Utilizing 3D Models to Unravel the Dynamics of Myeloma Plasma Cells’ Escape from the Bone Marrow Microenvironment
Abstract
Simple Summary
Abstract
1. Introduction
2. Present Status of Therapeutic Advancements in Multiple Myeloma Treatment
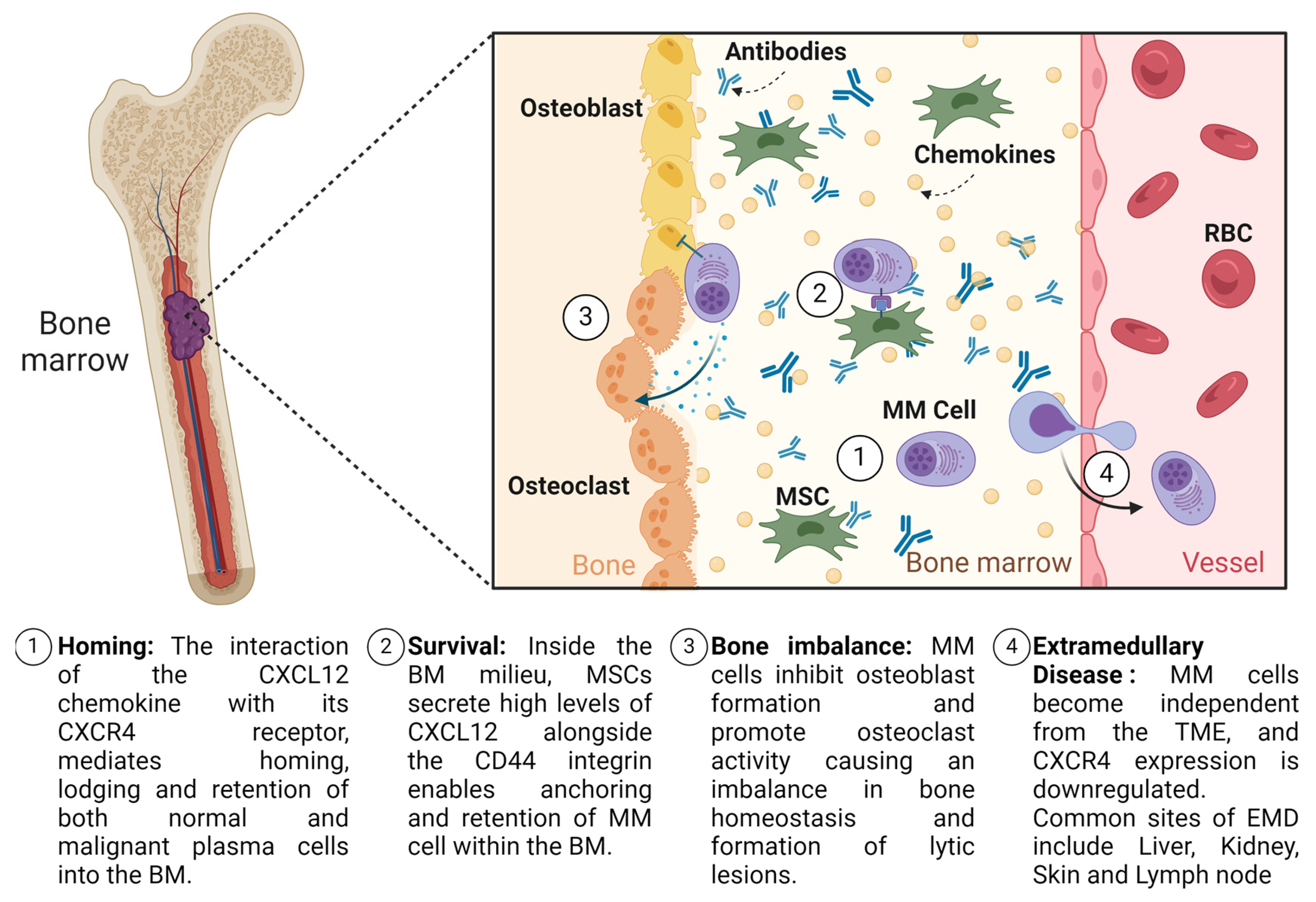
Extramedullary Disease in Multiple Myeloma: Unravelling the Intricacies and Clinical Implications
3. Three-Dimensional Models of Multiple Myeloma
3.1. In Silico MM Bone Marrow Models
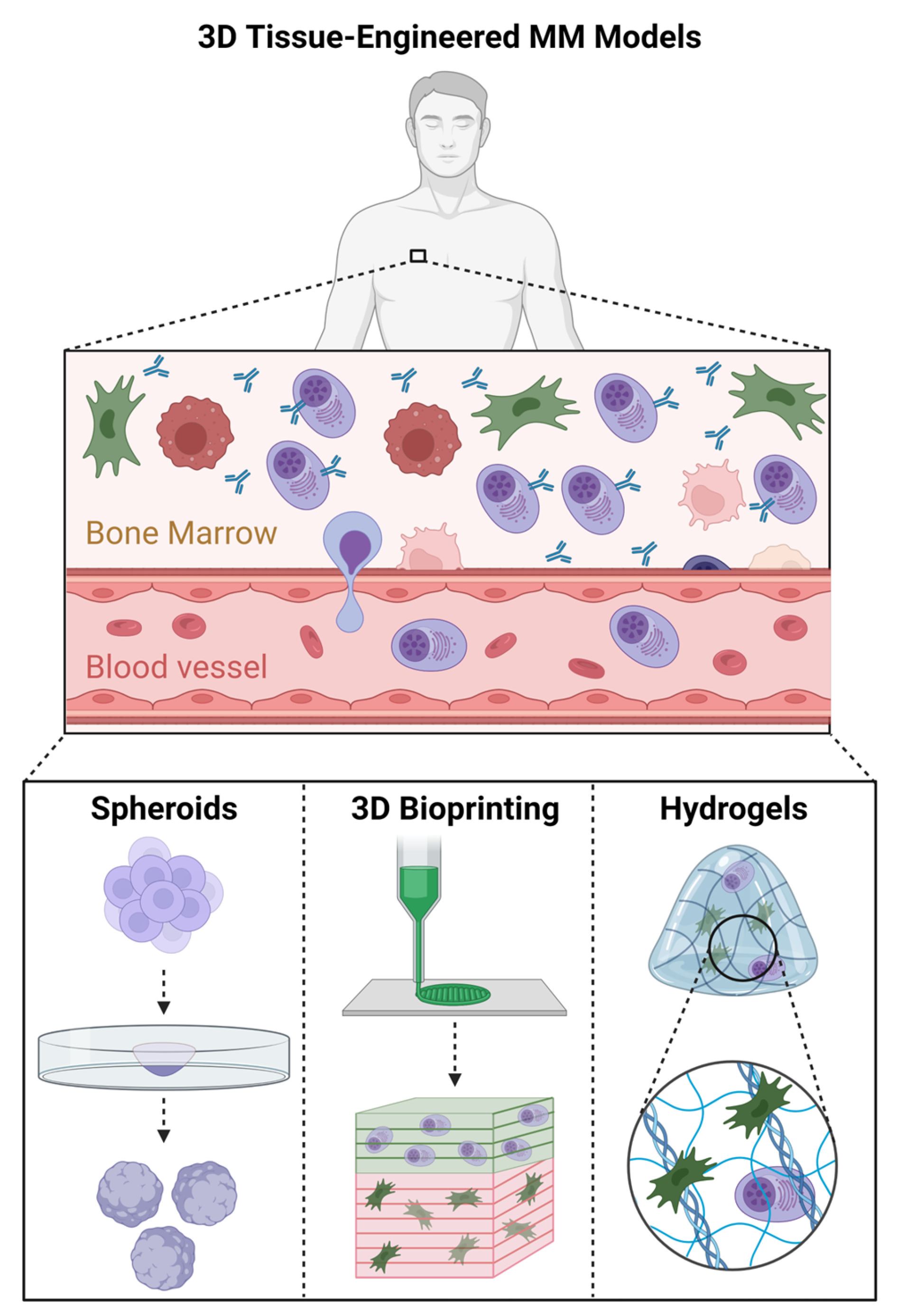
3.2. Three-Dimensional Tissue-Engineered MM Models
3.2.1. Three-Dimensional Spheroid Models of MM
3.2.2. Bioprinted MM Bone Marrow Models
3.2.3. Hydrogel-Based MM Bone Marrow Models
| 3D Model | Cells | Biomaterials | In Vitro Viability | Advantages | Limitations | Refs |
|---|---|---|---|---|---|---|
| Spheroids | CD14+ Monocytes and MM cells | Matrigel | 3 days |
|
| [54] |
| Spheroids + hydrogel | iPSC, MSCs, MM cells | Matrigel and Collagen Type I | 12 days |
|
| [55] |
| Bioprinting | O-MSCs, MSCs and EPCs, and CD138+ MM cells | Matrigel and Calcium Phosphate Cement | 28 days |
|
| [56] |
| Bioprinting | MM cells and fibroblasts | Gelman, nHA, Alginate, and PEGDA | 9 days |
|
| [57] |
| Hydrogel | MSCs and EPCs, and CD138+ MM cells | Matrigel | 14 days |
|
| [58] |
| Hydrogel | MSCs, CD138+-selected MM patients’ cells, and patient-derived plasma | Collagen-I | 5 days |
|
| [58,62,63] |
| Hydrogel | MM cells | Matrigel, Fibronectin, and Collagen Type IV | 6 days |
|
| [60] |
| Hydrogel | MSCs and MM cells | Parametric | 7 days |
|
| [62] |
| Hydrogel | MM cells | Matrigel, Fibronectin, and Collagen Type I | 25 days |
|
| [61] |
| Hydrogel | MM cells | Hyaluronic Acid | 21 days |
|
| [64] |
| Hydrogel | MM cells, MSCs, and EPCs | Fibrinogen | 7 days |
|
| [65] |
| Hydrogel | O-MSCs, HUVECs, and MM cells | Silk Fibroin | 1 month |
|
| [66] |
| Hydrogel | MM and MSCs | PGMA52–PHPMA122 diblock copolymer | 7 days |
|
| [71] |
| Hydrogel | MSCs and MM cells | Gelatine Sponge | 3 days |
|
| [67] |
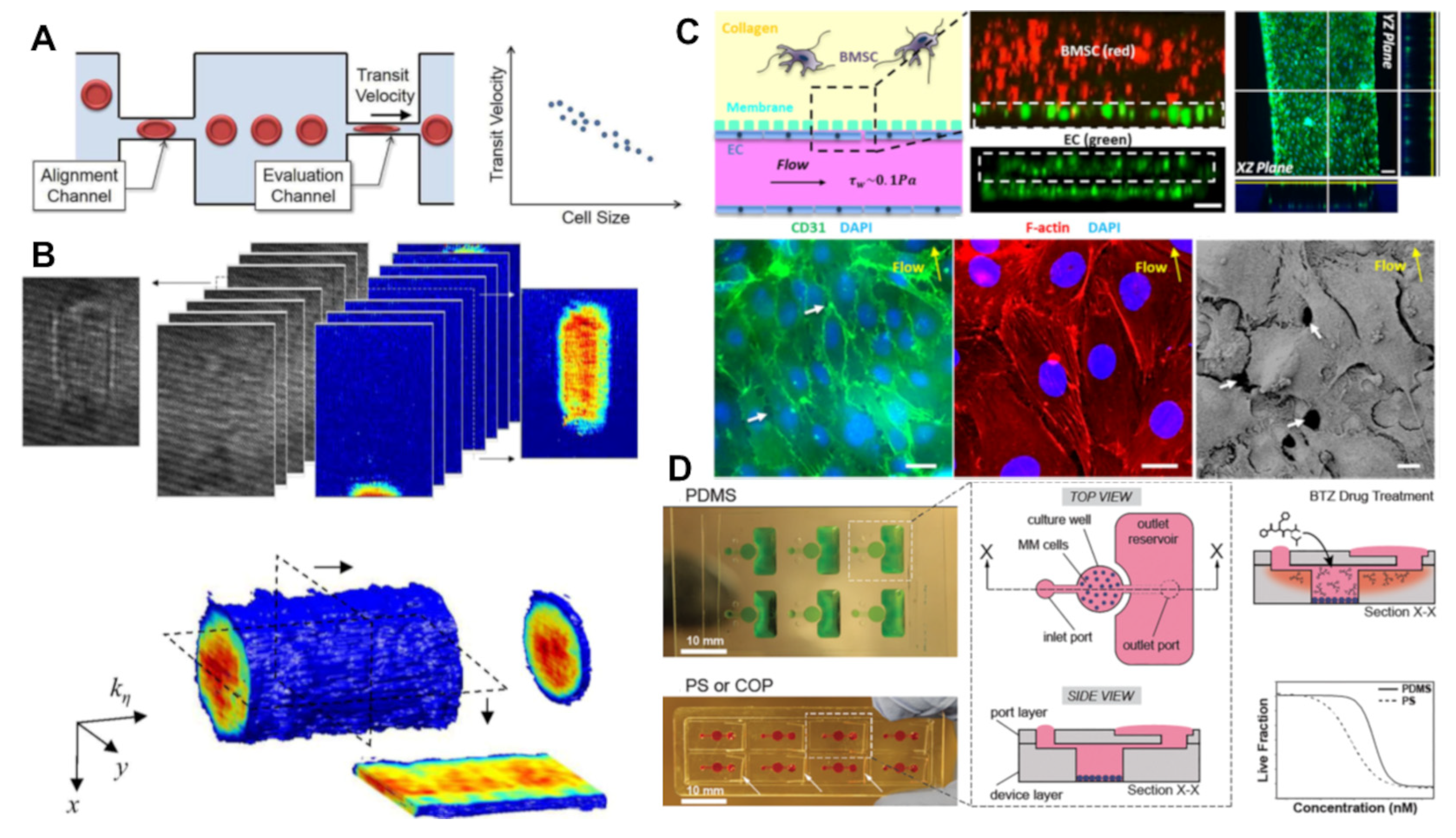
3.3. Microfluidic and Organ-on-a-Chip Models of MM
3.3.1. Circulating Tumour Cell Analysis
3.3.2. Tumour Microenvironment Modelling
3.3.3. Drug Screening and Personalized Medicine
4. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- SEER: Surveillance, Epidemiology, and End Results Program; Cancerstat Facts: Myeloma. Available online: https://seer.cancer.gov/explorer/application.html?site=89&data_type=1&graph_type=3&compareBy=sex&chk_sex_1=1&rate_type=2&race=1&advopt_precision=1&advopt_show_ci=on (accessed on 22 September 2021).
- He, H.; Li, Z.; Lu, J.; Qiang, W.; Jiang, S.; Xu, Y.; Fu, W.; Zhai, X.; Zhou, L.; Qian, M.; et al. Single-cell RNA-seq reveals clonal diversity and prognostic genes of relapsed multiple myeloma. Clin. Transl. Med. 2022, 12, e757. [Google Scholar] [CrossRef]
- Rasche, L.; Chavan, S.S.; Stephens, O.W.; Patel, P.H.; Tytarenko, R.; Ashby, C.; Bauer, M.; Stein, C.; Deshpande, S.; Wardell, C.; et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat. Commun. 2017, 8, 268. [Google Scholar] [CrossRef]
- Jones, J.R.; Weinhold, N.; Ashby, C.; Walker, B.A.; Wardell, C.; Pawlyn, C.; Rasche, L.; Melchor, L.; Cairns, D.A.; Gregory, W.M.; et al. Clonal evolution in myeloma: The impact of maintenance lenalidomide and depth of response on the genetics and sub-clonal structure of relapsed disease in uniformly treated newly diagnosed patients. Haematologica 2019, 104, 1440–1450. [Google Scholar] [CrossRef] [PubMed]
- Misund, K.; Hofste op Bruinink, D.; Coward, E.; Hoogenboezem, R.M.; Rustad, E.H.; Sanders, M.A.; Rye, M.; Sponaas, A.-M.; van der Holt, B.; Zweegman, S.; et al. Clonal evolution after treatment pressure in multiple myeloma: Heterogenous genomic aberrations and transcriptomic convergence. Leukemia 2022, 36, 1887–1897. [Google Scholar] [CrossRef] [PubMed]
- Mey, U.J.M.; Leitner, C.; Driessen, C.; Cathomas, R.; Klingbiel, D.; Hitz, F. Improved survival of older patients with multiple myeloma in the era of novel agents. Hematol. Oncol. 2016, 34, 217–223. [Google Scholar] [CrossRef]
- Bladé, J.; Beksac, M.; Caers, J.; Jurczyszyn, A.; von Lilienfeld-Toal, M.; Moreau, P.; Rasche, L.; Rosiñol, L.; Usmani, S.Z.; Zamagni, E.; et al. Extramedullary disease in multiple myeloma: A systematic literature review. Blood Cancer J. 2022, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.K.; Rajkumar, S.V.; Dispenzieri, A.; Lacy, M.Q.; Hayman, S.R.; Buadi, F.K.; Zeldenrust, S.R.; Dingli, D.; Russell, S.J.; Lust, J.A.; et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood 2008, 111, 2516–2520. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.L.F.; Turesson, I.; Genell, A.; Klausen, T.W.; Knut-Bojanowska, D.; Redder, L.; Sverrisdottir, I.; Thorsen, J.; Vangsted, A.J.; Blimark, C.H. Improved survival in myeloma patients-a nationwide registry study of 4,647 patients ≥75 years treated in Denmark and Sweden. Haematologica 2023, 108, 1640–1651. [Google Scholar] [CrossRef] [PubMed]
- Sonneveld, P.; Dimopoulos, M.A.; Boccadoro, M.; Quach, H.; Ho, P.J.; Beksac, M.; Hulin, C.; Antonioli, E.; Leleu, X.; Mangiacavalli, S.; et al. Daratumumab, Bortezomib, Lenalidomide, and Dexamethasone for Multiple Myeloma. N. Engl. J. Med. 2024, 390, 301–313. [Google Scholar] [CrossRef]
- Hansen, D.K.; Sidana, S.; Peres, L.C.; Leitzinger, C.C.; Shune, L.; Shrewsbury, A.; Gonzalez, R.; Sborov, D.W.; Wagner, C.; Dima, D.; et al. Idecabtagene Vicleucel for Relapsed/Refractory Multiple Myeloma: Real-World Experience from the Myeloma CAR T Consortium. J. Clin. Oncol. 2023, 41, 2087–2097. [Google Scholar] [CrossRef]
- Munshi, N.C.; Anderson, L.D.; Shah, N.; Madduri, D.; Berdeja, J.; Lonial, S.; Raje, N.; Lin, Y.; Siegel, D.; Oriol, A.; et al. Idecabtagene Vicleucel in Relapsed and Refractory Multiple Myeloma. N. Engl. J. Med. 2021, 384, 705–716. [Google Scholar] [CrossRef]
- Oswald, L.B.; Gudenkauf, L.M.; Li, X.; De Avila, G.; Peres, L.C.; Kirtane, K.; Gonzalez, B.D.; Hoogland, A.I.; Nguyen, O.; Rodriguez, Y.; et al. Patient-Reported Outcomes among Multiple Myeloma Patients Treated with Standard of Care Idecabtagene Vicleucel. Cancers 2023, 15, 4711. [Google Scholar] [CrossRef]
- Sidana, S.; Peres, L.C.; Hashmi, H.; Hosoya, H.; Ferreri, C.; Khouri, J.; Dima, D.; Atrash, S.; Voorhees, P.; Simmons, G.; et al. Idecabtagene vicleucel chimeric antigen receptor T-cell therapy for relapsed/refractory multiple myeloma with renal impairment. Haematologica 2022, 140, 10377–10379. [Google Scholar] [CrossRef]
- Berdeja, J.G.; Madduri, D.; Usmani, S.Z.; Jakubowiak, A.; Agha, M.; Cohen, A.D.; Stewart, A.K.; Hari, P.; Htut, M.; Lesokhin, A.; et al. Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): A phase 1b/2 open-label study. Lancet 2021, 398, 314–324. [Google Scholar] [CrossRef]
- Hansen, D.K.; Patel, K.K.; Peres, L.C.; Kocoglu, M.H.; Shune, L.; Simmons, G.; Ferreri, C.J.; Atrash, S.; Parrondo, R.D.; Chhabra, S.; et al. Safety and efficacy of standard of care (SOC) ciltacabtagene autoleucel (Cilta-cel) for relapsed/refractory multiple myeloma (RRMM). J. Clin. Oncol. 2023, 41, 8012. [Google Scholar] [CrossRef]
- Moreau, P.; Garfall, A.L.; van de Donk, N.W.C.J.; Nahi, H.; San-Miguel, J.F.; Oriol, A.; Nooka, A.K.; Martin, T.; Rosinol, L.; Chari, A.; et al. Teclistamab in Relapsed or Refractory Multiple Myeloma. N. Engl. J. Med. 2022, 387, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Chari, A.; Minnema, M.C.; Berdeja, J.G.; Oriol, A.; van de Donk, N.W.C.J.; Rodríguez-Otero, P.; Askari, E.; Mateos, M.-V.; Costa, L.J.; Caers, J.; et al. Talquetamab, a T-Cell–Redirecting GPRC5D Bispecific Antibody for Multiple Myeloma. N. Engl. J. Med. 2022, 387, 2232–2244. [Google Scholar] [CrossRef]
- Lesokhin, A.M.; Tomasson, M.H.; Arnulf, B.; Bahlis, N.J.; Miles Prince, H.; Niesvizky, R.; Rodrίguez-Otero, P.; Martinez-Lopez, J.; Koehne, G.; Touzeau, C.; et al. Elranatamab in relapsed or refractory multiple myeloma: Phase 2 MagnetisMM-3 trial results. Nat. Med. 2023, 29, 2259–2267. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Rakshit, S.; Kumar, S. Extramedullary disease in multiple myeloma. Blood Cancer J. 2021, 11, 161. [Google Scholar] [CrossRef]
- Rosiñol, L.; Beksac, M.; Zamagni, E.; Van de Donk, N.W.C.J.; Anderson, K.C.; Badros, A.; Caers, J.; Cavo, M.; Dimopoulos, M.-A.; Dispenzieri, A.; et al. Expert review on soft-tissue plasmacytomas in multiple myeloma: Definition, disease assessment and treatment considerations. Br. J. Haematol. 2021, 194, 496–507. [Google Scholar] [CrossRef]
- Gagelmann, N.; Eikema, D.J.; Iacobelli, S.; Koster, L.; Nahi, H.; Stoppa, A.M.; Masszi, T.; Caillot, D.; Lenhoff, S.; Udvardy, M.; et al. Impact of extramedullary disease in patients with newly diagnosed multiple myeloma undergoing autologous stem cell transplantation: A study from the Chronic Malignancies Working Party of the EBMT. Haematologica 2018, 103, 890–897. [Google Scholar] [CrossRef]
- Hathi, D.; Chanswangphuwana, C.; Cho, N.; Fontana, F.; Maji, D.; Ritchey, J.; O’Neal, J.; Ghai, A.; Duncan, K.; Akers, W.J.; et al. Ablation of VLA4 in multiple myeloma cells redirects tumor spread and prolongs survival. Sci. Rep. 2022, 12, 30. [Google Scholar] [CrossRef]
- Bou Zerdan, M.; Nasr, L.; Kassab, J.; Saba, L.; Ghossein, M.; Yaghi, M.; Dominguez, B.; Chaulagain, C.P. Adhesion molecules in multiple myeloma oncogenesis and targeted therapy. Int. J. Hematol. Oncol. 2022, 11, Ijh39. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Mishima, Y.; Sacco, A.; Moschetta, M.; Tai, Y.T.; Shi, J.; Zhang, Y.; Reagan, M.R.; Huynh, D.; Kawano, Y.; et al. CXCR4 Regulates Extra-Medullary Myeloma through Epithelial-Mesenchymal-Transition-like Transcriptional Activation. Cell Rep. 2015, 12, 622–635. [Google Scholar] [CrossRef]
- Demchenko, Y.N.; Glebov, O.K.; Zingone, A.; Keats, J.J.; Bergsagel, P.L.; Kuehl, W.M. Classical and/or alternative NF-κB pathway activation in multiple myeloma. Blood 2010, 115, 3541–3552. [Google Scholar] [CrossRef]
- de Haart, S.J.; Willems, S.M.; Mutis, T.; Koudijs, M.J.; van Blokland, M.T.; Lokhorst, H.M.; de Weger, R.A.; Minnema, M.C. Comparison of intramedullary myeloma and corresponding extramedullary soft tissue plasmacytomas using genetic mutational panel analyses. Blood Cancer J. 2016, 6, e426. [Google Scholar] [CrossRef]
- Long, X.; Xu, Q.; Lou, Y.; Li, C.; Gu, J.; Cai, H.; Wang, D.; Xu, J.; Li, T.; Zhou, X.; et al. The utility of non-invasive liquid biopsy for mutational analysis and minimal residual disease assessment in extramedullary multiple myeloma. Br. J. Haematol. 2020, 189, e45–e48. [Google Scholar] [CrossRef] [PubMed]
- Freeman, C.L.L.; Zhao, X.; Meads, M.B.; Noble, J.; Alugubelli, R.R.; Renatino-Canevarolo, R.; Hampton, O.; Sudalagunta, P.R.; Siqueira Silva, A.; Baz, R.; et al. Single Cell RNA Sequencing of Sequential Samples before and after BCMA-Directed CAR-T Reveal Features Associated with Non-Durable Response, Exhausted T-Cells and Decreased Expression of Genes Encoding Key Surface Targets in Particular in Patients with Extramedullary Disease. Blood 2023, 142, 3304. [Google Scholar] [CrossRef]
- Oshima, K.; Kanda, Y.; Nannya, Y.; Kaneko, M.; Hamaki, T.; Suguro, M.; Yamamoto, R.; Chizuka, A.; Matsuyama, T.; Takezako, N.; et al. Clinical and pathologic findings in 52 consecutively autopsied cases with multiple myeloma. Am. J. Hematol. 2001, 67, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Mailankody, S.; Devlin, S.M.; Landa, J.; Nath, K.; Diamonte, C.; Carstens, E.J.; Russo, D.; Auclair, R.; Fitzgerald, L.; Cadzin, B.; et al. GPRC5D-Targeted CAR T Cells for Myeloma. N. Engl. J. Med. 2022, 387, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Dima, D.; Davis, J.A.; Rashid, A.; Rice, M.; DeJarnette, S.; Shune, L.; Khouri, J.; Raza, S.; McGuirk, J.P.; Anwer, F.; et al. Outcomes of BCMA-Directed Chimeric Antigen Receptor T-Cell (CART) Therapy in Patients with Relapse-Refractory Multiple Myeloma with Extramedullary Disease. Blood 2023, 142, 4882. [Google Scholar] [CrossRef]
- Gagelmann, N.; Ayuk, F.A.; Klyuchnikov, E.; Wolschke, C.; Berger, S.C.; Kröger, N. Impact of high-risk disease on the efficacy of chimeric antigen receptor T-cell therapy for multiple myeloma: A meta-analysis of 723 patients. Haematologica 2023, 108, 2799–2802. [Google Scholar] [CrossRef]
- Jurczyszyn, A.; Olszewska-Szopa, M.; Hungria, V.; Crusoe, E.; Pika, T.; Delforge, M.; Leleu, X.; Rasche, L.; Nooka, A.K.; Druzd-Sitek, A.; et al. Cutaneous involvement in multiple myeloma: A multi-institutional retrospective study of 53 patients. Leuk. Lymphoma 2016, 57, 2071–2076. [Google Scholar] [CrossRef]
- Meral, B.; Guldane Cengiz, S.; Nicholas, K.; Daniel, C.; Laura, R.; Gulsum, O.; Vesselina, G.-M.; Ali, U.; Jelena, B.; Hayri, O.; et al. A real world multicenter retrospective study on extramedullary disease from Balkan Myeloma Study Group and Barcelona University: Analysis of parameters that improve outcome. Haematologica 2020, 105, 201–208. [Google Scholar] [CrossRef]
- Yanamandra, U.; Deo, P.; Sahu, K.K.; Nampoothiri, R.V.; Gupta, N.; Prabhakaran, A.; Dhibhar, D.P.; Khadwal, A.; Prakash, G.; Sachdeva, M.U.S.; et al. Clinicopathological Profile of Myelomatous Pleural Effusion: Single-center Real-world Experience and Review of Literature. Clin. Lymphoma Myeloma Leuk. 2019, 19, 183–189.e181. [Google Scholar] [CrossRef] [PubMed]
- Egan, P.A.; Elder, P.T.; Deighan, W.I.; O’Connor, S.J.M.; Alexander, H.D. Multiple myeloma with central nervous system relapse. Haematologica 2020, 105, 1780–1790. [Google Scholar] [CrossRef] [PubMed]
- Calura, E.; Bisognin, A.; Manzoni, M.; Todoerti, K.; Taiana, E.; Sales, G.; Morgan, G.J.; Tonon, G.; Amodio, N.; Tassone, P.; et al. Disentangling the microRNA regulatory milieu in multiple myeloma: Integrative genomics analysis outlines mixed miRNA-TF circuits and pathway-derived networks modulated in t(4;14) patients. Oncotarget 2016, 7, 2367–2378. [Google Scholar] [CrossRef]
- Vandyke, K.; Zeissig, M.N.; Hewett, D.R.; Martin, S.K.; Mrozik, K.M.; Cheong, C.M.; Diamond, P.; To, L.B.; Gronthos, S.; Peet, D.J.; et al. HIF-2α Promotes Dissemination of Plasma Cells in Multiple Myeloma by Regulating CXCL12/CXCR4 and CCR1. Cancer Res. 2017, 77, 5452–5463. [Google Scholar] [CrossRef] [PubMed]
- Jacqueline, E.N.; Sharon, A.W.; Christine, M.T.; Hongsheng, W.; Julie, M.Q.; Louise, E.P.; Katherine, P.; Luen, B.T.; Andreas, E.; Stan, G.; et al. Myeloma plasma cells alter the bone marrow microenvironment by stimulating the proliferation of mesenchymal stromal cells. Haematologica 2014, 99, 163–171. [Google Scholar] [CrossRef]
- Macauda, A.; Piredda, C.; Clay-Gilmour, A.I.; Sainz, J.; Buda, G.; Markiewicz, M.; Barington, T.; Ziv, E.; Hildebrandt, M.A.T.; Belachew, A.A.; et al. Expression quantitative trait loci of genes predicting outcome are associated with survival of multiple myeloma patients. Int. J. Cancer 2021, 149, 327–336. [Google Scholar] [CrossRef]
- Leng, H.; Ersek, A.; Morris, E.; Molina, B.G.; Edwards, C.M.; Horwood, N. Altering Glycosphingolipid Composition to Improve Multiple Myeloma Bone Complication. Blood 2018, 132, 1942. [Google Scholar] [CrossRef]
- Suyal, S.; Singh, M.P.; Shekhar, H.; Srivastava, S. In silico screening of proteins targeting circulating miRNAs for improved diagnosis of multiple myeloma. Biochem. Biophys. Res. Commun. 2018, 497, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, D.; Agnelli, L.; Taiana, E.; Galletti, S.; Manzoni, M.; Todoerti, K.; Musto, P.; Strozzi, F.; Neri, A. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget 2016, 7, 14814–14830. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, D.; Riillo, C.; Juli, G.; Scionti, F.; Todoerti, K.; Polerà, N.; Grillone, K.; Fiorillo, L.; Arbitrio, M.; Di Martino, M.T.; et al. miR-22 Modulates Lenalidomide Activity by Counteracting MYC Addiction in Multiple Myeloma. Cancers 2021, 13, 4365. [Google Scholar] [CrossRef]
- Roseth Aass, K.; Nedal, T.M.V.; Anshushaug Bouma, S.; Tryggestad, S.S.; Haukås, E.; Slørdahl, T.S.; Waage, A.; Standal, T.; Mjelle, R. Comprehensive small RNA-sequencing of primary myeloma cells identifies miR-105-5p as a predictor of patient survival. Br. J. Cancer 2023, 128, 656–664. [Google Scholar] [CrossRef]
- Ghosal, S.; Banerjee, S. In silico bioinformatics analysis for identification of differentially expressed genes and therapeutic drug molecules in Glucocorticoid-resistant Multiple myeloma. Med. Oncol. 2022, 39, 53. [Google Scholar] [CrossRef]
- Perumal, D.; Imai, N.; Laganà, A.; Finnigan, J.; Melnekoff, D.; Leshchenko, V.V.; Solovyov, A.; Madduri, D.; Chari, A.; Cho, H.J.; et al. Mutation-derived Neoantigen-specific T-cell Responses in Multiple Myeloma. Clin. Cancer Res. 2020, 26, 450–464. [Google Scholar] [CrossRef]
- de Jong, M.M.E.; Kellermayer, Z.; Papazian, N.; Tahri, S.; Hofste op Bruinink, D.; Hoogenboezem, R.; Sanders, M.A.; van de Woestijne, P.C.; Bos, P.K.; Khandanpour, C.; et al. The multiple myeloma microenvironment is defined by an inflammatory stromal cell landscape. Nat. Immunol. 2021, 22, 769–780. [Google Scholar] [CrossRef]
- Musolino, C.; Oteri, G.; Allegra, A.; Mania, M.; D’Ascola, A.; Avenoso, A.; Innao, V.; Allegra, A.G.; Campo, S. Altered microRNA expression profile in the peripheral lymphoid compartment of multiple myeloma patients with bisphosphonate-induced osteonecrosis of the jaw. Ann. Hematol. 2018, 97, 1259–1269. [Google Scholar] [CrossRef]
- Urdeitx, P.; Clara-Trujillo, S.; Gomez Ribelles, J.L.; Doweidar, M.H. Multiple Myeloma Cell Simulation Using an Agent-Based Framework Coupled with a Continuous Fluid Model. Mathematics 2023, 11, 1824. [Google Scholar] [CrossRef]
- Giorgi, M.; Verbruggen, S.W.; Lacroix, D. In silico bone mechanobiology: Modeling a multifaceted biological system. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 485–505. [Google Scholar] [CrossRef] [PubMed]
- Freeman, F.E.; Burdis, R.; Mahon, O.R.; Kelly, D.J.; Artzi, N. A Spheroid Model of Early and Late-Stage Osteosarcoma Mimicking the Divergent Relationship between Tumor Elimination and Bone Regeneration. Adv. Healthc. Mater. 2022, 11, 2101296. [Google Scholar] [CrossRef] [PubMed]
- Cucè, M.; Gallo Cantafio, M.E.; Siciliano, M.A.; Riillo, C.; Caracciolo, D.; Scionti, F.; Staropoli, N.; Zuccalà, V.; Maltese, L.; Di Vito, A.; et al. Trabectedin triggers direct and NK-mediated cytotoxicity in multiple myeloma. J. Hematol. Oncol. 2019, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.O.; Rodriguez-Romera, A.; Reyat, J.S.; Olijnik, A.A.; Colombo, M.; Wang, G.; Wen, W.X.; Sousos, N.; Murphy, L.C.; Grygielska, B.; et al. Human Bone Marrow Organoids for Disease Modeling, Discovery, and Validation of Therapeutic Targets in Hematologic Malignancies. Cancer Discov. 2023, 13, 364–385. [Google Scholar] [CrossRef] [PubMed]
- Braham, M.V.J.; Ahlfeld, T.; Akkineni, A.R.; Minnema, M.C.; Dhert, W.J.A.; Oner, F.C.; Robin, C.; Lode, A.; Gelinsky, M.; Alblas, J. Endosteal and Perivascular Subniches in a 3D Bone Marrow Model for Multiple Myeloma. Tissue Eng. Part C Methods 2018, 24, 300–312. [Google Scholar] [CrossRef]
- Wu, D.; Wang, Z.; Li, J.; Song, Y.; Perez, M.E.M.; Wang, Z.; Cao, X.; Cao, C.; Maharjan, S.; Anderson, K.C.; et al. A 3D-Bioprinted Multiple Myeloma Model. Adv. Healthc. Mater. 2022, 11, e2100884. [Google Scholar] [CrossRef]
- Braham, M.V.; Deshantri, A.K.; Minnema, M.C.; Öner, F.C.; Schiffelers, R.M.; Fens, M.H.; Alblas, J. Liposomal drug delivery in an in vitro 3D bone marrow model for multiple myeloma. Int. J. Nanomed. 2018, 13, 8105–8118. [Google Scholar] [CrossRef]
- Braham, M.V.J.; Minnema, M.C.; Aarts, T.; Sebestyen, Z.; Straetemans, T.; Vyborova, A.; Kuball, J.; Oner, F.C.; Robin, C.; Alblas, J. Cellular immunotherapy on primary multiple myeloma expanded in a 3D bone marrow niche model. Oncoimmunology 2018, 7, e1434465. [Google Scholar] [CrossRef]
- Huang, Y.H.; Almowaled, M.; Li, J.; Venner, C.; Sandhu, I.; Peters, A.; Lavasanifar, A.; Lai, R. Three-Dimensional Reconstructed Bone Marrow Matrix Culture Improves the Viability of Primary Myeloma Cells In-Vitro via a STAT3-Dependent Mechanism. Curr. Issues Mol. Biol. 2021, 43, 313–323. [Google Scholar] [CrossRef]
- Kirshner, J.; Thulien, K.J.; Martin, L.D.; Debes Marun, C.; Reiman, T.; Belch, A.R.; Pilarski, L.M. A unique three-dimensional model for evaluating the impact of therapy on multiple myeloma. Blood 2008, 112, 2935–2945. [Google Scholar] [CrossRef] [PubMed]
- Jakubikova, J.; Cholujova, D.; Hideshima, T.; Gronesova, P.; Soltysova, A.; Harada, T.; Joo, J.; Kong, S.Y.; Szalat, R.E.; Richardson, P.G.; et al. A novel 3D mesenchymal stem cell model of the multiple myeloma bone marrow niche: Biologic and clinical applications. Oncotarget 2016, 7, 77326–77341. [Google Scholar] [CrossRef]
- Silva, A.; Silva, M.C.; Sudalagunta, P.; Distler, A.; Jacobson, T.; Collins, A.; Nguyen, T.; Song, J.; Chen, D.-T.; Chen, L.; et al. An Ex Vivo Platform for the Prediction of Clinical Response in Multiple Myeloma. Cancer Res. 2017, 77, 3336–3351. [Google Scholar] [CrossRef]
- Narayanan, N.K.; Duan, B.; Butcher, J.T.; Mazumder, A.; Narayanan, B.A. Characterization of multiple myeloma clonal cell expansion and stromal Wnt/β-catenin signaling in hyaluronic acid-based 3D hydrogel. In Vivo 2014, 28, 67–73. [Google Scholar]
- de la Puente, P.; Muz, B.; Gilson, R.C.; Azab, F.; Luderer, M.; King, J.; Achilefu, S.; Vij, R.; Azab, A.K. 3D tissue-engineered bone marrow as a novel model to study pathophysiology and drug resistance in multiple myeloma. Biomaterials 2015, 73, 70–84. [Google Scholar] [CrossRef]
- Reagan, M.R.; Mishima, Y.; Glavey, S.V.; Zhang, Y.; Manier, S.; Lu, Z.N.; Memarzadeh, M.; Zhang, Y.; Sacco, A.; Aljawai, Y.; et al. Investigating osteogenic differentiation in multiple myeloma using a novel 3D bone marrow niche model. Blood 2014, 124, 3250–3259. [Google Scholar] [CrossRef]
- Zdzisińska, B.; Roliński, J.; Piersiak, T.; Kandefer-Szerszeń, M. A comparison of cytokine production in 2-dimensional and 3-dimensional cultures of bone marrow stromal cells of multiple myeloma patients in response to RPMI8226 myeloma cells. Folia Histochem. Cytobiol. 2009, 47, 69–74. [Google Scholar] [CrossRef]
- Garcia-Ortiz, A.; Rodriguez-Garcia, Y.; Encinas, J.; Maroto-Martin, E.; Castellano, E.; Teixido, J.; Martinez-Lopez, J. The Role of Tumor Microenvironment in Multiple Myeloma Development and Progression. Cancers 2021, 13, 217. [Google Scholar] [CrossRef] [PubMed]
- Belloni, D.; Heltai, S.; Ponzoni, M.; Villa, A.; Vergani, B.; Pecciarini, L.; Marcatti, M.; Girlanda, S.; Tonon, G.; Ciceri, F.; et al. Modeling multiple myeloma-bone marrow interactions and response to drugs in a 3D surrogate microenvironment. Haematologica 2018, 103, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Ferrarini, M.; Steimberg, N.; Ponzoni, M.; Belloni, D.; Berenzi, A.; Girlanda, S.; Caligaris-Cappio, F.; Mazzoleni, G.; Ferrero, E. Ex-vivo dynamic 3-D culture of human tissues in the RCCS bioreactor allows the study of Multiple Myeloma biology and response to therapy. PLoS ONE 2013, 8, e71613. [Google Scholar] [CrossRef]
- Spelat, R.; Ferro, F.; Contessotto, P.; Warren, N.J.; Marsico, G.; Armes, S.P.; Pandit, A. A worm gel-based 3D model to elucidate the paracrine interaction between multiple myeloma and mesenchymal stem cells. Mater. Today Bio 2020, 5, 100040. [Google Scholar] [CrossRef] [PubMed]
- Nolan, J.; Pearce, O.M.T.; Screen, H.R.C.; Knight, M.M.; Verbruggen, S.W. Organ-on-a-Chip and Microfluidic Platforms for Oncology in the UK. Cancers 2023, 15, 635. [Google Scholar] [CrossRef]
- Keats, J.J.; Maxwell, C.A.; Taylor, B.J.; Hendzel, M.J.; Chesi, M.; Bergsagel, P.L.; Larratt, L.M.; Mant, M.J.; Reiman, T.; Belch, A.R.; et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood 2005, 105, 4060–4069. [Google Scholar] [CrossRef] [PubMed]
- VanDijken, J.; Kaigala, G.V.; Lauzon, J.; Atrazhev, A.; Adamia, S.; Taylor, B.J.; Reiman, T.; Belch, A.R.; Backhouse, C.J.; Pilarski, L.M. Microfluidic chips for detecting the t(4;14) translocation and monitoring disease during treatment using reverse transcriptase-polymerase chain reaction analysis of IgH-MMSET hybrid transcripts. J. Mol. Diagn. 2007, 9, 358–367. [Google Scholar] [CrossRef]
- Dylan Tsai, C.-H.; Sakuma, S.; Arai, F.; Taniguchi, T.; Ohtani, T.; Sakata, Y.; Kaneko, M. Geometrical alignment for improving cell evaluation in a microchannel with application on multiple myeloma red blood cells. RSC Adv. 2014, 4, 45050–45058. [Google Scholar] [CrossRef]
- Sung, Y.; Lue, N.; Hamza, B.; Martel, J.; Irimia, D.; Dasari, R.R.; Choi, W.; Yaqoob, Z.; So, P. Three-Dimensional Holographic Refractive-Index Measurement of Continuously Flowing Cells in a Microfluidic Channel. Phys. Rev. Appl. 2014, 1, 014002. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, D.; Li, Y.; He, W.; Lin, W.; Hu, L.; Wang, C.; Xu, L.; Park, J.; You, L. Mechanical segregation and capturing of clonal circulating plasma cells in multiple myeloma using micropillar-integrated microfluidic device. Biomicrofluidics 2019, 13, 064114. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Z.; Kang, D.; Li, K.; Liang, G.; Liu, Z.; Mai, Q.; Chen, Q.; Yao, C.; Wei, R.; Tan, X.; et al. DEPTOR exacerbates bone–fat imbalance in osteoporosis by transcriptionally modulating BMSC differentiation. Biomed. Pharmacother. 2022, 151, 113164. [Google Scholar] [CrossRef] [PubMed]
- Sui, C.; Zilberberg, J.; Lee, W. Microfluidic device engineered to study the trafficking of multiple myeloma cancer cells through the sinusoidal niche of bone marrow. Sci. Rep. 2022, 12, 1439. [Google Scholar] [CrossRef] [PubMed]
- Moore, T.A.; Brodersen, P.; Young, E.W.K. Multiple Myeloma Cell Drug Responses Differ in Thermoplastic vs PDMS Microfluidic Devices. Anal. Chem. 2017, 89, 11391–11398. [Google Scholar] [CrossRef]
- Qasaimeh, M.A.; Wu, Y.C.; Bose, S.; Menachery, A.; Talluri, S.; Gonzalez, G.; Fulciniti, M.; Karp, J.M.; Prabhala, R.H.; Karnik, R. Isolation of Circulating Plasma Cells in Multiple Myeloma Using CD138 Antibody-Based Capture in a Microfluidic Device. Sci. Rep. 2017, 7, 45681. [Google Scholar] [CrossRef]
- Zeng, Y.; Gao, L.; Luo, X.; Chen, Y.; Kabeer, M.H.; Chen, X.; Stucky, A.; Loudon, W.G.; Li, S.C.; Zhang, X.; et al. Microfluidic enrichment of plasma cells improves treatment of multiple myeloma. Mol. Oncol. 2018, 12, 1004–1011. [Google Scholar] [CrossRef]
- Liu, Y.; Su, R.; Song, J.; Yu, X.; Lin, S.; Zhu, Z.; Yang, Y.; Zhang, M.; Yang, L.; Zhang, H.; et al. Stimulus-Responsive Microfluidic Interface Enables Efficient Enrichment and Cytogenetic Profiling of Circulating Myeloma Cells. ACS Appl. Mater. Interfaces 2021, 13, 14920–14927. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.K.; Alberge, J.B.; Lightbody, E.D.; Boehner, C.J.; Dunford, A.; Sklavenitis-Pistofidis, R.; Mouhieddine, T.H.; Cowan, A.N.; Su, N.K.; Horowitz, E.M.; et al. MinimuMM-seq: Genome Sequencing of Circulating Tumor Cells for Minimally Invasive Molecular Characterization of Multiple Myeloma Pathology. Cancer Discov. 2023, 13, 348–363. [Google Scholar] [CrossRef]
- Foulk, B.; Schaffer, M.; Gross, S.; Rao, C.; Smirnov, D.; Connelly, M.C.; Chaturvedi, S.; Reddy, M.; Brittingham, G.; Mata, M.; et al. Enumeration and characterization of circulating multiple myeloma cells in patients with plasma cell disorders. Br. J. Haematol. 2018, 180, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Bian, S.; Zhang, X.; Lin, L.; Sun, L.; Guo, Z.; Pan, J.; Cui, J.; Yao, H.; Xu, J.; Hao, Z.; et al. Exosomal MiR-4261 mediates calcium overload in RBCs by downregulating the expression of ATP2B4 in multiple myeloma. Front. Oncol. 2022, 12, 978755. [Google Scholar] [CrossRef]
- Zhang, W.; Lee, W.Y.; Siegel, D.S.; Tolias, P.; Zilberberg, J. Patient-Specific 3D Microfluidic Tissue Model for Multiple Myeloma. Tissue Eng. Part C Methods 2014, 20, 663–670. [Google Scholar] [CrossRef]
- Zhang, W.; Gu, Y.; Sun, Q.; Siegel, D.S.; Tolias, P.; Yang, Z.; Lee, W.Y.; Zilberberg, J. Ex Vivo Maintenance of Primary Human Multiple Myeloma Cells through the Optimization of the Osteoblastic Niche. PLoS ONE 2015, 10, e0125995. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Sabhachandani, P.; Stroopinsky, D.; Palmer, K.; Cohen, N.; Rosenblatt, J.; Avigan, D.; Konry, T. Dynamic analysis of immune and cancer cell interactions at single cell level in microfluidic droplets. Biomicrofluidics 2016, 10, 054115. [Google Scholar] [CrossRef]
- Pak, C.; Callander, N.S.; Young, E.W.; Titz, B.; Kim, K.; Saha, S.; Chng, K.; Asimakopoulos, F.; Beebe, D.J.; Miyamoto, S. MicroC(3): An ex vivo microfluidic cis-coculture assay to test chemosensitivity and resistance of patient multiple myeloma cells. Integr. Biol. 2015, 7, 643–654. [Google Scholar] [CrossRef]
- Carreras, P.; Gonzalez, I.; Gallardo, M.; Ortiz-Ruiz, A.; Martinez-Lopez, J. Droplet Microfluidics for the Ex Vivo Expansion of Human Primary Multiple Myeloma Cells. Micromachines 2020, 11, 261. [Google Scholar] [CrossRef]
- Khin, Z.P.; Ribeiro, M.L.C.; Jacobson, T.; Hazlehurst, L.; Perez, L.; Baz, R.; Shain, K.; Silva, A.S. A Preclinical Assay for Chemosensitivity in Multiple Myeloma. Cancer Res. 2014, 74, 56–67. [Google Scholar] [CrossRef]
- Silva, A.; Jacobson, T.; Meads, M.; Distler, A.; Shain, K. An Organotypic High Throughput System for Characterization of Drug Sensitivity of Primary Multiple Myeloma Cells. J. Vis. Exp. 2015, 101, e53070. [Google Scholar] [CrossRef]
- Sarkar, S.; McKenney, S.; Sabhachandani, P.; Adler, J.; Hu, X.; Stroopinksy, D.; Rosenblatt, J.; Avigan, D.; Konry, T. Anti-myeloma activity and molecular logic operation by Natural Killer cells in microfluidic droplets. Sens. Actuators B Chem. 2019, 282, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, D.; Lopes, R.; Pestana, C.; Queirós, A.C.; João, C.; Carneiro, E.A. Patient-Derived Multiple Myeloma 3D Models for Personalized Medicine-Are We There Yet? Int. J. Mol. Sci. 2022, 23, 12888. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, L.; Lam, T.; Andersch, L.; Klaus, A.; Schwiebert, S.; Winkler, A.; Gauert, A.; Heeren-Hagemann, A.I.; Astrahantseff, K.; Klironomos, F.; et al. A Reproducible Bioprinted 3D Tumor Model Serves as a Preselection Tool for CAR T Cell Therapy Optimization. Front. Immunol. 2021, 12, 689697. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, I.D.; Patino-Escobar, B.; Tuomivaara, S.T.; Lin, Y.T.; Nix, M.A.; Leung, K.K.; Kasap, C.; Ramos, E.; Nieves Vasquez, W.; Talbot, A.; et al. The surfaceome of multiple myeloma cells suggests potential immunotherapeutic strategies and protein markers of drug resistance. Nat. Commun. 2022, 13, 4121. [Google Scholar] [CrossRef] [PubMed]
- Sudalagunta, P.; Silva, M.C.; Canevarolo, R.R.; Alugubelli, R.R.; DeAvila, G.; Tungesvik, A.; Perez, L.; Gatenby, R.; Gillies, R.; Baz, R.; et al. A pharmacodynamic model of clinical synergy in multiple myeloma. EBioMedicine 2020, 54, 102716. [Google Scholar] [CrossRef]
| IMiDs | Proteasome Inhibitors | Chemotherapy Anthracyclines | Chemotherapy Alkylators | Steroids | mABs | Other Small Molecules | Complex Immunotherapies |
|---|---|---|---|---|---|---|---|
| Thalidomide (Thalomid) | Bortezomib (Velcade) | Adriamycin | Cyclophosphamide (Cytoxan) | Dexamethasone | Daratumumab/ anti-CD38 (Darzalex) | XPO1 inhibitor (Selinexor) | BCMA targeting
|
| Lenalidomide (Revlimid) | Carfilzomib (Kyprolis) | Doxil (liposomal doxorubicin) | Bendamustine | Prednisone | Isatuximab /anti-CD38 (Sarclisa) | GPRC5D targeting
| |
| Pomalidomide (Pomalyst) | Ixazomib (Ninlaro) | Mephalan | Elotuzumab/ anti-SLAMF7 (Empliciti) |
| Paraskeletal Plasmacytomas (PSP), % | Extramedullary Plasmacytomas (EMP), % | |
|---|---|---|
| At diagnosis | 7–34.4 | 1.75–4.5 |
| At relapse | 6–34.2 | 3.4–48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verbruggen, S.W.; Freeman, C.L.; Freeman, F.E. Utilizing 3D Models to Unravel the Dynamics of Myeloma Plasma Cells’ Escape from the Bone Marrow Microenvironment. Cancers 2024, 16, 889. https://doi.org/10.3390/cancers16050889
Verbruggen SW, Freeman CL, Freeman FE. Utilizing 3D Models to Unravel the Dynamics of Myeloma Plasma Cells’ Escape from the Bone Marrow Microenvironment. Cancers. 2024; 16(5):889. https://doi.org/10.3390/cancers16050889
Chicago/Turabian StyleVerbruggen, Stefaan W., Ciara L. Freeman, and Fiona E. Freeman. 2024. "Utilizing 3D Models to Unravel the Dynamics of Myeloma Plasma Cells’ Escape from the Bone Marrow Microenvironment" Cancers 16, no. 5: 889. https://doi.org/10.3390/cancers16050889
APA StyleVerbruggen, S. W., Freeman, C. L., & Freeman, F. E. (2024). Utilizing 3D Models to Unravel the Dynamics of Myeloma Plasma Cells’ Escape from the Bone Marrow Microenvironment. Cancers, 16(5), 889. https://doi.org/10.3390/cancers16050889







