HCC | 7/19/2022 | SBRT | 09/26/22–10/10/22: 4 treatments | PR | 12/30/2022 | No | |||||||
| 10 | HCC/CCA | 5/4/2021 | - | 7/7/2022 | No | |||||||||
| 11 | CCA | 7/14/2021 | Chemoradiation | 08/29/22–09/16/22: capecitabine | 08/29/22–09/16/22 | SD | 2/7/2023, 07/13/23 | No | ||||||
| 12 | CCA | 7/3/2020 | SBRT | 09/26/2019–09/27/2019 | CR | 8/6/2020 | 11/3/2021 | Extrahepatic; multifocal—liver, bone | Chemotherapy, radiation | 9/6/22–9/21/22: radiation; 12/1/21–7/1/22: gemcitabine/oxaliplatin; 7/26/22–8/1/22: FOLFIRI; 10/1/22–12/1/22: gemcitabine/abraxane x 3 with PR | 3/15/23: cardiovascular event during dialysis; CCA | |||
| 13 | CCA | 11/25/2022 | Chemoradiation, SBRT | 1/10/23–2/3/23: capecitabine | 1/10/23–2/3/23 | CR | 3/1/2023, adjuvant capecitabine x 4 cycles (6/5/23) | No | ||||||
| 14 | CRLM | 2015 | Chemotherapy, surgery, microwave ablation | 7/11/17–9/20/17, 8/2019–5/8/2018: FOLFOX/cetuximab; 8/19–2/20: capecitabine, 8 cycles; 4/19/21–9/21: capecitabine | 5/18/22: microwave ablation; 2/2/23: SBRT 30 Gy in 1 fraction | 12/12/2017: open wedge resection (segments 4–8); 1/18/19: segment 4b lesion resection; 7/2/19: segment 8 lesion resection; 2/23/21: segments 7/8 liver resection | CR | 10/11/23 | No | |||||
| 15 | CRLM | 2/20/2020 | Chemotherapy, immunotherapy, radiation therapy | 3/20–8/18/20: CAPOX, bevacizumab; 10/2020–early 2021: 5FU, bevacizumab; 07–08/21: 5FU only; 10/21–01/22: 5FU, bevacizumab | 01–06/2021 | CR | 9/14/2022 | No | ||||||
| 16 | CRLM | 10/5/2017 | Chemotherapy, TARE | 10/2017–02/2018: FOLFOX, Avastin x 9 cycles; 02/18–12/11/19: FOLFIRI/panitumumab | 4 rounds | PR | 1/12/2020 | No | ||||||
| 17 | CRLM | 2019 | Chemotherapy, radiation therapy, SBRT | 09–11/11/2020: FOLFOX, Avastin x 12 cycles; 12/2020–05/2021: Avastin | SBRT: 9/20/2020 | CR | 11/1/2022 | No | ||||||
| 18 | CRLM | 11/12/2011 | Chemotherapy, radiation therapy, surgery, TACE, RFA | 10/18/2011–04/2012: Xeloda, FOLFIRI x 3 cycles; 07/2017: FOLFIRI, Erbitux; 02/25/15–03/2015: HAI pump infusion therapy | Hepatic resection 02/25/2015 and 09/2016 | PR | 9/13/2020 | No | ||||||
| 19 | CRLM | 4/1/2016 | Surgery, TACE, chemotherapy | 1/17/2015: HAI FUDR; 8/26/2016: FOLFIRI w/ panitumumab x 6 cycles, FOLFOX Avastin x 3 cycles | Wedge resection segments 2 and 3, caudate lobe removal, R hepatectomy | CR | 4/22/2018 | No | ||||||
| 20 | CRLM | 6/16/2012 | Chemotherapy, ablation, TACE, radiotherapy | 08–10/2013: FOLFIRI; 12/2013: hepatic resection, HAI pump; until 10/2014: FUDR; 01–04/2014: 5FU; 05–01/2016: irinotecan, cetuximab; 02/2016–11/2017: 5FU cetuximab, 3/7/2018: FOLFOX x 13 cycles | 12/2017: proton beam radiotherapy | 07/2013: Ablation | * | 5/27/2018 | 9/19/2019 | Extrahepatic; unifocal, right upper lobe of lung | 0.9 | Chemotherapy, surgery | Right upper lobe metastectomy; 12/16/2019–7/27/2020: FOLFIRI, bevacizumab with complete response | |
| 21 | CRLM | 11/9/2020 | Chemotherapy, TACE | 5/2021: FOLFOX x 7 cycles; 6/28/22–11/7/22: irinotecan; 9/28/22–1/4/23: panitumumab; 3/2/22: infusional 5FU | PR | 2/6/23 | 9/25/23 | Intrahepatic and extrahepatic—lung nodule | Chemotherapy, plan for surgery | 10/17/23: irinotecan, panitumumab |
| Patient # | Cancer Type | Time From Pre-op Testing to Surgery (Days) | Pre-op Somatic Alterations Detected | Pre-Transplant ctDNA | Time from Surgery to Post-op Testing (Days) | Post-op Somatic Alterations Detected | Post-Transplant ctDNA |
|---|---|---|---|---|---|---|---|
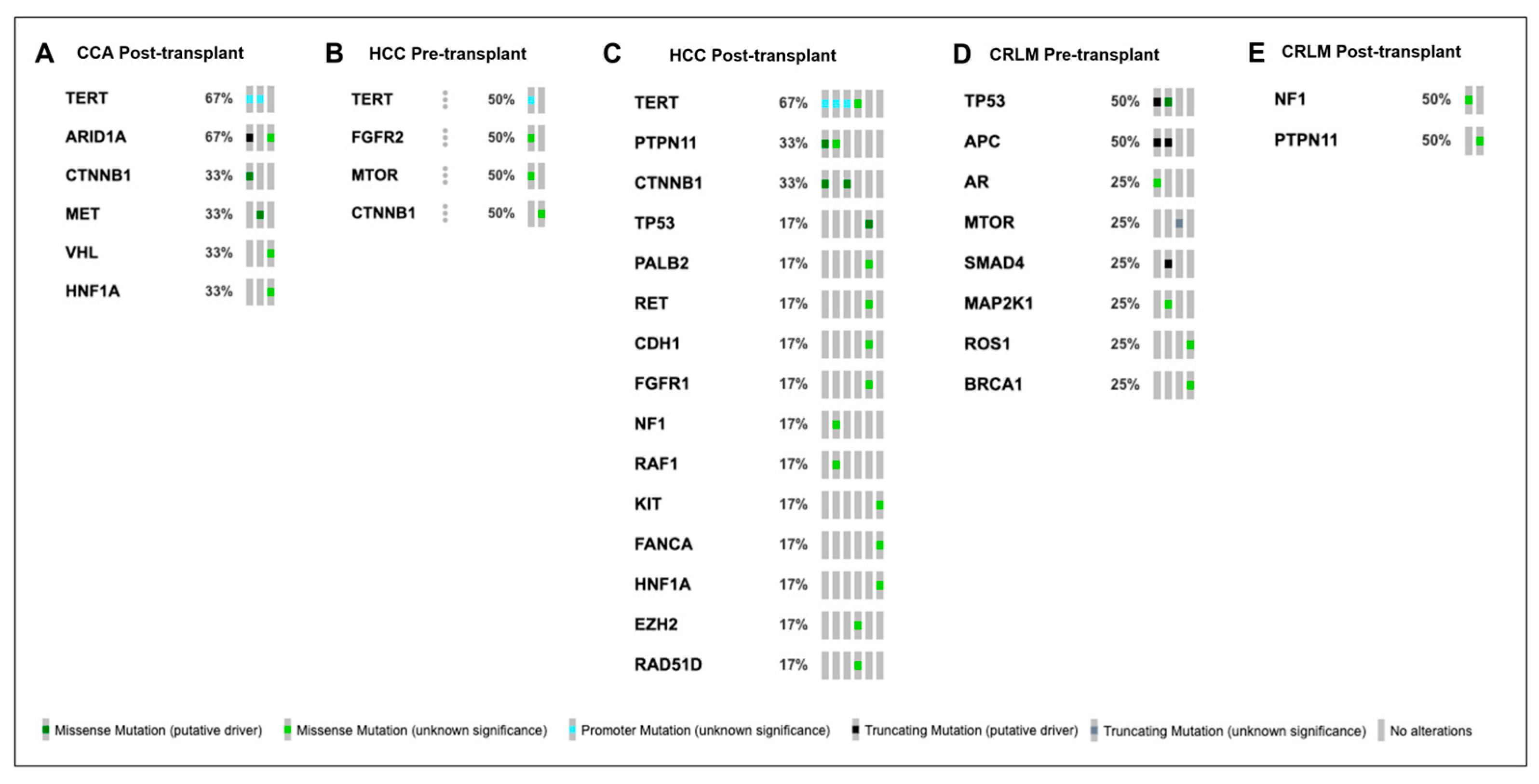
| 2 | HCC | 51 | Yes | CTNNB1 L31V 0.20% | 11 | Yes | CTNNB1 D32V N/A |
| 9 | HCC | 137 | Yes | TERT Promoter SNV 0.80% FGFR2 K509E 2.00% | 245 | No | Not Identified |
| 11 | HCC | 148 | Yes | Not Identified | 233 | No | Not Identified |
| 13 | CCA | 78 | No | Not Identified | 153 | No | Not Identified |
| 14 | CRLM | 16 | No | Not Identified | 21 | Yes | ROS1 L1899F 0.2% |
| 15 | CRLM | 26 | Yes | MTOR Q1715 0.40% | 62 | No | Not Identified |
| 16 | CRLM | 62 | Yes | APC E1064 * 0.50% TP53 R248Q 0.10% SMAD4 A418fs 0.06% MAP2K1 K84R 0.20% | 731 | No | Not Identified |
| 17 | CRLM | 0 | Yes | NF1 A706V 0.10% MLH1 I191 0.20% | 37 | Yes | FGFR3 T317A 1.80% PALB2 N241D 1.60% BRCA2 C1290Y 1.50% ROS1 T632N 1.20% MET V378I 0.10% |
| 18 | CRLM | 293 | Yes | ROS1 A2106T 0.20% BRCA1 K22E 0.10% | 421 | No | Not Identified |
| 21 | CRLM | 184 | Yes | APC S1415fs 1% TP53 S149fs 1.3% | 266 | Yes | APC S1415fs 0.2% TP53 S149fs 0.2% |
| Pt | Cancer Type | DxNumber of Tumors | Dx-Largest Tumor Size (cm) | Pathologic Tumor Numbers | Pathologic Largest Tumor Size (Viable) (cm) | % Viable Tumor Explanted Liver | Pathologic Vascular Invasion | Pathologic Perineural Invasion | Pathologic Liver Capsule Involvement | Histologic Grade of Differentiation | MSI | Pathologic TNM Staging from Transplant |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HCC | 1 | 3.6 | 3 | 0.8 | 20% | Small vessel | Absent | Absent | G2 | T2 | |
| 2 | HCC | 1 | 6.6 | 1 | 2.5 | 100% | Absent | Absent | Absent | G2 | T1b | |
| 3 | HCC | 1 | 4 | 1 | 3 | 0% | Absent | Absent | Absent | G2 | T1bN0 | |
| 4 | HCC | 3 | 2.8 | 1 | 2.3 | 100% | Absent | Absent | Absent | G2 | T2 | |
| 5 | HCC | 5 | 1.7 | 5 | 1.7 | 100% | Small vessel | Absent | Absent | G2–3 | T2 | |
| 6 | HCC | 1 | 4.9 | 4 | 2.7 | 5% | Small vessel | Absent | Absent | G2 | T2N0 | |
| 7 | HCC | 1 | 4.2 | Multiple | 4.3 | 50% | Small and large vessel | Absent | Abuts | G2 | T4 | |
| 8 | HCC | 1 | 2.6 | 1 | 0.8 | 50% | Absent | Absent | Absent | G2 | T1a | |
| 9 | HCC | 1 | 8 | 1 | 2.3 | 20% | Absent | Absent | Absent | G2 | T1b | |
| 10 | HCC/CCA | 3 | 2.3 | 3 (2-HCC, 1-CCA) | 2-HCC, 10-CCA | 0%, 5%, 95% | Present | Present | Posterior capsule | G2–3 | T2 | |
| 11 | CCA | 1 | 1 | 1 | 0.1 | 100% | Absent | Absent | Absent | G1 | T2aN0 | |
| 12 | CCA | 1 | 1 | 0 | 0 | N/A | N/A | N/A | N/A | N/A | ||
| 13 | CCA | 1 | 1 | 1 (residual) | No gross lesion visible | G2 | T1N0 | |||||
| 14 | CRLM | Numerous | 7.6 | 0 | 0 | N/A | N/A | N/A | N/A | N/A | Stable | T0N1aM1 |
| 15 | CRLM | Numerous | 7.7 | 21 | 4.1 | 20% | Absent | Absent | Absent | Stable | T3N1M1a | |
| 16 | CRLM | 3 | 5.8 | 3 | 4 | 100%, 0% | Absent | Absent | Absent | Stable | T3N1aM1 | |
| 17 | CRLM | * | * | 1 | 8.5 | 0% | Absent | Absent | Absent | Stable | T3N1aM1 | |
| 18 | CRLM | 3 | * | 1 | 4 | 0% | Absent | Absent | Absent | Stable | ||
| 19 | CRLM | 2 | * | 0 | Stable | T3N1aM1 | ||||||
| 20 | CRLM | * | * | 4 | 1.7 | 100% | Absent | Absent | Absent | G2 | Unknown | |
| 21 | CRLM | 2 | 1.4 | 6 | 3.3 | 100% | Absent | Absent | Absent | G2 | Stable | T3N0M1 |
| Patient Number | Cancer Type | Date Pre-Transplant ctDNA Collected | Pre-op Somatic Alterations Detected | Pre-Transplant ctDNA | Date Post-Transplant ctDNA Collected | Post-op Somatic Alterations Detected | Post-Transplant ctDNA | Date of Recurrence |
|---|---|---|---|---|---|---|---|---|
| 3 | HCC | 12/19/2019 | Yes | CTNNB1 T41A 3.70% TERT Promoter 2.00% | 12/16/2019 | |||
| 6 | HCC | 11/12/21 | Yes | ARID1A S696fs 0.70% CTNNB1 S33A 16.50% TERT promoter 13.30% | 10/23/2020 | |||
| 7 | HCC | 11/14/2022 | Yes | TP53 R248Q 0.10% FGFR1 V247V 6.00% | 10/4/2023 | |||
| 12 | CCA | 12/3/22 | No | Not identified | 8/1/23 | No | Not identified | 11/3/2021 |
| 20 | CRLM | 7/28/23 | No | Not Identified | 9/19/2019 | |||
| 21 | CRLM | 8/6/22 | Yes | TP53 S149fs 1.30% APC S1415fs 1.00% AR R780W 0.50% | 10/31/23 | Yes | APC S1415fs 0.2% TP53 S149fs 0.2% | 9/25/23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, H.; Wehrle, C.J.; Zhang, M.; Fares, S.; Stitzel, H.; Garib, D.; Estfan, B.; Kamath, S.; Krishnamurthi, S.; Ma, W.W.; et al. Circulating Tumor DNA Profiling in Liver Transplant for Hepatocellular Carcinoma, Cholangiocarcinoma, and Colorectal Liver Metastases: A Programmatic Proof of Concept. Cancers 2024, 16, 927. https://doi.org/10.3390/cancers16050927
Hong H, Wehrle CJ, Zhang M, Fares S, Stitzel H, Garib D, Estfan B, Kamath S, Krishnamurthi S, Ma WW, et al. Circulating Tumor DNA Profiling in Liver Transplant for Hepatocellular Carcinoma, Cholangiocarcinoma, and Colorectal Liver Metastases: A Programmatic Proof of Concept. Cancers. 2024; 16(5):927. https://doi.org/10.3390/cancers16050927
Chicago/Turabian StyleHong, Hanna, Chase J. Wehrle, Mingyi Zhang, Sami Fares, Henry Stitzel, David Garib, Bassam Estfan, Suneel Kamath, Smitha Krishnamurthi, Wen Wee Ma, and et al. 2024. "Circulating Tumor DNA Profiling in Liver Transplant for Hepatocellular Carcinoma, Cholangiocarcinoma, and Colorectal Liver Metastases: A Programmatic Proof of Concept" Cancers 16, no. 5: 927. https://doi.org/10.3390/cancers16050927
APA StyleHong, H., Wehrle, C. J., Zhang, M., Fares, S., Stitzel, H., Garib, D., Estfan, B., Kamath, S., Krishnamurthi, S., Ma, W. W., Kuzmanovic, T., Azzato, E., Yilmaz, E., Modaresi Esfeh, J., Linganna, M. W., Khalil, M., Pita, A., Schlegel, A., Kim, J., ... Aucejo, F. (2024). Circulating Tumor DNA Profiling in Liver Transplant for Hepatocellular Carcinoma, Cholangiocarcinoma, and Colorectal Liver Metastases: A Programmatic Proof of Concept. Cancers, 16(5), 927. https://doi.org/10.3390/cancers16050927






