Simple Summary
In this review we summarize recent articles on the role of specific biotypes of non-coding RNAs in different cancer types. We will focus on micro RNAs that has been characterized in the cell nucleus, as well as long non-coding RNAs and circular RNAs that are bound by components of the polycomb repressive complex 2 or by the protein Fused in Sarcoma. We will describe mechanistic aspects offering comprehensive insights into specific diagnostic and therapeutic implications of the non-coding RNAs in the cancer types described in the review.
Abstract
The eukaryotic genome is mainly transcribed into non-coding RNAs (ncRNAs), including different RNA biotypes, such as micro RNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), among others. Although miRNAs are assumed to act primarily in the cytosol, mature miRNAs have been reported and functionally characterized in the nuclei of different cells. Further, lncRNAs are important regulators of different biological processes in the cell nucleus as part of different ribonucleoprotein complexes. CircRNAs constitute a relatively less-characterized RNA biotype that has a circular structure as result of a back-splicing process. However, circRNAs have recently attracted attention in different scientific fields due to their involvement in various biological processes and pathologies. In this review, we will summarize recent studies that link to cancer miRNAs that have been functionally characterized in the cell nucleus, as well as lncRNAs and circRNAs that are bound by core components of the polycomb repressive complex 2 (PRC2) or the protein fused in sarcoma (FUS), highlighting mechanistic aspects and their diagnostic and therapeutic potential.
1. Introduction
The genome in a eukaryotic cell is mainly transcribed into RNAs that are not translated into proteins. These so-called non-coding RNAs (ncRNAs) include long non-coding RNAs (lncRNAs; >200 nucleotides long) and micro RNAs (miRNAs; 21–25 nucleotides long) [1]. LncRNAs are important regulators of different biological processes in the nucleus [2,3]. Together with other factors, lncRNAs provide a framework for the assembly of defined chromatin structures at specific loci, thereby silencing repetitive DNA elements, modulating centromere function, and regulating gene expression [2,3,4,5]. On the other hand, miRNAs are assumed to act primarily in the cytosol by inhibiting translation [6,7]. Currently, the miRBase database (www.mirbase.org; accessed in 1 December 2023) lists over 2588 distinct miRNAs that are involved in nearly every cellular process, highlighting the vast and complex nature of this field of study [7,8]. Moreover, alterations in miRNA expression have been implicated in a myriad of diseases, emphasizing their importance in maintaining cellular homeostasis [9,10,11,12,13,14,15,16,17].
The biogenesis of miRNAs begins with the transcription of their genes by RNA polymerase II, generating primary miRNAs (pri-miRNAs). These pri-miRNAs are then processed in the nucleus by the DROSHA-DGCR8 complex into preliminary miRNAs (pre-miRNAs) [18], which are transported from the nucleus to the cytosol by Exportin-5 in a Ran-GTP-dependent manner [19]. In the cytosol, the pre-miRNAs undergo further cleavage by the enzyme DICER, resulting in a small double-stranded RNA [20,21]. One strand of this double-stranded RNA, determined by its thermodynamic properties, is then incorporated into the RNA-induced silencing complex (RISC), where it guides the complex to target messenger RNAs (mRNAs) to reduce gene expression, either through mRNA degradation or inhibition of translation [22]. Even though miRNAs are supposed to act mainly in the cytosol [6,7], mature miRNAs have been also detected in the nuclei of different cells [23,24,25,26,27,28]. Moreover, a hexanucleotide element has been reported to direct miRNA nuclear import [29]. However, the function of miRNAs in the cell nucleus has barely been investigated. In this review, we will summarize the cancer-related findings of miRNAs that have been detected in the cell nucleus.
We have demonstrated in previous studies that a nuclear miRNA, lethal 7d (Mirlet7d, also known as let-7d), is part of the multicomponent ncRNA–protein complex MiCEE, which mediates epigenetic repression of bidirectionally transcribed genes [23,30]. In this context, nuclear Mirlet7d binds ncRNAs expressed from these bidirectionally transcribed genes. The miRNA-lncRNA duplexes are bound by the exosome-associated protein C1D, which in turn serves as a dock for the RNA-degrading exosome complex [31,32,33] and the polycomb repressive complex 2 (PRC2) [34,35]. While the exosome RNA-degrading complex mediates degradation of the ncRNA, PRC2 induces heterochromatin at the promoter of the gene, thereby reducing the transcription of the protein-coding RNA. The name of the MiCEE complex is due to its components, the nuclear Mirlet7d, C1D, the nuclear-specific exosome subunit EXOSC10, and the histone methyl transferase EZH2 from PRC2 (Mirlet7d-C1D-EXOSC10-EZH2). Whereas the function of the exosome RNA-degrading complex is well documented in virtually all aspects of RNA biology [31,32,33], the role of the RNA-binding activity of various components of PRC2 is less documented [36,37]. The PRC2 core consists of four subunits: EZH2 (enhancer of zeste homolog 2) or its closely related homolog EZH1, SUZ12 (suppressor of zeste 12), EED (embryonic ectoderm development), and RBBP4 (retinoblastoma binding protein 4) [35]. We will discuss in the present review the RNA-binding activity of the subunits of the PRC2 core within the context of different cancer types.
The human FUS protein family is made up of more than 25 proteins [3,38]. The first member of this protein family was FUS [39]. In addition to FUS, EWSR1 and TAF15, known together as FET proteins, TDP-43, and hnRNPA1 have been reported to mediate liquid-liquid phase separation [40]. Common characteristics of FET proteins are long, intrinsically disordered domains; domains with repeated motifs; and domains for interaction with other proteins, RNA, or DNA [41,42]. FUS is an RNA-binding protein involved in various aspects of RNA metabolism. In addition, FUS and different translocations of FUS have been related to various pathologies, including amyotrophic lateral sclerosis [43], frontotemporal lobar degeneration [44], and liposarcoma [39], among others. We have unpublished results demonstrating the functional interaction between FUS and various components of the MiCEE complex during transcriptional regulation and three-dimensional (3D) genome organization. Motivated by our findings, we included in this review recent publications focusing on the RNA-binding activity of FUS, which has been related to different cancer types.
2. Nuclear miRNAs and Their Implications in Cancer
Nuclear miRNAs have been identified and profiled across various cell types, including different cancer cell lines, through advanced techniques such as next-generation sequencing (NGS), RNA fluorescence in situ hybridization (FISH), Northern blot, and quantitative polymerase chain reaction after reverse transcription (qRT-PCR), showing their localization not just to the cytoplasm but also to the nucleus [23,24,25,26,27,28,29]. These discoveries broaden the understanding of miRNA functions beyond post-transcriptional regulation in the cytoplasm to include the regulation of processes in the cell nucleus, such as chromatin structure, transcriptional control, and RNA processing, among others [45]. This is further evidenced by the study on the HCT116 colon cancer cell line, where mature miRNAs were found within the nucleus, hinting at nuclear functions beyond the established cytoplasmic post-transcriptional regulation [24]. These combined insights suggest a revised understanding of miRNA function, indicating their broader regulatory potential within cellular biology.
The human miRNA 29b (miR-29b) is predominantly localized in the cell nucleus [29]. This study suggests that miR-29b participates in the regulation of transcription or the splicing of target transcripts within the cell nucleus, rather than the miRNA-canonical translation regulatory functions in the cytosol. The nuclear localization of miR-29b is directed by a distinctive hexanucleotide 3′-terminal motif with the nucleotide sequence 5′-AGUGUU-3′ (Figure 1, top), which is not present in miR-29a. Remarkably, the hexanucleotide 3′-terminal motif of miR-29b acts as a transferable nuclear localization element that directs nuclear enrichment of other miRNAs or small interfering RNAs to which it is attached. Moreover, we found by sequence alignment analysis that a partially conserved and extended version of this nuclear shuttling motif is present in mouse and human miRNAs that have been functionally characterized in the cell nucleus [46]. The presence of a nuclear shuttling motif in various miRNAs, which in turn is conserved across species, suggests the existence of a common mechanism regulating the enrichment of these miRNAs in the cell nucleus, thereby indicating that even miRNAs with common seed sequences can have diverse functions and regulatory capacities due to distinct subcellular localizations, underscoring the complexity of miRNA-mediated regulation.
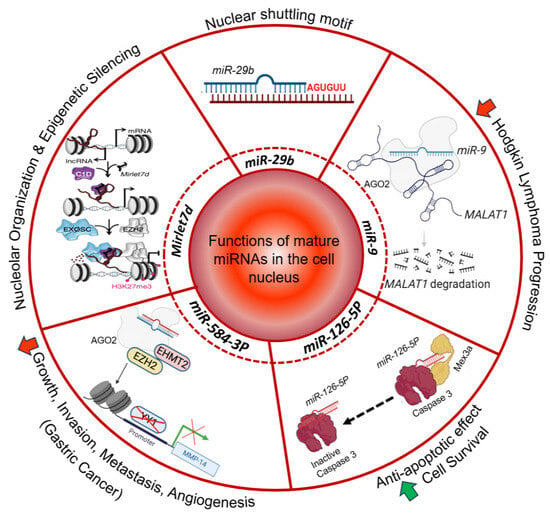
Figure 1.
Functions of mature miRNAs in the cell nucleus. The nuclear shuttling motif of miR-29b is highlighted [29]. The diagram further indicates the participation of Mirlet7d in nucleolar organization and epigenetic silencing [23]. miR-9 is associated with a decrease in Hodgkin lymphoma progression [27]. miR-126-5p is linked to apoptotic control and cell survival [47]. miR-584-3p is linked to gastric cancer [48]. Red arrow, decrease; green arrow, increase.
The function of miR-29b has been reported in various cancer types [49]. The specific role of miR-29b in melanoma has been shown by targeting transcripts like LAMC1 and LASP1 to reduce cell invasiveness, which could be crucial for stopping disease progression [50] (Figure 2, left top). In colorectal cancer, miR-29b overexpression is associated with suppression of the ERK/EGFR signaling pathway, leading to apoptosis and a decrease in angiogenesis and metastasis [51]. Furthermore, in acute myeloid leukemia, miR-29b reduces the levels of human antigen R (HuR, also known as ELAVL1 for embryonic lethal abnormal vision 1) by targeting its transcript [52], resulting in decreased levels of RELA (v-rel avian reticuloendotheliosis viral oncogene homolog A, also known as p65 or NFKB3) in the cell nucleus and reduced phosphorylation levels of RELA, IκBα, STAT1, STAT3, and STAT5. All these proteins are components of the NF-κB and JAK/STAT signaling pathways. The inhibition of these key oncogenic pathways mediated by miR-29b is accompanied by cell cycle arrest, cell viability decrease, apoptosis increase, and reduction of invasion and migration. These findings highlight the multifaceted roles of miR-29b in cancer, where its nuclear regulatory functions could be intrinsically linked to its ability to control key processes implicated in oncogenesis and cancer progression (Figure 2, left).
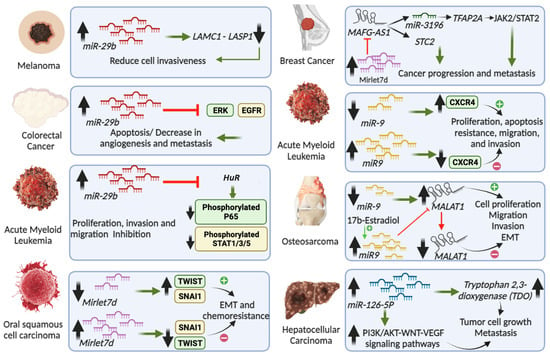
Figure 2.
Role in cancers of miRNAs that have been characterized in the cell nucleus. The figure depicts miRNAs’ involvement in various cancer types. For example, miR-29b in melanoma [50], colorectal cancer [51], and acute myeloid leukemia [52]; MIRLET7D in oral squamous cell carcinoma [53] and breast cancer [23,54,55]; miR-9 in acute myeloid leukemia [56] and osteosarcoma [57]; and miR-126-5p in hepatocellular carcinoma [58].
The miRNA lethal 7 (Mirlet7, also known as let-7) was together with lin-4 one of the first miRNAs discovered. Mirlet7 was initially discovered in Caenorhabditis elegans, where it plays a critical role in proliferation and differentiation of stem cells [59]. Subsequently, MIRLET7 was the first miRNA identified in humans and its sequence is highly conserved across species. In vertebrates, the Mirlet7 family consists of 10 miRNAs, including Mirlet7a, Mirlet7b, Mirlet7c, Mirlet7d, Mirlet7e, Mirlet7f, Mirlet7g, Mirlet7i, miR-98, miR-202, which are encoded by 13 genes, underlining the genetic complexity and significance of this miRNA family. In addition to its sequences, the functions of the Mirlet7 family members seem to be conserved across species, since they are integral to various biological processes such as development, stem cell biology, aging, and metabolism [60,61]. Reduced levels of Mirlet7 family members have been related to less-differentiated cellular stages and to different cancers [60,61]. Interestingly, LIN28B and its paralog, LIN28A, interact with the primary and/or preliminary transcripts of various members of the Mirlet7 family during development and oncogenesis and suppress the biogenesis of the mature miRNAs [62,63,64,65,66]. LIN28A together with Musashi1 (MSI1) sequester pri-Mirlet7 in the cell nucleus [66]. In addition, LIN28A also binds pre-Mirlet7 in the cytoplasm to block processing by DICER and recruits TUT4/7 (terminal uridylyltransferase 4/7), which adds uridyl groups to the 3′ end of pre-Mirlet7, thereby tagging it for degradation [67,68,69,70]. In contrast, LIN28B sequesters pri-Mirlet7 transcripts in the nucleolus, preventing the biogenesis of mature Mirlet7 [71]. As commented above, mature Mirlet7d participates as part of the MiCEE complex during epigenetic silencing of bidirectionally transcribed genes (Figure 1, left). In addition, Mirlet7d is involved in the structure and function of the nucleolus, the cellular hub for ribosomal RNA synthesis and ribosome biogenesis [23]. By tethering specific genomic regions to the nucleolus periphery, Mirlet7d contributes to the establishment of perinucleolar heterochromatin-associated domains (PNHADs), thus actively participating in the dynamic interface between 3D genome organization and gene expression regulation [23]. The role of mature Mirlet7d in nucleolar organization complements and adds another layer of complexity to our understanding of the functions of Mirlet7 family members [61,72]. The integrated view of all these molecular mechanisms underscores the multifaceted nature of non-coding RNAs and their regulatory proteins in both normal cellular processes and disease states. Recent studies have revealed that reduced Mirlet7d levels in oral squamous cell carcinoma are linked to enhanced epithelial-mesenchymal transition (EMT) and increased migratory and invasive capabilities of cancer cells [53]. Additionally, Mirlet7d downregulation correlated with heightened chemoresistance in oral squamous cell carcinoma, underlining its crucial role in cancer progression and treatment response (Figure 2, left bottom). Conversely, increased Mirlet7d levels reversed EMT, reducing invasiveness and enhancing chemosensitivity in cancer cells. In breast cancer, reduced levels of Mirlet7d have been related to enhanced cancer hallmarks [73,74]. On the other hand, the lncRNA MAFG-AS1 (MAF BZIP transcription factor G antisense RNA 1) promotes proliferation and metastasis of breast cancer by different mechanisms (Figure 2, right top), such as modulating the STC2 (Stanniocalcin 2) pathway and activating the JAK2/STAT3 (Janus kinase 2/signal transducer and activator of transcription 3) pathway through the miR-3196/TFAP2A axis [54,55]. Interestingly, we have shown that Mirlet7d induced degradation of MAFG-AS1 in the cell nucleus through the MiCEE complex [23]. Moreover, we have unpublished results demonstrating that Mirlet7d reduced cancer hallmarks in breast cancer cells by degradation of MAFG-AS1 and suppression of the JAK2/STAT2 signaling pathway. All these reports together suggest the development of therapies for oral squamous cell carcinoma and breast cancer using Mirlet7d.
The function of miR-9 in the cell nucleus illustrates different aspects of nuclear miRNA [27]. It has been reported that miR-9 exerts regulatory functions within the nucleus by targeting the lncRNA MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), which plays a role in cancer progression (Figure 1, right). The authors demonstrated that miR-9 in the cell nucleus binds directly to MALAT1 via specific binding motifs, leading to its degradation in an AGO2 (Argonaute 2)-dependent manner. The degradation of MALAT1 mediated by miR-9 was shown in osteosarcoma cells MG-63 that were treated with 17β-estradiol, which increased miR-9 levels, degraded MALAT1, and reduced several cancer hallmarks, including cell proliferation, colony formation, migration, and invasion (Figure 2, right) [57]. These findings not only shed light on the intricate regulatory mechanisms involving ncRNAs in the nucleus but also suggest a broader functional landscape for miR-9 in gene regulation beyond its traditional cytoplasmic activities.
Highlighting the role of miR-9 in the cell nucleus, we recently published a sequencing experiment after chromatin isolation by miRNA purification (ChIRP-seq) in mouse lung fibroblasts showing that miR-9 is enriched at promoters and super-enhancers of genes that are responsive to tissue growth factor beta 1 (TGFB1) signaling [46]. Further, we found that nuclear miR-9 is required for chromatin features related to increased transcriptional activity, including broad domains of the euchromatin histone mark histone 3 tri-methylated lysine 4 (H3K4me3) and the non-canonical DNA secondary structures G-quadruplex (G4). Moreover, we showed by chromosome conformation capture-based methods that nuclear miR-9 is required for promoter-super-enhancer looping. Our study places nuclear miR-9 in the same functional context as G4s and promoter-enhancer interactions during 3D genome organization and transcriptional activation induced by TGFB1 signaling, a critical regulator of proliferation programs in cancer [75].
A recent study underscores the function of miR-9 as a tumor suppressor in acute myeloid leukemia [56]. The authors found that miR-9 and CXCR4 (C-X-C chemokine receptor 4) were differentially expressed in acute myeloid leukemia samples, showing an inverse correlation between miR-9 and CXCR4 (Figure 2, right). The acute myeloid leukemia patients with high levels of CXCR4 and low expressions of miR-9 showed poor prognosis. Further, miR-9 overexpression inhibited proliferation, apoptosis resistance, migration, and invasion of acute myeloid leukemia cells. The authors also demonstrated that miR-9 directly targets CXCR4, with the miR-9/CXCR4 axis emerging as a critical factor and potential therapeutic target in acute myeloid leukemia pathology.
Another miRNA that has been functionally characterized in the cell nucleus is miR-126-5p [47]. Santovito et al. reported that nuclear miR-126-5p inhibits caspase-3 and confers endothelial protection by autophagy in atherosclerosis (Figure 1, right bottom). Mechanistically, miR-126-5p, in association with AGO2 and Mex3a (Mex-3 RNA-b Binding Family Member A), is transported to the nucleus via autophagic vesicles. In the cell nucleus, the authors showed that miR-126-5p dissociates from Ago2 and binds to caspase-3 with its seed sequence, preventing dimerization of the caspase and inhibiting its activity to block apoptosis. The non-canonical inhibition of caspase-3 by nuclear miR-126-5p is a mechanism by which miRNAs can modulate protein function.
Hepatocellular carcinoma is one of the most common gastrointestinal malignancies, with the third-highest mortality rate [76]. In the context of this cancer type, it has been reported that miR-126-5p promotes tumor cell proliferation, metastasis, and invasion by targeting tryptophan 2,3-dioxygenase (TDO2), which is a key enzyme in the tryptophan–kynurenine metabolic pathway (Figure 2, right bottom) [58]. By RNA immunoprecipitation (RIP) using TDO2-specific antibodies and sequencing of the precipitated RNAs, the authors identified 645 known miRNAs and 138 novel miRNAs that can bind to TDO2 in human hepatocellular carcinoma (HCCLM3) cells. By gain-of-function experiments after transfection of miR-126-5p mimics in HCCLM3 cells, the authors detected significantly increased expression levels of PI3K, AKT, WNTI1, and CTNNB1 (β-catenin). Loss-of-function experiments after transfection of miR-126-5p inhibitors in HCCLM3 cells reduced the expression levels of these transcripts. These results suggested that the PI3K/AKT pathway and the WNT pathway may be involved in miR-126-5p-related promotion of proliferation and metastasis. Although this study supports that the interaction between TDO2 and miR-126-5p plays a role in hepatocellular carcinoma, it is not clear whether the observed effects take place in specific cellular compartments due to limitations in the experimental systems that were used. For example, RIP was performed with whole-cell lysate without cellular fractionation. In ovarian cancer cells, the expression of miR-126-5p was reported to be high and to correlate with increased proliferation, migration, and invasion, while inhibiting apoptosis [77]. Mechanistically, miR-126-5p targets the PTEN transcript in the cytosol, thereby activating the PI3K/Akt/mTOR pathway. Further, the RNA methyltransferase-like 3 (METTL3) promotes miR-126-5p maturation through N6-methyladenosine (m6A) modification in the cell nucleus, thereby enhancing its activity. Silencing METTL3 disrupts this mechanism, offering therapeutic potential against ovarian cancer by targeting miR-126-5p and its regulatory mechanisms.
In line with the roles of miRNAs in the cell nucleus, miR-584-3p has been shown to participate in transcription regulation of the gene coding for matrix metalloproteinase 14 (MMP-14) in the nucleus of gastric cancer cells [48]. The transcription factor Yin Yang 1 (YY1) directly binds the promoter and mediates the transcription of the MMP-14 gene (Figure 1, left bottom). On the other hand, the authors found that miR-584-3p binds to sites that are adjacent to the YY1 binding site at the MMP-14 promoter, interacts with AGO2, and recruits EZH2 and euchromatic histone lysine methyltransferase 2 (EHMT2). The formation of this RNA–protein complex results in enrichment of repressive epigenetic markers at the MMP-14 promoter, decreased binding of YY1, and reduced MMP-14 transcription, thereby inhibiting the tumorigenesis and aggressiveness of gastric cancer.
3. Non-Coding RNAs Bound by PRC2 and Their Implications in Cancer
In addition to nuclear Mirlet7d as a component of the MiCEE complex, we will discuss the RNA-binding activity of other components of this ncRNA–protein complex, namely the subunits of the PRC2 core. The recruitment of PRC2 to specific genomic sites is a pertinent question that needs to be answered. A growing body of literature supports that different RNA molecules interact in vivo with PRC2 proteins [78,79,80,81]. The interaction of PRC2 components with specific RNA molecules contributes to the recruitment of the whole complex to specific genomic loci to regulate chromatin structure and transcription. For example, it has been reported that PRC2 members interact with nascent RNA at transcriptionally active genes, which determines the recruitment of PRC2 components to chromatin [80]. EZH2 is the enzymatic catalytic subunit of PRC2 that can alter downstream target gene expression by trimethylation of Lys-27 in histone 3 (H3K27me3). In addition, EZH2 can regulate gene expression in other ways besides H3K27me3 [82,83,84]. Earlier studies have revealed that EZH2 possesses an RNA-binding domain in the amino acid residues 342–368, and the phosphorylation of specific residues is cell cycle regulated and increases the binding to ncRNAs [85]. Further, it has been reported that promiscuous RNA binding by PRC2 members depends on the length of the RNA, where shorter RNAs are bound with lower affinity [86]. The same group proposed that RNA binding by EZH2 is not random but selective to maintain the repressed state of chromatin of those target genes that have escaped repression.
EZH2 is actively involved in transcription regulation of genes that are crucial for various biological mechanisms including cell fate, cell cycle, cell differentiation, DNA damage repair, lineage specification, autophagy, apoptosis, and tumorigenesis [87,88,89]. Further, studies show the involvement of EZH2 in the regulation of tumor microenvironment and antitumor immune response in solid cancers, directly affecting immunotherapy efficacy [90,91,92,93]. Notably, in a clinical study involving 696 patients, high EZH2 levels were positively correlated with increased tumor cell proliferation in four major cancer types including cutaneous melanoma and cancers of the endometrium, prostate, and breast [94]. Interestingly, the authors further demonstrated that high EZH2 levels led to poor survival in all four cancer types studied in a population-based setting. This study suggests that high EZH2 levels can be used as a predictive factor to identify increased tumor cell proliferation and aggressive subgroups in several cancers, and that it may be used as target for the development of therapies. The regulatory effects of ncRNAs binding to EZH2 have significant therapeutic potential in different cancers. However, they have received less attention. Seminal contributions focusing on ncRNAs related to EZH2 in major cancers are discussed below (see also Figure 3, top).
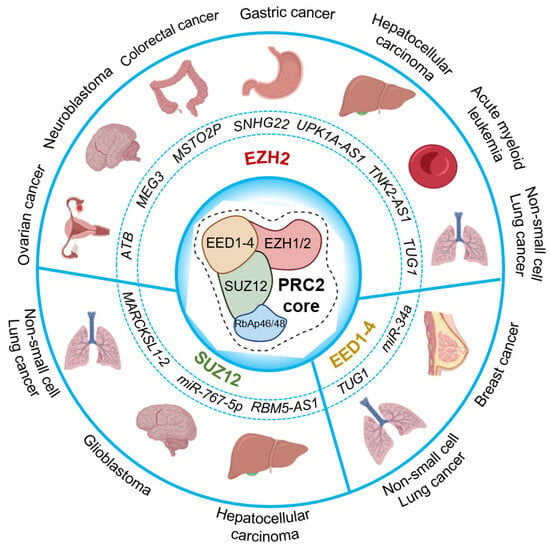
Figure 3.
Cancer-related RNA-binding activity of PRC2 core components. The circle chart above illustrates several long non-coding RNAs and micro-RNAs that are reported to bind the core components of PRC2, implicated in different cancer types. PRC2 core in the center; non-coding RNAs that bind to EZH2 (top); SUZ12 (bottom left); and EED1-4 (bottom right) are represented.
It has been reported that EZH2 is highly expressed in various cancer types [95,96,97] and mediates epigenetic silencing by catalyzing the heterochromatin histone mark H3K27me3 at specific loci, which in turn induces changes in gene expression leading to abnormal biological functions [82,83,84,98]. Increasing evidence has reported that the lncRNA ATB (lnc-ATB) promoted tumor progression in breast [99], prostate [100], and colon cancers [101], therefore suggesting lnc-ATB as a potential prognostic marker and therapeutic target in human cancers [102]. Chen et al. have reported that levels of lnc-ATB were increased in ovarian cancer tissue and high levels of lnc-ATB were associated with poor outcomes of patients with ovarian cancer [103]. Further, the authors demonstrated that lnc-ATB directly interacts with EZH2 by RIP and RNA pull-down assays in human ovarian cancer cell lines SKOV3 and A2780. Interestingly, lnc-ATB loss-of-function in SKOV3 and A2780 cells reduced cancer hallmarks, such as cell proliferation, invasion, and migration. In addition, chromatin immunoprecipitation (ChIP) assay after lnc-ATB loss-of-function demonstrated that lnc-ATB is required for enrichment of EZH2 and heterochromatic histone mark H3K27me3 at promoters of tumor suppressor genes, such as CDX1, FOXC1, LATS2, CDH1, and DAB2IP. Accordingly, the expression of these tumor suppressor genes was increased after lnc-ATB loss-of-function. These results suggest lnc-ATB as a potential biomarker for ovarian cancer diagnosis.
Neuroblastoma is a common extracranial solid tumor type with poor prognosis in children. The lncRNA MEG3 (maternally expressed 3) is found to be downregulated in neuroblastoma [104]. MEG3 overexpression in neuroblastoma cell lines attenuated autophagy through inhibition of FOXO1 and EMT via the mTOR pathway. Moreover, in a second article the authors showed that MEG3 and EZH2 negatively regulate each other [105]. RIP and co-immunoprecipitation (Co-IP) experiments were used to demonstrate the interaction of MEG3 and EZH2. Further, MEG3 exerted anti-cancer effects by mediating ubiquitination of EZH2, thereby leading to its degradation. Conversely, EZH2 interacted with DNMT1 and HDAC1 to induce silencing of MEG3. Summarizing both articles, reduced MEG3 levels and increased EZH2 levels form a feedback loop that promotes the development of neuroblastoma. Combined blockage of EZH2 and HDAC1 with the appropriate inhibitors may be an effective treatment strategy for neuroblastoma cases with low MEG3 and high EZH2 levels. Recently, a novel therapeutic approach using polymer nanoparticle-based administration of the lncRNA MEG3 was designed to control the progression of hepatocellular carcinoma in HepG2 cells [106].
It has been reported that the lncRNA pseudogene misato family member 2 (MSTO2P) affects cell proliferation, apoptosis, metastasis and invasion in hepatocellular carcinoma through the PI3K/AKT/mTOR pathway [107], thereby supporting the diagnostic and prognostic value of MSTO2P. A recent study explored the link between EZH2-mediated epigenetic silencing and ncRNAs, specifically the lncRNA MSTO2P in colorectal cancer [108]. MSTO2P levels were high in colorectal cancer tissues and cells. Further, experiments performed in colon cancer cells (HT-29 and SW480) revealed that loss-of-function of MSTO2P suppressed cell proliferation and invasion, promoting cell cycle arrest and apoptosis. In addition, the group also demonstrated that MSTO2P interacts with EZH2 in the cell nucleus and mediates epigenetic silencing of the tumor suppressor gene cyclin-dependent kinase inhibitor 1A (CDKN1A).
The lncRNA SNHG22 (small nucleolar RNA host gene 22) is highly expressed in gastric cancer cells and tissues and is correlated with poor prognosis in patients with gastric cancer [109]. The authors of this study found that the transcription factor ELK4 (ETS transcription factor ELK4) binds to the promoter region of the SNHG22 and promotes its expression in gastric cancer cells. Further, RNA pull-down using biotinylated SNHG22 and nuclear protein extracts from gastric cancer cells followed by mass spectrometry analysis of enriched proteins showed that SNHG22 directly interacts with EZH2. Consistently, RIP using EZH2-specific antibodies showed specific enrichment of SNHG22. Further, ChIP assays in BGC-823 and MGC-803 cells showed that EZH2 and H3K27me3 are enriched at promoters of various tumor suppressor genes (CDH1, EAF2, ADRB2, RUNX3, and RAP1GAP) in a SNHG22-dependent manner, since SNHG22 loss-of-function reduced the levels of EZH2 and H3K27me3 and increased the expression of all tumor suppressor genes analyzed, which in turn also inhibited the proliferation and invasion ability of gastric cancer cells. Another study reported that SNHG22 induces invasion, migration, and angiogenesis via the miR-361-3p/HMGA1/Wnt axis in gastric cancer cells [109]. Therefore, targeting the biomarker candidate SNHG22 could be a promising strategy for the diagnosis and prognosis of gastric cancer.
A newly identified lncRNA named UPK1A antisense RNA 1 (UPK1A-AS1) was found to promote cellular proliferation and tumor growth by accelerating cell cycle progression in hepatocellular carcinoma cells [110]. Moreover, cell cycle-related genes including CCND1, CDK2, CDK4, CCNB1, and CCNB2 were observed to be upregulated in hepatocellular carcinoma cells overexpressing UPK1A-AS1. Mechanistically, UPK1A-AS1 directly interacted with EZH2 to mediate its nuclear translocation, therefore leading to increased H3K27me3 levels. The authors reported that targeting EZH2 with specific small interfering RNA impaired UPK1A-AS1-mediated upregulation of cell cycle-related genes. Additionally, UPK1A-AS1 was significantly increased in hepatocellular carcinoma tissue, and increased levels of UPK1A-AS1 in hepatocellular carcinoma patients predicted poor prognosis. Thus, UPK1A-AS1 exhibits potential as a novel biomarker for the prognosis and therapy of hepatocellular carcinoma.
In a clinical study involving 89 patients with non-small cell lung cancer, the expression of the lncRNA taurine-upregulated gene 1 (TUG1) was determined to be significantly downregulated [111]. Using TUG1 4C sequencing and bioinformatic analysis, the authors found CELF1 (CUGBP and Elav-like family member 1) to be a potential target of TUG1 in-trans regulation. Subsequent experiments by RIP showed that TUG1 was bound to two components of the PRC2 core, namely EZH2 and EED. Moreover, ChIP assays showed that EZH2 and EDD were enriched at the promotor of the CELF1 gene in a TUG1-dependent manner. The study showed that TUG1 could bind to PRC2 at the promoter region of CELF1 and negatively regulate CELF1 expressions in lung squamous cell carcinoma H520 cells. These results may facilitate the development of new treatment modalities targeting TUG1/PRC2/CELF1 interactions in patients with lung squamous cell carcinoma.
Elevated expression of the lncRNA tyrosine kinase non receptor 2 antisense RNA 1 (TNK2-AS1) is reported in acute myeloid leukemia cell lines and is negatively correlated with patients’ survival [112]. The authors found that TNK2-AS1 exerted tumor-promoting activity in acute myeloid leukemia cells. Mechanistically, increased levels of TNK2-AS1 in acute myeloid leukemia cells were mediated by transcription factor ELK1 (ETS domain-containing protein-1). Further, EZH2 bound to TNK2-AS1 silenced the gene coding for CELF2 (CUGBP Elav-like family member 2) and exerted tumor-promoting effects through activation of the PI3K/Akt pathway. In contrast, knockdown of TNK2-AS1 markedly reduced cell proliferation and promoted apoptosis and differentiation in acute myeloid leukemia cells, suggesting TNK2-AS1 as a potential therapeutic target and prognostic marker for patients with acute myeloid leukemia. Another study reported the positive feedback loop between STAT3 and TNK2-AS1-mediated dysregulated STAT3 signaling by elevating VEGFA expression to facilitate angiogenesis in non-small cell lung cancer, indicating TNK2-AS1 as a potential target for therapeutic intervention [113].
EZH2-mediated epigenetic silencing of tumor suppressor genes leading to cancer proliferation has been reported earlier [114]. The transcriptional programs controlled by EZH2 were investigated in normal germinal center B cells of lymphomagenesis [115]. This study uncovered EZH2 binding sites at approximately 1800 promoter regions and identified key cell cycle-related tumor suppressor genes that are specifically downregulated in germinal center B cells of lymphomagenesis. In addition, the involvement of various long and short non-coding RNAs modulating the function of EZH2 in different cancer types was comprehensively reviewed by Mirzaei et al. [116].
In addition to EZH2, other PRC2 core proteins that have RNA-binding activity are SUZ12 and EED [80]. Recent literature that reports RNA-binding activity of SUZ12 and EED implicated in cancers is summarized below (see also Figure 3, bottom).
It has been reported that the lncRNA MARCKSL1-2 (MARCKSL1-transcript variant 2, NR_052852.1) promoted tumor progression by regulating epithelial–mesenchymal transition [117], thereby supporting the prognostic and therapeutic value of this lncRNA. In addition, the function of MARCKSL1-2 in docetaxel resistance of lung adenocarcinoma cells was investigated [118]. The authors found that SUZ12 is recruited by MARCKSL1-2 to the promoter of HDAC1 and increases H3K27me3 levels, thereby reducing HDAC1 expression. The reduced levels of HDAC1 blocked the suppressive effect of HDAC1 on histone acetylation modification at the miR-200b promoter, which in turn resulted in increased miR-200b expression. In summary, MARCKSL1-2 increases miR-200b expression by repressing HDAC1 expression. This hypothesis was further validated by in vivo experiments using a mouse xenograft tumor model that supported MARCKSL1-2 overexpression leading to attenuated docetaxel resistance in lung adenocarcinoma tumors.
The expression of miR-767-5p was identified to be significantly upregulated in glioblastoma tissues and cell lines. The authors demonstrated SUZ12 as a putative target of miR-767-5p, and the possibility that miR-767-5p acts by regulating SUZ12 expression [119]. It was revealed that the inhibitory effects of miR-767-5p on glioblastoma cell phenotypes were reversed by overexpression of SUZ12, indicating that the forced upregulation of miR-767-5p may represent a novel therapeutic strategy for glioma patients by targeting SUZ12. In vitro experiments suggest that the ectopic expression of miR-767-5p led to reduced proliferation, promoting cell cycle arrest and apoptosis in glioblastoma cell lines and inhibiting glioblastoma tumor growth in a mouse xenograft model.
Hepatocellular carcinoma is regarded as one of the most common malignancies leading to cancer-related death worldwide. The function of the lncRNA RBM5-AS1 (RBM5 antisense RNA 1) in the development of hepatocellular carcinoma was studied [120]. RBM5-AS1 levels are high in hepatocellular carcinoma tissues and cell lines, especially in Hep3B and HepG2 cells. Further, RBM5-AS1 loss-of-function reduced cell proliferation, invasion, and migration of hepatocellular carcinoma cells. Mechanistically, RBM5-AS1 was shown to recruit PRC2 core components (EZH2, SUZ12, EED) to the miR-132/212 promoter, elevate H3K27me3 levels, and reduce miR-132/212 expression. In summary, RBM5-AS1 was shown to act together with PRC2 as epigenetic regulator by repressing miR-132/212 expressions, thereby promoting hepatocellular carcinoma progression.
4. Non-Coding RNAs Bound by FUS and Their Implications in Cancer
Various lncRNAs have been associated with the severity and prognosis of hepatocellular carcinoma [121]. Among these transcripts are MALAT1, HOTAIR, HOTTIP, H19, and GTL2 (also known as MEG3) (Figure 4, top). A metadata study looking for RNA-binding proteins (RBP) that are potentially binding these lncRNAs identified FUS together with eIF4AIII and PTB as the proteins binding to the majority of the previously identified lncRNAs [122]. In addition, we have unpublished results demonstrating the functional interaction between FUS and components of the MiCEE complex during transcriptional regulation and 3D genome organization. Even though the mechanism by which FUS affects the development of hepatocellular carcinoma is unknown, the data suggest that FUS might play a role in the pathology by interacting with the identified lncRNAs.
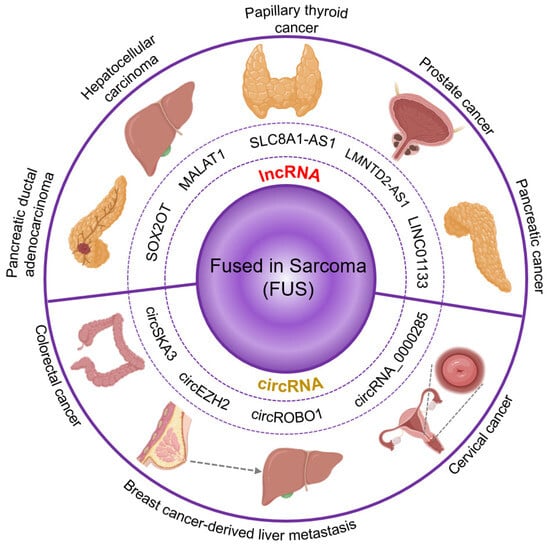
Figure 4.
Cancer-related RNA-binding activity of fused in sarcoma. FUS has been related to several types of cancer by binding to different biotypes of RNA. In the lncRNAs already reported to have an effect in cancer that also interact with FUS, we found SOX2OT, associated with pancreatic ductal adenocarcinoma. MALAT1, associated with hepatocellular carcinoma. SCL8A1-AS1, associated with papillary thyroid cancer. LMNTD2-AS1, associated with prostate cancer. LINC01133, associated with pancreatic cancer. Another RNA biotype we found to be frequently bound by FUS and also associated with cancer was circular RNA. Among them, we found circSKA3, associated with colorectal cancer; circRNAs_0000285 associated with cervical cancer; and circEZH2 and circROBO1 associated with breast cancer-derived liver metastasis.
The lncRNA SLC8A1-AS1 was shown to be downregulated in papillary thyroid cancer clinical samples. Moreover, the overexpression of SLC8A1-AS1 led to reduced proliferation and increased apoptosis in papillary thyroid cancer cells. A recent study demonstrated that the observed effect is due to SLC8A1-AS1 binding FUS as well as the mRNA of NUMB-like endocytic adaptor protein (Numbl) [123]. Furthermore, a loss-of-function of FUS showed similar results as the SLC8A1-AS1 downregulation, leading to decreased levels of Numbl and increased cell proliferation, even when overexpression of SLC8A1-AS1 was induced, suggesting that FUS is a key part of the mechanism. This lncRNA-FUS-mRNA interaction leads to stability and the maintained expression of Numbl, which in turn is an inhibitor of the Notch signaling pathway. The Notch pathway regulates cell proliferation and differentiation, and it has been reported to be hyperactive in several human cancers. Therefore, the evidence suggesting that SLC8A1-AS1-FUS interaction stabilizes Numbl expression, a Notch inhibitor, fits with the observations of decreased proliferation and increased apoptosis [124].
In a recent metadata study performed to identify lncRNAs associated with prostate cancer, LMNTD2-AS1 was found to be overexpressed. Increased levels of this transcript correlated with poor overall survival as well as poor progression-free interval in the TCGA database [125]. Furthermore, the levels of LMNTD2-AS1 correlated with the outcome of primary therapy. For example, patients with low levels of LMNTD2-AS1 showed a complete positive response to the therapy, whereas patients with high levels of LMNTD2-AS1 showed partial response to the therapy. Interestingly, LMNTD2-AS1 loss-of-function experiments showed a significant reduction in the proliferation, migration, and invasion of prostate cancer cells. LMNTD2-AS1 is reported to be bound by several RBPs, for example, FUS, CSTF2, SMNDC1, and RANGAP1. Strikingly, FUS is the only RBP protein also showing strong correlation with poor overall survival and progression-free interval in the TCGA database. Further, FUS gain-of-function experiments showed an increase in the proliferation of prostate cancer cells, rescuing the effect observed by the LMNTD2-AS1 loss-of-function.
Non-coding RNAs and RNA-binding proteins can affect cancer progression not only by influencing cell migration and proliferation but also by promoting drug resistance. High levels of the lncRNA LINC01133 have been associated with resistance to treatment with sorafenib in patients with pancreatic cancer [126]. The mechanism describes LINC01133 increasing the resistance of pancreatic cancer cells to ferroptosis. This process occurs by LINC01133 stabilizing FSP1 mRNA. Such stabilization is achieved through the binding of FUS, forming a LINC01133-FUS-FSP1 complex.
FUS has also been associated with tumor suppressor roles. High levels of the lncRNA SOX2OT have been correlated with poor prognosis in patients with pancreatic ductal adenocarcinoma [127]. The authors found that SOX2OT directly binds to FUS implementing ChIRP and RIP assays that were performed in in pancreatic ductal adenocarcinoma cells. In another manuscript, the authors showed that the binding of SOX2OT to FUS promoted the ubiquitination and degradation of FUS [128]. Downstream, FUS regulates cell cycle-associated factors, such as CCND1 and p27. Thus, the degradation of FUS mediated by SOX2OT leads to cell cycle disruption. The SOX2OT–FUS regulatory axis promotes migration, invasion, tumor growth, and metastasic abilities of pancreatic cancer cells. Pancreatic cancer is an extremely aggressive cancer type. Only 8% of patients with pancreatic cancer are alive five years after the disease is diagnosed. Therefore, the urge to find effective therapeutic target is pressing. A better understanding of the epigenetic mechanism of the disease would offer the opportunity to develop novel therapeutic approaches.
Circular RNA (circRNA) is a biotype of single-stranded RNA that shows a circular structure, a product of a back-splicing process. CircRNAs have gained importance in recent years since they have been reported to participate in the regulation of several biological processes [129]. CircRNA_0000285 is highly expressed in cervical cancer tissues when compared to control tissue [130]. Proliferation and migration of cervical cancer cells were significantly reduced after circRNA_0000285 loss-of-function experiments. Likewise, FUS levels decreased after circRNA_0000285 loss-of-function. Further results indicated that the expression level of FUS was positively correlated with the expression of circRNA_0000285 in cervical cancer tissues. In addition, the knockdown of circRNA_0000285 significantly inhibited the formation and metastasis of cervical cancer in nude mice. In summary, circRNA_0000285 was able to enhance the proliferation and metastasis of cervical cancer by increasing FUS levels (Figure 4, bottom), which might be a potential therapeutic target for cervical cancer treatment.
Interactions between FUS and circRNA have been reported to be involved in metastatic processes of other cancer types. For example, circROBO1 was found to be upregulated in liver metastasis cells derived from breast cancer [131]. The overexpression of circROBO1 was associated with a high rate of proliferation and migration in breast cancer cells. Loss-of-function of circROBO1 showed a decrease in both proliferation and migration of breast cancer cells. In contrast, gain-of-function of circROBO1 promoted liver metastasis and tumor growth in murine models. In addition, circROBO1 increased KLF5 levels by sponging miR-217-5p. In turn, KLF5 regulates the expression of FUS. Interestingly, FUS is involved in the back-splicing process of circROBO1, thereby establishing a positive feedback loop. Thus, the circROBO1/KLF5/FUS axis becomes both, a potential biomarker and a therapeutic target for breast cancer metastatic cells. In a similar case, circEZH2 was found to be upregulated in liver metastasic cells derived from breast cancer [132]. The overexpression of circEZH2 is associated with poor prognosis in breast cancer. In addition, overexpression of circEZH2 promoted in vivo liver metastasis in murine models. In vitro loss-of-function experiments targeting circEZH2 showed decreased cell migration and invasion. Similar to circROBO1, circEZH2 also absorbs miR-217-5p, as well leading to increased KLF5 levels and FUS transcription activation. FUS is also involved in the back-splicing of circEZH2, therefore showing a positive feedback loop.
CircRNAs have also been associated with tumor suppressor roles. In colorectal cancer, tumor-suppressive circRNAs are selectively secreted in exosomes to maintain cancer cell viability [133]. However, while the levels of circSKA3 were significantly high in colorectal cancer tissue, it was not found in serum from patients with colorectal cancer. CircSKA3 was retained in colorectal cancer cells via a specific intra-cellular motif, which the authors called cellmotif element. Furthermore, the zinc-finger transcription factor snail 2 (SNAI2, also known as SLUG) bound circSKA3 by the cellmotif element, which stabilized SNAI2 by inhibiting its ubiquitination and degradation [134]. SNAI2 has already been reported to promote EMT in patients with colorectal cancer. Interestingly, FUS plays a key role in the back-splicing and circularization of circSKA3, binding also to the cellmotif element of circSKA3.
5. Conclusions and Future Directions
The discovery of mature miRNAs in the cell nucleus opens an intriguing chapter in cancer research, underscoring the complexity and versatility of miRNA functions. The presence of miRNAs like miR-29b, Mirlet7d, miR-9, miR-126-5p, and miR-584-3p in the nucleus and their diverse regulatory roles, ranging from direct gene silencing to influencing intricate signaling pathways in cancer, signify a paradigm shift in our understanding of miRNA biology. These findings challenge the traditional view of miRNAs as predominantly cytoplasmic entities and highlight their potential in nuclear processes such as transcriptional regulation, epigenetic modification, and chromatin organization.
Clinical studies have shown that high levels of the core components of PRC2 correlated with increased cancer hallmarks and the poor survival of patients with various cancer types [94]. Thus, high levels of PRC2 core components can be used as a predictive factor to identify increased tumor cell proliferation and aggressive subgroups in several cancers, and they may be used as target for the development of therapies. Mechanistically, in most of the studies presented here, PRC2 mediates the heterochromatic histone mark H3K27me3 at promoters of tumor suppressor genes. Further, the interaction of PRC2 core components with specific RNA molecules contributes to the recruitment of the whole complex to specific genomic loci to regulate chromatin structure and transcription. The regulatory effects of ncRNA binding on PRC2 core components, especially on EZH2 as the enzymatic catalytic subunit of the complex, have significant therapeutic potential in different cancers.
The studies presented here suggest that the role played by FUS in specific cancer types may depend on the RNAs with which it is interacting in the cancer cells. Interestingly, FUS interacts with several circRNAs within the context of different cancer types, which may be a result of the role that FUS plays during the back-splicing process of specific circRNAs. The regulatory effects of circRNAs on the function of FUS and other FET proteins may have potential for the development of therapies in various cancer types.
Author Contributions
G.B. designed the manuscript; G.S., D.G.R.-A., A.A. and G.B. wrote the manuscript. All authors discussed and commented on the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Guillermo Barreto was funded by the “Centre National de la Recherche Scientifique” (CNRS, France), “Délégation Centre-Est” (CNRS-DR6) and the “Lorraine Université” (LU, France) through the initiative “Lorraine Université d’Excellence” (LUE) and the dispositive “Future Leader”. Guruprasadh Swaminathan receives a doctoral fellowship through the initiative “Lorraine Université d’Excellence” (LUE) (32131861). Diana G. Rogel-Ayala receives a doctoral fellowship from the DAAD (57552340).
Acknowledgments
We thank Karla Rubio and the members of Team 2 at the IMoPA for their comments. The figures were created in PowerPoint implementing BioRender.com.
Conflicts of Interest
The authors declare no conflict of interest.
References
- ENCODE Project Consortium. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Rubio, K.; Dobersch, S.; Barreto, G. Functional Interactions between Scaffold Proteins, Noncoding RNAs, and Genome Loci Induce Liquid-Liquid Phase Separation as Organizing Principle for 3-Dimensional Nuclear Architecture: Implications in Cancer. FASEB J. 2019, 33, 5814–5822. [Google Scholar] [CrossRef]
- Postepska-Igielska, A.; Giwojna, A.; Gasri-Plotnitsky, L.; Schmitt, N.; Dold, A.; Ginsberg, D.; Grummt, I. LncRNA Khps1 Regulates Expression of the Proto-Oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol. Cell 2015, 60, 626–636. [Google Scholar] [CrossRef]
- Rubio, K.; Castillo-Negrete, R.; Barreto, G. Non-Coding RNAs and Nuclear Architecture during Epithelial-Mesenchymal Transition in Lung Cancer and Idiopathic Pulmonary Fibrosis. Cell Signal. 2020, 70, 109593. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of Post-Transcriptional Regulation by microRNAs: Are the Answers in Sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of Small RNAs in Animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Wang, X.; Yin, S.; Liang, B.; Zhang, Y.; Fan, M.; Fu, Z.; Shen, C.; Han, Y.; et al. Function of microRNA-124 in the Pathogenesis of Cancer (Review). Int. J. Oncol. 2024, 64, 6. [Google Scholar] [CrossRef]
- Letelier, P.; Saldías, R.; Loren, P.; Riquelme, I.; Guzmán, N. MicroRNAs as Potential Biomarkers of Environmental Exposure to Polycyclic Aromatic Hydrocarbons and Their Link with Inflammation and Lung Cancer. Int. J. Mol. Sci. 2023, 24, 16984. [Google Scholar] [CrossRef]
- Jouve, M.; Carpentier, R.; Kraiem, S.; Legrand, N.; Sobolewski, C. MiRNAs in Alcohol-Related Liver Diseases and Hepatocellular Carcinoma: A Step toward New Therapeutic Approaches? Cancers 2023, 15, 5557. [Google Scholar] [CrossRef]
- Michas, A.; Michas, V.; Anagnostou, E.; Galanopoulos, M.; Tolia, M.; Tsoukalas, N. The Clinical Significance of MicroRNAs in Colorectal Cancer Signaling Pathways: A Review. Glob. Med. Genet. 2023, 10, 315–323. [Google Scholar] [CrossRef]
- Wang, L.; Shui, X.; Diao, Y.; Chen, D.; Zhou, Y.; Lee, T.H. Potential Implications of miRNAs in the Pathogenesis, Diagnosis, and Therapeutics of Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 16259. [Google Scholar] [CrossRef]
- Improta-Caria, A.C.; Rodrigues, L.F.; Joaquim, V.H.A.; De Sousa, R.A.L.; Fernandes, T.; Oliveira, E.M. MicroRNAs Regulating Signaling Pathways in Cardiac Fibrosis: Potential Role of the Exercise Training. Am. J. Physiol. Heart Circ. Physiol. 2023, 326, H497–H510. [Google Scholar] [CrossRef]
- Chao, C.-M.; Moiseenko, A.; Kosanovic, D.; Rivetti, S.; El Agha, E.; Wilhelm, J.; Kampschulte, M.; Yahya, F.; Ehrhardt, H.; Zimmer, K.-P.; et al. Impact of Fgf10 Deficiency on Pulmonary Vasculature Formation in a Mouse Model of Bronchopulmonary Dysplasia. Hum. Mol. Genet. 2019, 28, 1429–1444. [Google Scholar] [CrossRef]
- Shrestha, A.; Carraro, G.; Nottet, N.; Vazquez-Armendariz, A.I.; Herold, S.; Cordero, J.; Singh, I.; Wilhelm, J.; Barreto, G.; Morty, R.; et al. A Critical Role for miR-142 in Alveolar Epithelial Lineage Formation in Mouse Lung Development. Cell. Mol. Life Sci. 2019, 76, 2817–2832. [Google Scholar] [CrossRef]
- Chao, C.-M.; Carraro, G.; Rako, Z.A.; Kolck, J.; Sedighi, J.; Zimmermann, V.; Moiseenko, A.; Wilhelm, J.; Young, B.M.; Chong, L.; et al. Failure to Down-Regulate miR-154 Expression in Early Postnatal Mouse Lung Epithelium Suppresses Alveologenesis, with Changes in Tgf-β Signaling Similar to Those Induced by Exposure to Hyperoxia. Cells 2020, 9, 859. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The Nuclear RNase III Drosha Initiates microRNA Processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of microRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A Cellular Function for the RNA-Interference Enzyme Dicer in the Maturation of the Let-7 Small Temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeon, K.; Lee, J.-T.; Kim, S.; Kim, V.N. MicroRNA Maturation: Stepwise Processing and Subcellular Localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef]
- Sontheimer, E.J. Assembly and Function of RNA Silencing Complexes. Nat. Rev. Mol. Cell Biol. 2005, 6, 127–138. [Google Scholar] [CrossRef]
- Singh, I.; Contreras, A.; Cordero, J.; Rubio, K.; Dobersch, S.; Günther, S.; Jeratsch, S.; Mehta, A.; Krüger, M.; Graumann, J.; et al. MiCEE Is a ncRNA-Protein Complex That Mediates Epigenetic Silencing and Nucleolar Organization. Nat. Genet. 2018, 50, 990–1001. [Google Scholar] [CrossRef]
- Park, C.W.; Zeng, Y.; Zhang, X.; Subramanian, S.; Steer, C.J. Mature microRNAs Identified in Highly Purified Nuclei from HCT116 Colon Cancer Cells. RNA Biol. 2010, 7, 606–614. [Google Scholar] [CrossRef]
- Rasko, J.E.J.; Wong, J.J.-L. Nuclear microRNAs in Normal Hemopoiesis and Cancer. J. Hematol. Oncol. 2017, 10, 8. [Google Scholar] [CrossRef]
- Huang, V. Endogenous miRNAa: miRNA-Mediated Gene Upregulation. Adv. Exp. Med. Biol. 2017, 983, 65–79. [Google Scholar] [CrossRef]
- Leucci, E.; Patella, F.; Waage, J.; Holmstrøm, K.; Lindow, M.; Porse, B.; Kauppinen, S.; Lund, A.H. microRNA-9 Targets the Long Non-Coding RNA MALAT1 for Degradation in the Nucleus. Sci. Rep. 2013, 3, 2535. [Google Scholar] [CrossRef]
- Wong, J.J.L.; Ritchie, W.; Gao, D.; Lau, K.A.; Gonzalez, M.; Choudhary, A.; Taft, R.J.; Rasko, J.E.J.; Holst, J. Identification of Nuclear-Enriched miRNAs during Mouse Granulopoiesis. J. Hematol. Oncol. 2014, 7, 42. [Google Scholar] [CrossRef]
- Hwang, H.-W.; Wentzel, E.A.; Mendell, J.T. A Hexanucleotide Element Directs microRNA Nuclear Import. Science 2007, 315, 97–100. [Google Scholar] [CrossRef]
- Rubio, K.; Singh, I.; Dobersch, S.; Sarvari, P.; Günther, S.; Cordero, J.; Mehta, A.; Wujak, L.; Cabrera-Fuentes, H.; Chao, C.-M.; et al. Inactivation of Nuclear Histone Deacetylases by EP300 Disrupts the MiCEE Complex in Idiopathic Pulmonary Fibrosis. Nat. Commun. 2019, 10, 2229. [Google Scholar] [CrossRef]
- Meola, N.; Domanski, M.; Karadoulama, E.; Chen, Y.; Gentil, C.; Pultz, D.; Vitting-Seerup, K.; Lykke-Andersen, S.; Andersen, J.S.; Sandelin, A.; et al. Identification of a Nuclear Exosome Decay Pathway for Processed Transcripts. Mol. Cell 2016, 64, 520–533. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Jensen, T.H. The Exosome: A Multipurpose RNA-Decay Machine. Trends Biochem. Sci. 2008, 33, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Tomecki, R.; Jensen, T.H.; Dziembowski, A. The Eukaryotic RNA Exosome: Same Scaffold but Variable Catalytic Subunits. RNA Biol. 2011, 8, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb Gene Silencing: Knowns and Unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Reinberg, D. The Polycomb Complex PRC2 and Its Mark in Life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, H.; Yin, M.; Cheng, Z.; Jiang, P.; Feng, M.; Liu, Z.; Liao, B. TGF-Β1/Smad3 Upregulates UCA1 to Promote Liver Fibrosis through DKK1 and miR18a. J. Mol. Med. 2022, 100, 1465–1478. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.M.; Lin, P.N.; Souroullas, G.P. Non-Canonical Functions of EZH2 in Cancer. Front. Oncol. 2023, 13, 1233953. [Google Scholar] [CrossRef] [PubMed]
- King, O.D.; Gitler, A.D.; Shorter, J. The Tip of the Iceberg: RNA-Binding Proteins with Prion-like Domains in Neurodegenerative Disease. Brain Res. 2012, 1462, 61–80. [Google Scholar] [CrossRef]
- Crozat, A.; Aman, P.; Mandahl, N.; Ron, D. Fusion of CHOP to a Novel RNA-Binding Protein in Human Myxoid Liposarcoma. Nature 1993, 363, 640–644. [Google Scholar] [CrossRef]
- Lin, Y.; Protter, D.S.W.; Rosen, M.K.; Parker, R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 2015, 60, 208–219. [Google Scholar] [CrossRef]
- Shapiro, D.M.; Ney, M.; Eghtesadi, S.A.; Chilkoti, A. Protein Phase Separation Arising from Intrinsic Disorder: First-Principles to Bespoke Applications. J. Phys. Chem. B 2021, 125, 6740–6759. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically Disordered Proteins in Overcrowded Milieu: Membrane-Less Organelles, Phase Separation, and Intrinsic Disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Kwiatkowski, T.J.; Bosco, D.A.; Leclerc, A.L.; Tamrazian, E.; Vanderburg, C.R.; Russ, C.; Davis, A.; Gilchrist, J.; Kasarskis, E.J.; Munsat, T.; et al. Mutations in the FUS/TLS Gene on Chromosome 16 Cause Familial Amyotrophic Lateral Sclerosis. Science 2009, 323, 1205–1208. [Google Scholar] [CrossRef]
- Ferguson, K.M.; Soydan, H.; Lee, S.-Y.; Yamanaka, A.; Freer, A.S.; Xie, B. Evaluation of the CSEC Community Intervention Project (CCIP) in Five U.S. Cities. Eval. Rev. 2009, 33, 568–597. [Google Scholar] [CrossRef]
- Liao, J.-Y.; Ma, L.-M.; Guo, Y.-H.; Zhang, Y.-C.; Zhou, H.; Shao, P.; Chen, Y.-Q.; Qu, L.-H. Deep Sequencing of Human Nuclear and Cytoplasmic Small RNAs Reveals an Unexpectedly Complex Subcellular Distribution of miRNAs and tRNA 3′ Trailers. PLoS ONE 2010, 5, e10563. [Google Scholar] [CrossRef] [PubMed]
- Cordero, J.; Swaminathan, G.; Rogel-Ayala, D.G.; Rubio, K.; Elsherbiny, A.; Günther, S.; Braun, T.; Dobreva, G.; Barreto, G. 3D Genome Organization during TGFB-Induced Transcription Requires Nuclear microRNA and G-Quadruplexes. bioRxiv 2023. [Google Scholar] [CrossRef]
- Santovito, D.; Egea, V.; Bidzhekov, K.; Natarelli, L.; Mourão, A.; Blanchet, X.; Wichapong, K.; Aslani, M.; Brunßen, C.; Horckmans, M.; et al. Noncanonical Inhibition of Caspase-3 by a Nuclear microRNA Confers Endothelial Protection by Autophagy in Atherosclerosis. Sci. Transl. Med. 2020, 12, eaaz2294. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Chen, Y.; Ye, L.; Jiao, W.; Song, H.; Mei, H.; Li, D.; Yang, F.; Li, H.; Huang, K.; et al. miRNA-584-3p Inhibits Gastric Cancer Progression by Repressing Yin Yang 1- Facilitated MMP-14 Expression. Sci. Rep. 2017, 7, 8967. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, B.; Lin, J.; Zhao, L. microRNA-29b: An Emerging Player in Human Cancer. Asian Pac. J. Cancer Prev. 2014, 15, 9059–9064. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Andrews, M.C.; Cursons, J.; Hurley, D.G.; Anaka, M.; Cebon, J.S.; Behren, A.; Crampin, E.J. Systems Analysis Identifies miR-29b Regulation of Invasiveness in Melanoma. Mol. Cancer 2016, 15, 72. [Google Scholar] [CrossRef]
- Leng, Y.; Chen, Z.; Ding, H.; Zhao, X.; Qin, L.; Pan, Y. Overexpression of microRNA-29b Inhibits Epithelial-Mesenchymal Transition and Angiogenesis of Colorectal Cancer through the ETV4/ERK/EGFR Axis. Cancer Cell Int. 2021, 21, 17. [Google Scholar] [CrossRef]
- Tang, Y.-J.; Wu, W.; Chen, Q.-Q.; Liu, S.-H.; Zheng, Z.-Y.; Cui, Z.-L.; Xu, J.-P.; Xue, Y.; Lin, D.-H. miR-29b-3p Suppresses the Malignant Biological Behaviors of AML Cells via Inhibiting NF-κB and JAK/STAT Signaling Pathways by Targeting HuR. BMC Cancer 2022, 22, 909. [Google Scholar] [CrossRef]
- Chang, C.-J.; Hsu, C.-C.; Chang, C.-H.; Tsai, L.-L.; Chang, Y.-C.; Lu, S.-W.; Yu, C.-H.; Huang, H.-S.; Wang, J.-J.; Tsai, C.-H.; et al. Let-7d Functions as Novel Regulator of Epithelial-Mesenchymal Transition and Chemoresistant Property in Oral Cancer. Oncol. Rep. 2011, 26, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Fu, Y.; Guo, F.; Chen, H.; Fu, X.; Tan, W.; Zhang, H. Long Non-Coding RNA MAFG-AS1 Knockdown Blocks Malignant Progression in Breast Cancer Cells by Inactivating JAK2/STAT3 Signaling Pathway via MAFG-AS1/miR-3196/TFAP2A Axis. Int. J. Clin. Exp. Pathol. 2020, 13, 2455–2473. [Google Scholar] [PubMed]
- Di, S.; Bai, R.; Lu, D.; Chen, C.; Ma, T.; Zou, Z.; Zhang, Z. Long Non-Coding RNA MAFG-AS1 Promotes Proliferation and Metastasis of Breast Cancer by Modulating STC2 Pathway. Cell Death Discov. 2022, 8, 249. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Xi, X.; Liu, Q.; Cheng, Y.; Yang, H. MiR-9 Functions as a Tumor Suppressor in Acute Myeloid Leukemia by Targeting CX Chemokine Receptor 4. Am. J. Transl. Res. 2019, 11, 3384–3397. [Google Scholar]
- Fang, D.; Yang, H.; Lin, J.; Teng, Y.; Jiang, Y.; Chen, J.; Li, Y. 17β-Estradiol Regulates Cell Proliferation, Colony Formation, Migration, Invasion and Promotes Apoptosis by Upregulating miR-9 and Thus Degrades MALAT-1 in Osteosarcoma Cell MG-63 in an Estrogen Receptor-Independent Manner. Biochem. Biophys. Res. Commun. 2015, 457, 500–506. [Google Scholar] [CrossRef]
- Ai, Y.; Luo, S.; Wang, B.; Xiao, S.; Wang, Y. MiR-126-5p Promotes Tumor Cell Proliferation, Metastasis and Invasion by Targeting TDO2 in Hepatocellular Carcinoma. Molecules 2022, 27, 443. [Google Scholar] [CrossRef]
- Roush, S.; Slack, F.J. The Let-7 Family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Sun, X.; Jiao, X.; Pestell, T.G.; Fan, C.; Qin, S.; Mirabelli, E.; Ren, H.; Pestell, R.G. MicroRNAs and Cancer Stem Cells: The Sword and the Shield. Oncogene 2014, 33, 4967–4977. [Google Scholar] [CrossRef]
- Su, J.-L.; Chen, P.-S.; Johansson, G.; Kuo, M.-L. Function and Regulation of Let-7 Family microRNAs. MicroRNA 2012, 1, 34–39. [Google Scholar] [CrossRef]
- Knowles, T.; Huang, T.; Qi, J.; An, S.; Burket, N.; Cooper, S.; Nazarian, J.; Saratsis, A.M. LIN28B and Let-7 in Diffuse Midline Glioma: A Review. Cancers 2023, 15, 3241. [Google Scholar] [CrossRef]
- Balzeau, J.; Menezes, M.R.; Cao, S.; Hagan, J.P. The LIN28/Let-7 Pathway in Cancer. Front. Genet. 2017, 8, 31. [Google Scholar] [CrossRef]
- Viswanathan, S.R.; Daley, G.Q. Lin28: A microRNA Regulator with a Macro Role. Cell 2010, 140, 445–449. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Kawahara, H.; Okada, Y.; Imai, T.; Iwanami, A.; Mischel, P.S.; Okano, H. Musashi1 Cooperates in Abnormal Cell Lineage Protein 28 (Lin28)-Mediated Let-7 Family microRNA Biogenesis in Early Neural Differentiation. J. Biol. Chem. 2011, 286, 16121–16130. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Cho, J.; Ha, M.; Han, J.; Kim, V.N. Lin28 Mediates the Terminal Uridylation of Let-7 Precursor MicroRNA. Mol. Cell 2008, 32, 276–284. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Kim, Y.-K.; Ha, M.; Yoon, M.-J.; Cho, J.; Yeom, K.-H.; Han, J.; Kim, V.N. TUT4 in Concert with Lin28 Suppresses microRNA Biogenesis through Pre-microRNA Uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.E.; Chang, H.-M.; Piskounova, E.; Gregory, R.I. Lin28-Mediated Control of Let-7 microRNA Expression by Alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 2012, 18, 1875–1885. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Nagaike, T.; Tomita, K. Crystal Structure of the Lin28-Interacting Module of Human Terminal Uridylyltransferase That Regulates Let-7 Expression. Nat. Commun. 2019, 10, 1960. [Google Scholar] [CrossRef] [PubMed]
- Piskounova, E.; Polytarchou, C.; Thornton, J.E.; LaPierre, R.J.; Pothoulakis, C.; Hagan, J.P.; Iliopoulos, D.; Gregory, R.I. Lin28A and Lin28B Inhibit Let-7 microRNA Biogenesis by Distinct Mechanisms. Cell 2011, 147, 1066–1079. [Google Scholar] [CrossRef]
- Díaz-Piña, G.; Rubio, K.; Ordoñez-Razo, R.M.; Barreto, G.; Montes, E.; Becerril, C.; Salgado, A.; Cabrera-Fuentes, H.; Aquino-Galvez, A.; Carlos-Reyes, A.; et al. ADAR1 Isoforms Regulate Let-7d Processing in Idiopathic Pulmonary Fibrosis. Int. J. Mol. Sci. 2022, 23, 9028. [Google Scholar] [CrossRef]
- Shahabi, A.; Naghili, B.; Ansarin, K.; Montazeri, M.; Dadashpour, M.; Zarghami, N. Let-7d and miR-185 Impede Epithelial-Mesenchymal Transition by Downregulating Rab25 in Breast Cancer. Asian Pac. J. Cancer Prev. 2021, 22, 305–313. [Google Scholar] [CrossRef]
- Wyss, C.B.; Duffey, N.; Peyvandi, S.; Barras, D.; Martinez Usatorre, A.; Coquoz, O.; Romero, P.; Delorenzi, M.; Lorusso, G.; Rüegg, C. Gain of HIF1 Activity and Loss of miRNA Let-7d Promote Breast Cancer Metastasis to the Brain via the PDGF/PDGFR Axis. Cancer Res. 2021, 81, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Syed, V. TGF-β Signaling in Cancer. J. Cell. Biochem. 2016, 117, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Lv, X.; Liu, D.; Guo, H.; Yao, G.; Wang, L.; Liang, X.; Yang, Y. METTL3-Mediated Maturation of miR-126-5p Promotes Ovarian Cancer Progression via PTEN-Mediated PI3K/Akt/mTOR Pathway. Cancer Gene Ther. 2021, 28, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.W.; Koseki, H.; Ito, S. In Preprints: Revisiting RNA in PRC2. Development 2023, 150, dev202440. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Kang, X.; Wolf, C.; Touma, M. Mapping Chromatin Occupancy of Ppp1r1b-lncRNA Genome-Wide Using Chromatin Isolation by RNA Purification (ChIRP)-Seq. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Dutta, B.; Hee, Y.T.; Chng, W.-J. Towards Understanding of PRC2 Binding to RNA. RNA Biol. 2019, 16, 176–184. [Google Scholar] [CrossRef] [PubMed]
- G Hendrickson, D.; Kelley, D.R.; Tenen, D.; Bernstein, B.; Rinn, J.L. Widespread RNA Binding by Chromatin-Associated Proteins. Genome Biol. 2016, 17, 28. [Google Scholar] [CrossRef]
- Guo, Y.; Yu, Y.; Wang, G.G. Polycomb Repressive Complex 2 in Oncology. Cancer Res. Treat. 2023, 190, 273–320. [Google Scholar] [CrossRef]
- Duan, R.; Du, W.; Guo, W. EZH2: A Novel Target for Cancer Treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef]
- Kim, K.H.; Roberts, C.W.M. Targeting EZH2 in Cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Li, G.; Son, J.; Xu, C.-F.; Margueron, R.; Neubert, T.A.; Reinberg, D. Phosphorylation of the PRC2 Component Ezh2 Is Cell Cycle-Regulated and up-Regulates Its Binding to ncRNA. Genes Dev. 2010, 24, 2615–2620. [Google Scholar] [CrossRef]
- Davidovich, C.; Zheng, L.; Goodrich, K.J.; Cech, T.R. Promiscuous RNA Binding by Polycomb Repressive Complex 2. Nat. Struct. Mol. Biol. 2013, 20, 1250–1257. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Rose, N.R.; Klose, R.J. Targeting Polycomb Systems to Regulate Gene Expression: Modifications to a Complex Story. Nat. Rev. Mol. Cell Biol. 2015, 16, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-R.; Lee, C.-H.; Oksuz, O.; Stafford, J.M.; Reinberg, D. PRC2 Is High Maintenance. Genes Dev. 2019, 33, 903–935. [Google Scholar] [CrossRef] [PubMed]
- Mas, G.; Di Croce, L. The Role of Polycomb in Stem Cell Genome Architecture. Curr. Opin. Cell Biol. 2016, 43, 87–95. [Google Scholar] [CrossRef]
- Sun, S.; Yu, F.; Xu, D.; Zheng, H.; Li, M. EZH2, a Prominent Orchestrator of Genetic and Epigenetic Regulation of Solid Tumor Microenvironment and Immunotherapy. Biochim. Biophys. Acta (BBA) Rev. Cancer 2022, 1877, 188700. [Google Scholar] [CrossRef]
- Kim, J.; Lee, Y.; Lu, X.; Song, B.; Fong, K.-W.; Cao, Q.; Licht, J.D.; Zhao, J.C.; Yu, J. Polycomb- and Methylation-Independent Roles of EZH2 as a Transcription Activator. Cell Rep. 2018, 25, 2808–2820.e4. [Google Scholar] [CrossRef]
- Tiffen, J.; Gallagher, S.J.; Filipp, F.; Gunatilake, D.; Emran, A.A.; Cullinane, C.; Dutton-Register, K.; Aoude, L.; Hayward, N.; Chatterjee, A.; et al. EZH2 Cooperates with DNA Methylation to Downregulate Key Tumor Suppressors and IFN Gene Signatures in Melanoma. J. Investig. Dermatol. 2020, 140, 2442–2454.e5. [Google Scholar] [CrossRef]
- Fratta, E.; Sigalotti, L.; Covre, A.; Parisi, G.; Coral, S.; Maio, M. Epigenetics of Melanoma: Implications for Immune-Based Therapies. Immunotherapy 2013, 5, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 Expression Is Associated with High Proliferation Rate and Aggressive Tumor Subgroups in Cutaneous Melanoma and Cancers of the Endometrium, Prostate, and Breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Han, H.D.; Mangala, L.S.; Ali-Fehmi, R.; Newton, C.S.; Ozbun, L.; Armaiz-Pena, G.N.; Hu, W.; Stone, R.L.; Munkarah, A.; et al. Regulation of Tumor Angiogenesis by EZH2. Cancer Cell 2010, 18, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.P.; Ling, K. EZH2 and Histone Deacetylase Inhibitors Induce Apoptosis in Triple Negative Breast Cancer Cells by Differentially Increasing H3 Lys27 Acetylation in the BIM Gene Promoter and Enhancers. Oncol. Lett. 2017, 14, 5735–5742. [Google Scholar] [CrossRef] [PubMed]
- Karakashev, S.; Zhu, H.; Wu, S.; Yokoyama, Y.; Bitler, B.G.; Park, P.-H.; Lee, J.-H.; Kossenkov, A.V.; Gaonkar, K.S.; Yan, H.; et al. CARM1-Expressing Ovarian Cancer Depends on the Histone Methyltransferase EZH2 Activity. Nat. Commun. 2018, 9, 631. [Google Scholar] [CrossRef] [PubMed]
- Dobersch, S.; Rubio, K.; Barreto, G. Pioneer Factors and Architectural Proteins Mediating Embryonic Expression Signatures in Cancer. Trends Mol. Med. 2019, 25, 287–302. [Google Scholar] [CrossRef]
- Shi, S.-J.; Wang, L.-J.; Yu, B.; Li, Y.-H.; Jin, Y.; Bai, X.-Z. LncRNA-ATB Promotes Trastuzumab Resistance and Invasion-Metastasis Cascade in Breast Cancer. Oncotarget 2015, 6, 11652–11663. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Yi, X.-M.; Tang, C.-P.; Ge, J.-P.; Zhang, Z.-Y.; Zhou, W.-Q. Long Non-Coding RNA ATB Promotes Growth and Epithelial-Mesenchymal Transition and Predicts Poor Prognosis in Human Prostate Carcinoma. Oncol. Rep. 2016, 36, 10–22. [Google Scholar] [CrossRef]
- Yue, B.; Qiu, S.; Zhao, S.; Liu, C.; Zhang, D.; Yu, F.; Peng, Z.; Yan, D. LncRNA-ATB Mediated E-Cadherin Repression Promotes the Progression of Colon Cancer and Predicts Poor Prognosis. J. Gastroenterol. Hepatol. 2016, 31, 595–603. [Google Scholar] [CrossRef]
- Fan, Y.-H.; Ji, C.-X.; Xu, B.; Fan, H.-Y.; Cheng, Z.-J.; Zhu, X.-G. Long Noncoding RNA Activated by TGF-β in Human Cancers: A Meta-Analysis. Clin. Chim. Acta 2017, 468, 10–16. [Google Scholar] [CrossRef]
- Chen, X.-J.; An, N. Long Noncoding RNA ATB Promotes Ovarian Cancer Tumorigenesis by Mediating Histone H3 Lysine 27 Trimethylation through Binding to EZH2. J. Cell. Mol. Med. 2021, 25, 37–46. [Google Scholar] [CrossRef]
- Ye, M.; Lu, H.; Tang, W.; Jing, T.; Chen, S.; Wei, M.; Zhang, J.; Wang, J.; Ma, J.; Ma, D.; et al. Downregulation of MEG3 Promotes Neuroblastoma Development through FOXO1-Mediated Autophagy and mTOR-Mediated Epithelial-Mesenchymal Transition. Int. J. Biol. Sci. 2020, 16, 3050–3061. [Google Scholar] [CrossRef]
- Ye, M.; Gao, R.; Chen, S.; Wei, M.; Wang, J.; Zhang, B.; Wu, S.; Xu, Y.; Wu, P.; Chen, X.; et al. Downregulation of MEG3 and Upregulation of EZH2 Cooperatively Promote Neuroblastoma Progression. J. Cell. Mol. Med. 2022, 26, 2377–2391. [Google Scholar] [CrossRef]
- Elzallat, M.; Hassan, M.; Elkramani, N.; Aboushousha, T.; AbdelLatif, A.; Helal, N.; Abu-Taleb, H.; El-Ahwany, E. Nanoconjugated Long Non-Coding RNA MEG3 as a New Therapeutic Approach for Hepatocellular Carcinoma. Heliyon 2023, 9, e15288. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yue, C.; Xu, Y.; Jiang, X.; Zhang, L.; Wu, J. Prognostic Value and Molecular Regulatory Mechanism of MSTO2P in Hepatocellular Carcinoma: A Comprehensive Study Based on Bioinformatics, Clinical Analysis and In Vitro Validation. OncoTargets Ther. 2020, 13, 2583–2598. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zhang, X. LncRNA MSTO2P Promotes Colorectal Cancer Progression through Epigenetically Silencing CDKN1A Mediated by EZH2. World J. Surg. Oncol. 2022, 20, 95. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Ji, T.; Liu, A.; Weng, Y. ELK4-Mediated lncRNA SNHG22 Promotes Gastric Cancer Progression through Interacting with EZH2 and Regulating miR-200c-3p/Notch1 Axis. Cell Death Dis. 2021, 12, 957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-Y.; Sun, Q.-C.; Zou, X.-J.; Song, Y.; Li, W.-W.; Guo, Z.-Q.; Liu, S.-S.; Liu, L.; Wu, D.-H. Long Noncoding RNA UPK1A-AS1 Indicates Poor Prognosis of Hepatocellular Carcinoma and Promotes Cell Proliferation through Interaction with EZH2. J. Exp. Clin. Cancer Res. 2020, 39, 229. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.-C.; Huang, H.-D.; Chang, C.-C.; Chang, Y.-S.; Yen, J.-C.; Lee, C.-C.; Chang, W.-H.; Liu, T.-C.; Chang, J.-G. Long Noncoding RNA TUG1 Is Downregulated in Non-Small Cell Lung Cancer and Can Regulate CELF1 on Binding to PRC2. BMC Cancer 2016, 16, 583. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Zhang, A.; Suo, M.; Wang, P.; Liang, Y. ELK1-Induced Upregulation of Long Non-Coding TNK2-AS1 Promotes the Progression of Acute Myeloid Leukemia by EZH2-Mediated Epigenetic Silencing of CELF2. Cell Cycle 2023, 22, 117–130. [Google Scholar] [CrossRef]
- Wang, Y.; Han, D.; Pan, L.; Sun, J. The Positive Feedback between lncRNA TNK2-AS1 and STAT3 Enhances Angiogenesis in Non-Small Cell Lung Cancer. Biochem. Biophys. Res. Commun. 2018, 507, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Béguelin, W.; Rivas, M.A.; Calvo Fernández, M.T.; Teater, M.; Purwada, A.; Redmond, D.; Shen, H.; Challman, M.F.; Elemento, O.; Singh, A.; et al. EZH2 Enables Germinal Centre Formation through Epigenetic Silencing of CDKN1A and an Rb-E2F1 Feedback Loop. Nat. Commun. 2017, 8, 877. [Google Scholar] [CrossRef]
- Velichutina, I.; Shaknovich, R.; Geng, H.; Johnson, N.A.; Gascoyne, R.D.; Melnick, A.M.; Elemento, O. EZH2-Mediated Epigenetic Silencing in Germinal Center B Cells Contributes to Proliferation and Lymphomagenesis. Blood 2010, 116, 5247–5255. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Gholami, M.H.; Hushmandi, K.; Hashemi, F.; Zabolian, A.; Canadas, I.; Zarrabi, A.; Nabavi, N.; Aref, A.R.; Crea, F.; et al. The Long and Short Non-Coding RNAs Modulating EZH2 Signaling in Cancer. J. Hematol. Oncol. 2022, 15, 18. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Gao, R.; Yang, M.; Wang, X.; Cheng, K.; Shi, X.; He, C.; Li, Y.; Wu, Y.; Shi, L.; et al. MARCKSL1 Promotes the Proliferation, Migration and Invasion of Lung Adenocarcinoma Cells. Oncol. Lett. 2020, 19, 2272–2280. [Google Scholar] [CrossRef]
- Jiang, M.; Qi, F.; Zhang, K.; Zhang, X.; Ma, J.; Xia, S.; Chen, L.; Yu, Z.; Chen, J.; Chen, D. MARCKSL1-2 Reverses Docetaxel-Resistance of Lung Adenocarcinoma Cells by Recruiting SUZ12 to Suppress HDAC1 and Elevate miR-200b. Mol. Cancer 2022, 21, 150. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, S.; Xu, J.; Li, Y.; Zhang, J.; Zhang, J.; Lu, X. miR-767-5p Inhibits Glioma Proliferation and Metastasis by Targeting SUZ12. Oncol. Rep. 2019, 42, 55–66. [Google Scholar] [CrossRef]
- Mu, J.-Y.; Tian, J.-X.; Chen, Y.-J. lncRNA RBM5-AS1 Promotes Cell Proliferation and Invasion by Epigenetically Silencing miR-132/212 in Hepatocellular Carcinoma Cells. Cell Biol. Int. 2021, 45, 2201–2210. [Google Scholar] [CrossRef]
- Mallela, V.R.; Rajtmajerová, M.; Trailin, A.; Liška, V.; Hemminki, K.; Ambrozkiewicz, F. miRNA and lncRNA as Potential Tissue Biomarkers in Hepatocellular Carcinoma. Non-Coding RNA Res. 2024, 9, 24–32. [Google Scholar] [CrossRef]
- Mohamadkhani, A. Long Noncoding RNAs in Interaction With RNA Binding Proteins in Hepatocellular Carcinoma. Hepat. Mon. 2014, 14, e18794. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Shang, X.; Sun, X.; Xu, G.; Liu, Y.; Liu, Y. SLC8A1 Antisense RNA 1 Suppresses Papillary Thyroid Cancer Malignant Progression via the FUS RNA Binding Protein (FUS)/NUMB like Endocytic Adaptor Protein (Numbl) Axis. Bioengineered 2022, 13, 12572–12582. [Google Scholar] [CrossRef]
- Kopan, R.; Ilagan, M.X.G. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Wu, H.; Liu, N.; He, A.; Li, H.; Liu, H.; Qian, J.; Mao, W.; Fu, G. LMNTD2-AS1 Regulates Immune Cell Infiltration and Promotes Prostate Cancer Progression by Targeting FUS to Regulate NRF2 Signal Pathway. Am. J. Cancer Res. 2023, 13, 3384–3400. [Google Scholar]
- Wang, S.; Chen, J.; Li, P.; Chen, Y. LINC01133 Can Induce Acquired Ferroptosis Resistance by Enhancing the FSP1 mRNA Stability through Forming the LINC01133-FUS-FSP1 Complex. Cell Death Dis. 2023, 14, 767. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, J.; Chen, Q.; Ge, W.; Meng, L.; Huang, X.; Shen, P.; Yuan, H.; Shi, G.; Miao, Y.; et al. Long Noncoding RNA SOX2OT Promotes the Proliferation of Pancreatic Cancer by Binding to FUS. Int. J. Cancer 2020, 147, 175–188. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.-F.; Wang, D.-Y.; Zhu, Y.; Chen, L.; Zhang, J.-J. Long Noncoding RNA SOX2OT Promotes Pancreatic Cancer Cell Migration and Invasion through Destabilizing FUS Protein via Ubiquitination. Cell Death Discov. 2021, 7, 261. [Google Scholar] [CrossRef]
- Khalilian, S.; Tabari, M.A.K.; Omrani, M.A.; Ghafouri-Fard, S. Emerging Functions and Significance of circCDYL in Human Disorders. Mol. Biol. Rep. 2023, 51, 7. [Google Scholar] [CrossRef]
- Chen, R.-X.; Liu, H.-L.; Yang, L.-L.; Kang, F.-H.; Xin, L.-P.; Huang, L.-R.; Guo, Q.-F.; Wang, Y.-L. Circular RNA circRNA_0000285 Promotes Cervical Cancer Development by Regulating FUS. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8771–8778. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L.; Wu, P.; Li, X.; Tang, Y.; Ou, X.; Zhang, Y.; Xiao, X.; Wang, J.; Tang, H. The circROBO1/KLF5/FUS Feedback Loop Regulates the Liver Metastasis of Breast Cancer by Inhibiting the Selective Autophagy of Afadin. Mol. Cancer 2022, 21, 29. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, Z.; Ou, X.; Wu, P.; Zhang, Y.; Wu, S.; Xiao, X.; Li, Y.; Ye, F.; Tang, H. The FUS/circEZH2/KLF5/Feedback Loop Contributes to CXCR4-Induced Liver Metastasis of Breast Cancer by Enhancing Epithelial-Mesenchymal Transition. Mol. Cancer 2022, 21, 198. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yu, H.; Han, F.; Lai, X.; Ye, K.; Lei, S.; Mai, M.; Lai, M.; Zhang, H. Tumor-Suppressive circRHOBTB3 Is Excreted out of Cells via Exosome to Sustain Colorectal Cancer Cell Fitness. Mol. Cancer 2022, 21, 46. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Liao, S.; Chen, C.; Han, F.; Lei, S.; Lai, X.; Ye, K.; Han, Q.; E, F.; Lu, C.; et al. Specific Intracellular Retention of circSKA3 Promotes Colorectal Cancer Metastasis by Attenuating Ubiquitination and Degradation of SLUG. Cell Death Dis. 2023, 14, 750. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).