Exploring Genomic Biomarkers for Pembrolizumab Response: A Real-World Approach and Patient Similarity Network Analysis Reveal DNA Response and Repair Gene Mutations as a Signature
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Real-World Cohort
- Cytological or histological diagnosis of aNSCLC (stage IIIB to IV)
- Receipt of at least one cycle of first-line pembrolizumab
- ECOG PS score of 0–2
- Availability of tumor tissue for next generation sequencing (NGS) analysis
2.2. POPLAR and OAK Population
2.3. Targeted Next-Generation Sequencing and Oncogenic Pathway Classification
2.4. PD-L1 Testing and (Blood) TMB Assessment
2.5. Clinical Outcomes
2.6. Statistical Analysis
2.7. Patient Similarity Network Analysis
3. Results
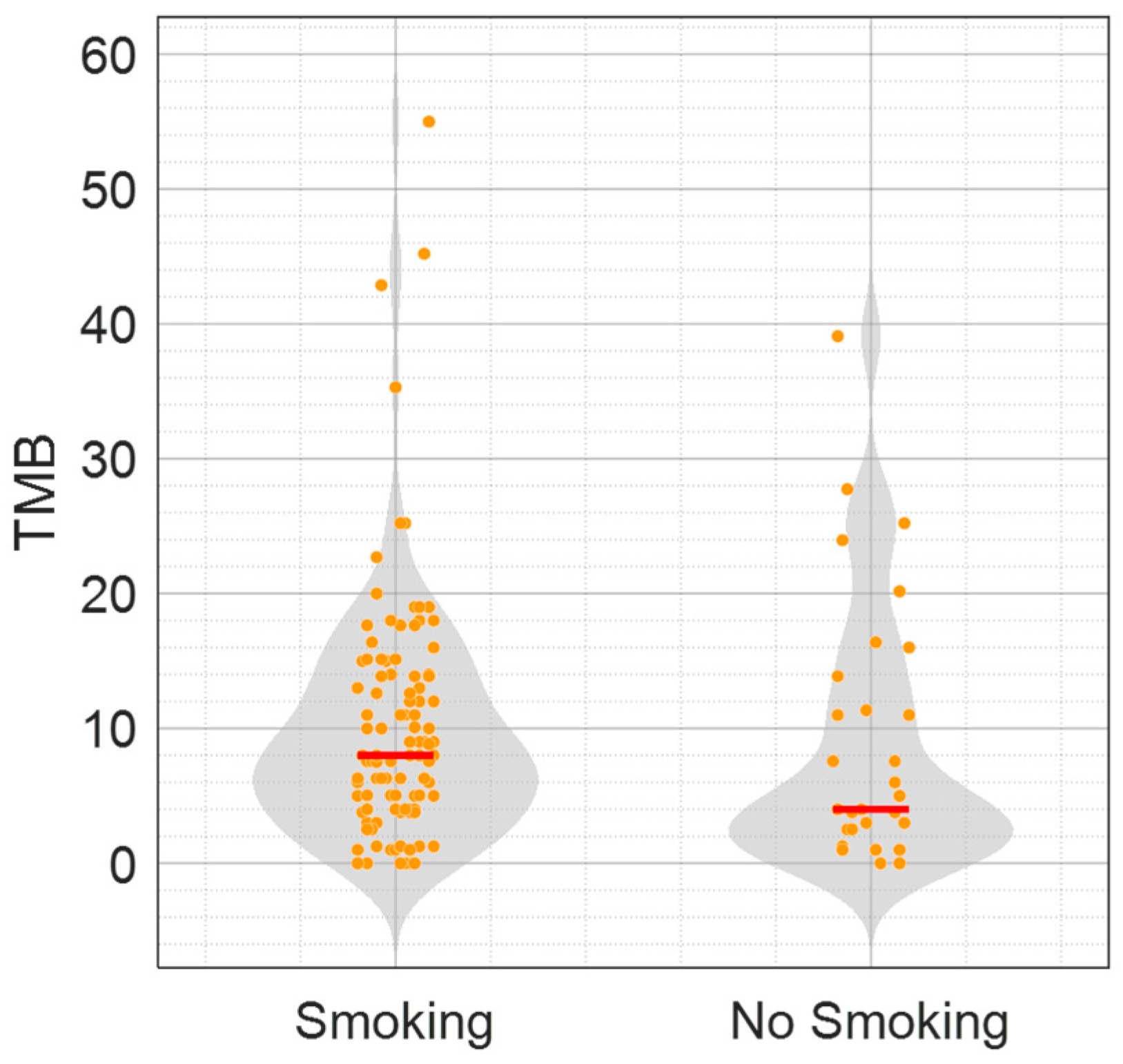
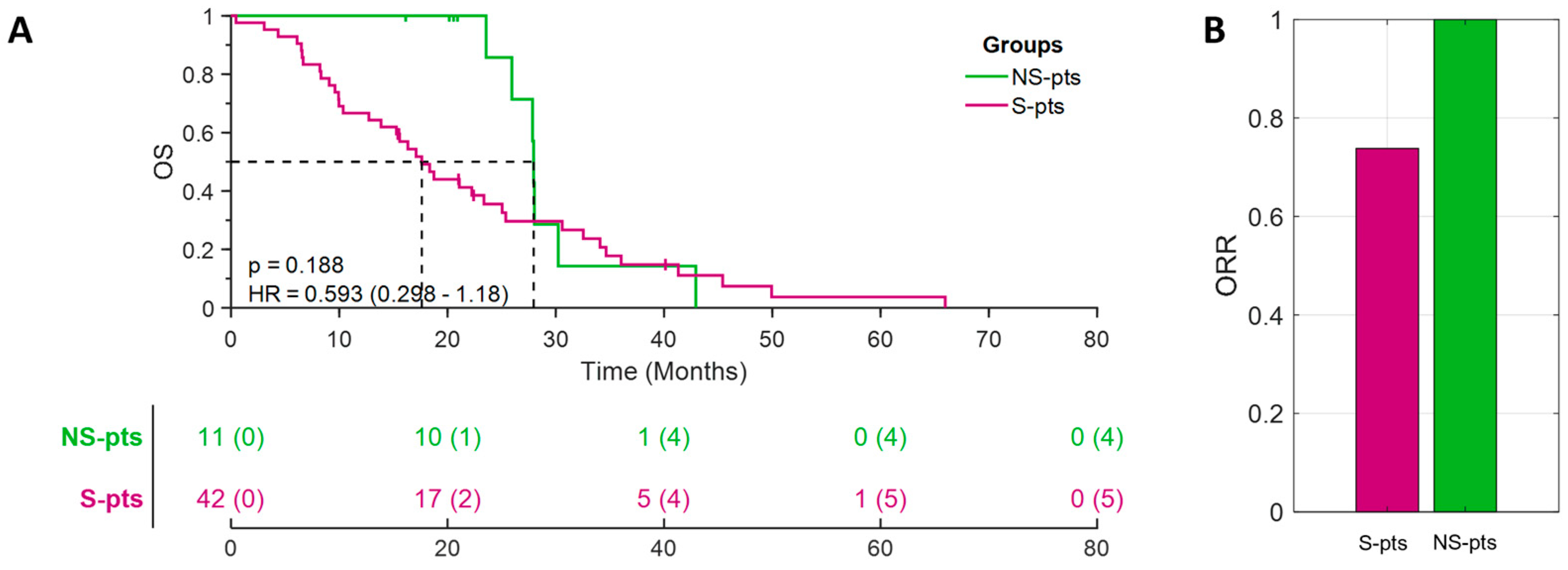
3.1. Non-Smoking Patients with H-TMB in a Real-World Cohort Displayed a Novel Genomic Profile Characterized by Mutations in the DDR Pathway, and a Better Outcome to Pembrolizumab Treatment
3.2. DDR Pathway Mutations as a Molecular Feature of NS/H-TMB Patients in POPLAR/OAK Populations
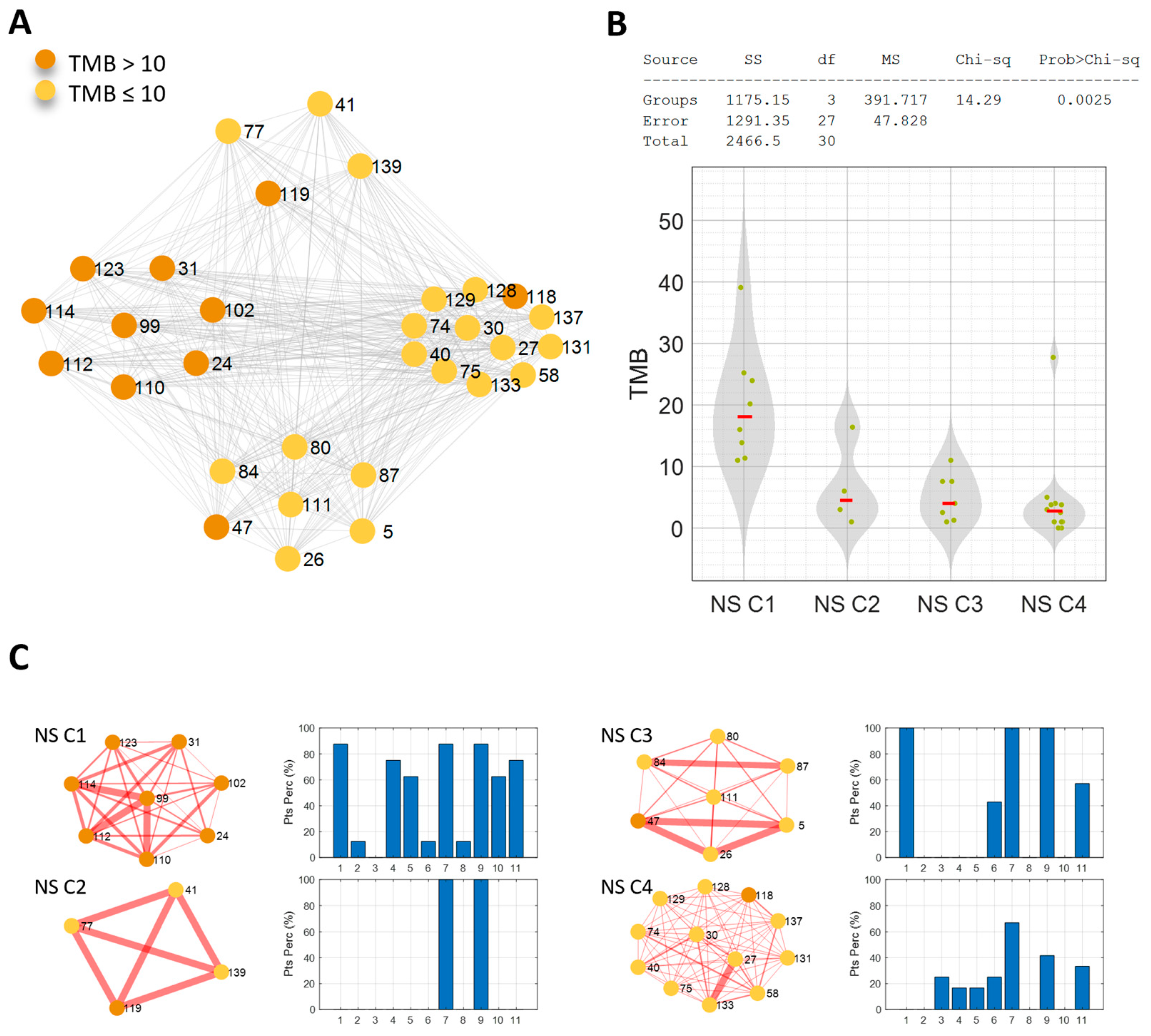
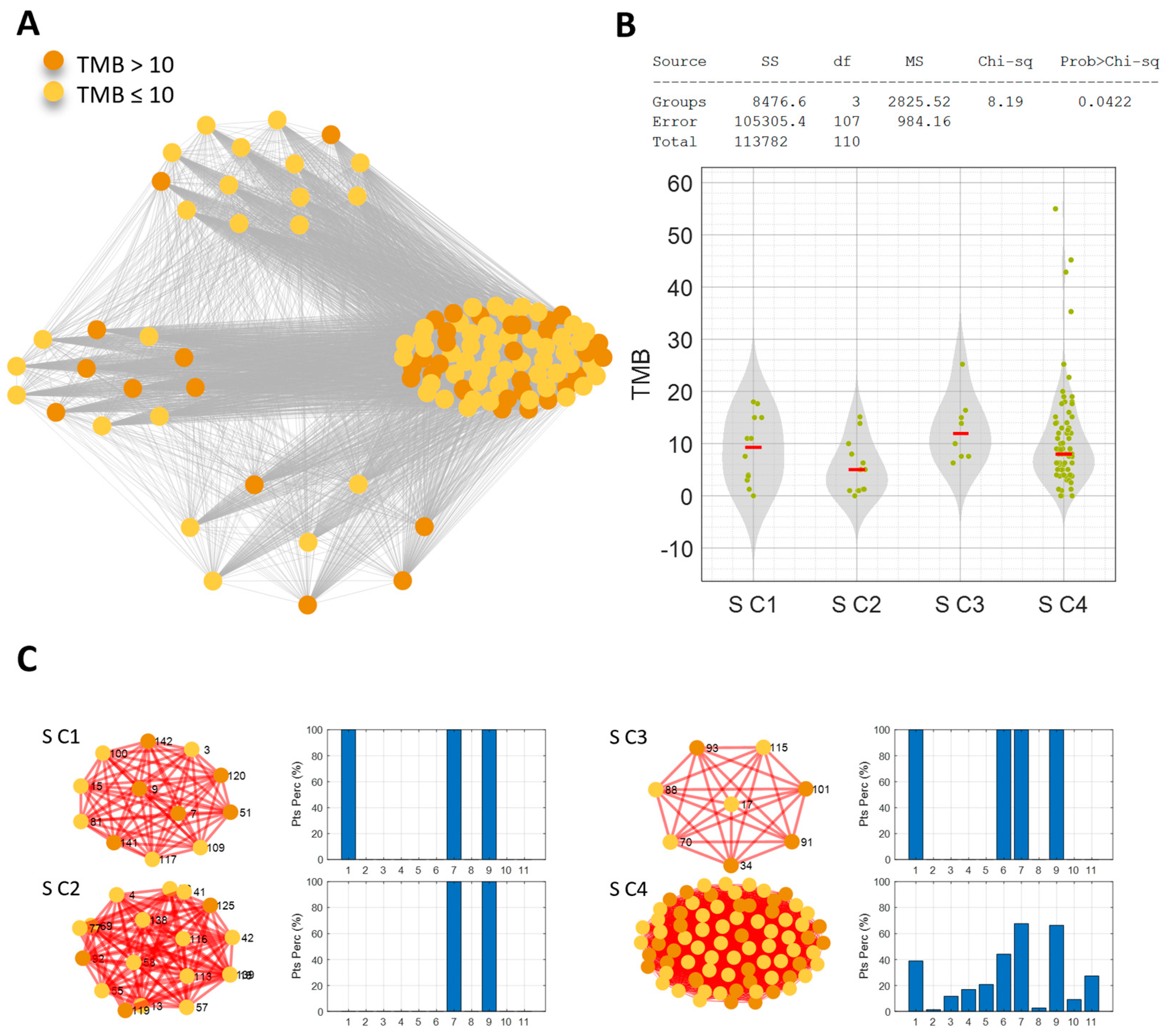
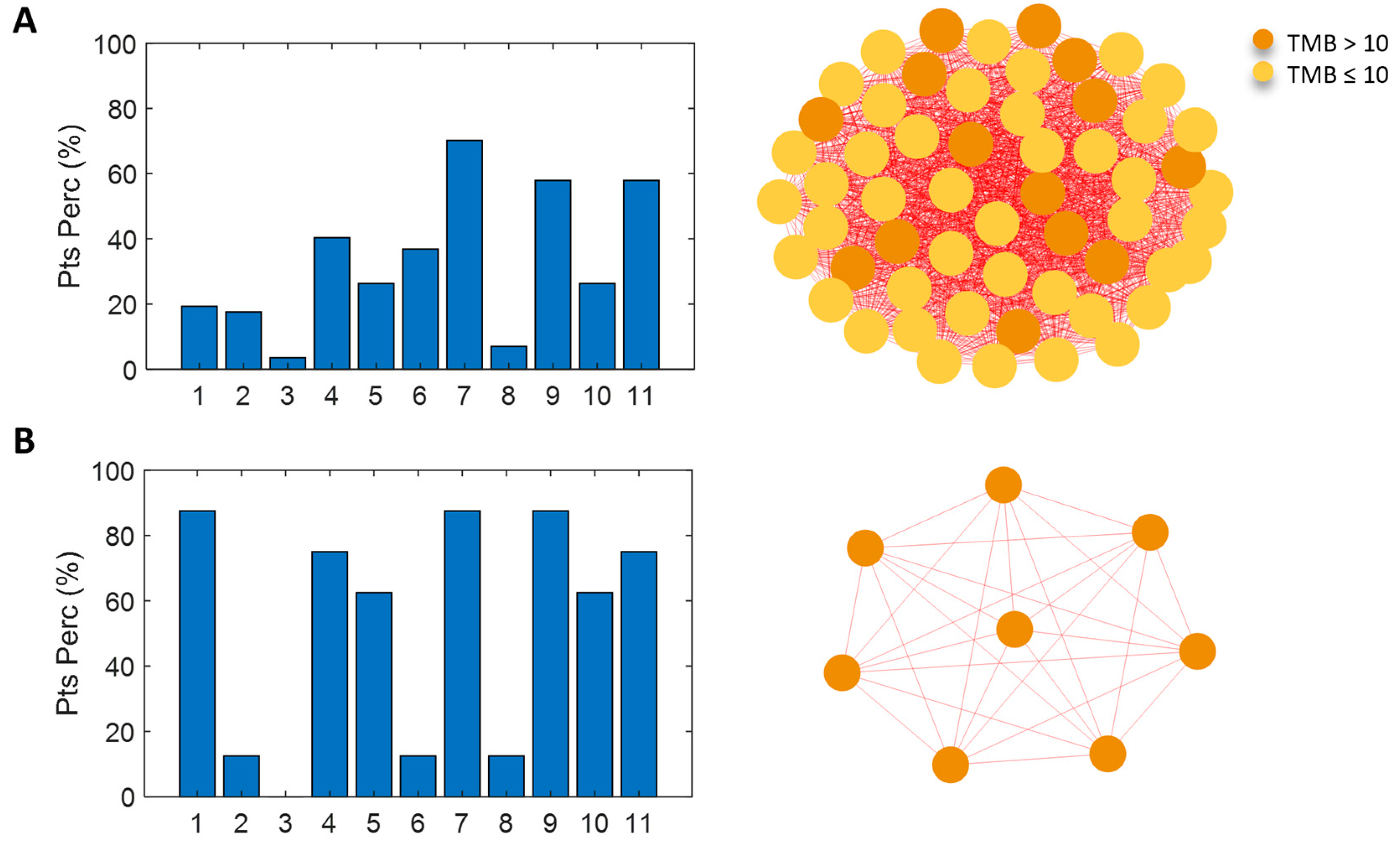
3.3. Network Analysis for Patient Similarity Evaluations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| aNSCLC | non-oncogene addicted advanced non-small cell lung cancer |
| PD-L1 | programmed death-ligand 1 |
| TMB | tumor mutation burden |
| IO | immunotherapy |
| H-TMB | high TMB |
| L-TMB | low TMB |
| S-pts | smoking patients |
| NS-pts | never-smoking patients |
| DDR | DNA damage response and repair |
| ECOG PS | Eastern Cooperative Oncology Group performance status score |
| NGS | next generation sequencing |
| PD | progressive disease |
| RECIST | Response Evaluation Criteria in Solid Tumors |
| CR | complete response |
| PR | partial response |
| SD | stable disease |
| ORR | objective response rate |
| bNGS | blood-based NGS |
| BEP | biomarker-evaluable population |
| OS | overall survival |
| PFS | progression-free survival |
| EGFR | epidermal growth factor receptor |
| ALK | anaplastic lymphoma kinase |
| COSMIC | Catalogue of Somatic Mutations in Cancer |
| Mb | megabase |
| bTMB | blood TMB (bTMB) |
| C | communities |
References
- Reck, M.; Rodríguez–Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Updated analysis of KEYNOTE-024: Pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J. Clin. Oncol. 2019, 37, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Samstein, R.M.; Lee, C.-H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Ricciuti, B.; Wang, X.; Alessi, J.V.; Rizvi, H.; Mahadevan, N.R.; Li, Y.Y.; Polio, A.; Lindsay, J.; Umeton, R.; Sinha, R.; et al. Association of high tumor mutation burden in non-small cell lung cancers with increased immune infiltration and improved clinical outcomes of PD-L1 blockade across PD-L1 expression levels. JAMA Oncol. 2022, 16, e221981. [Google Scholar] [CrossRef] [PubMed]
- Gainor, J.F.; Rizvi, H.; Jimenez Aguilar, E.; Skoulidis, F.; Yeap, B.Y.; Naidoo, J.; Khosrowjerdi, S.; Mooradian, M.; Lydon, C.; Illei, P.; et al. Clinical activity of programmed cell death 1 (PD-1) blockade in never, light, and heavy smokers with non-small-cell lung cancer and PD-L1 expression ≥50. Ann. Oncol. 2020, 31, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Zhou, H.; Shen, J.; Li, J.; Zhang, Y.; Hong, S.; Zhang, L. MDM2/4 amplification predicts poor response to immune checkpoint inhibitors: A pan-cancer analysis. ESMO Open 2020, 5, e000614. [Google Scholar] [CrossRef]
- Ugolini, A.; Zizzari, I.G.; Ceccarelli, F.; Botticelli, A.; Colasanti, T.; Strigari, L.; Rughetti, A.; Rahimi, H.; Conti, F.; Valesini, G.; et al. IgM-Rheumatoid factor confers primary resistance to anti-PD-1 immunotherapies in NSCLC patients by reducing CD137+T-cells. EBioMedicine 2020, 62, 103098. [Google Scholar] [CrossRef]
- Mouw, K.W.; Goldberg, M.S.; Konstantinopoulos, P.A.; D’Andrea, A.D. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017, 7, 675–693. [Google Scholar] [CrossRef]
- Strickland, K.C.; Howitt, B.E.; Shukla, S.A.; Rodig, S.; Ritterhouse, L.L.; Liu, J.F.; Garber, J.E.; Chowdhury, D.; Wu, C.J.; D’Andrea, A.D.; et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 2016, 7, 13587–13598. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef]
- Zizzari, I.G.; Di Filippo, A.; Botticelli, A.; Strigari, L.; Pernazza, A.; Rullo, E.; Pignataro, M.G.; Ugolini, A.; Scirocchi, F.; Di Pietro, F.R.; et al. Circulating CD137+ T Cells Correlate with Improved Response to Anti-PD1 Immunotherapy in Patients with Cancer. Clin. Cancer Res. 2022, 28, 1027–1037. [Google Scholar] [CrossRef]
- Scirocchi, F.; Strigari, L.; Di Filippo, A.; Napoletano, C.; Pace, A.; Rahimi, H.; Botticelli, A.; Rughetti, A.; Nuti, M.; Zizzari, I.G. Soluble PD-L1 as a Prognostic Factor for Immunotherapy Treatment in Solid Tumors: Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2022, 23, 14496. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.K.; Davis, A.A.; Raparia, K.; Agte, S.; Pan, A.; Mohindra, N.; Villaflor, V.; Giles, F. Association of tumor mutational burden with DNA repair mutations and response to anti–PD-1/PD-L1 therapy in non–small-cell lung cancer. Clin. Lung Cancer 2019, 20, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Ricciuti, B.; Recondo, G.; Spurr, L.F.; Li, Y.Y.; Lamberti, G.; Venkatraman, D.; Umeton, R.; Cherniack, A.D.; Nishino, M.; Sholl, L.M.; et al. Impact of DNA damage response and repair (DDR) gene mutations on efficacy of PD-(L)1 immune checkpoint inhibition in non-small cell lung cancer. Clin. Cancer Res. 2020, 26, 4135–4142. [Google Scholar] [CrossRef] [PubMed]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Fehrenbacher, L.; Spira, A.; Ballinger, M.; Kowanetz, M.; Vansteenkiste, J.; Mazieres, J.; Park, K.; Smith, D.; Artal-Cortes, A.; Lewanski, C.; et al. POPLAR Study Group. Atezolizumab vs. docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): A multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016, 387, 1837–1846. [Google Scholar] [CrossRef]
- Rittmeyer, A.; Barlesi, F.; Waterkamp, D.; Park, K.; Ciardiello, F.; von Pawel, J.; Gadgeel, S.M.; Hida, T.; Kowalski, D.M.; Dols, M.C.; et al. OAK Study Group. Atezolizumab vs. docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet 2017, 389, 255–265. [Google Scholar] [CrossRef]
- Fehrenbacher, L.; von Pawel, J.; Park, K.; Rittmeyer, A.; Gandara, D.R.; Ponce Aix, S.; Han, J.Y.; Gadgeel, S.M.; Hida, T.; Cortinovis, D.L.; et al. Updated efficacy analysis including secondary population results for OAK: A randomized phase III study of atezolizumab vs. docetaxel in patients with previously treated advanced non-small cell lung cancer. J. Thorac. Oncol. 2018, 13, 1156–1170. [Google Scholar] [CrossRef]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Hoover, J.; et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016, 44, D862–D868. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e10. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. RAS oncogenes: The first 30 years. Nat. Rev. Cancer 2003, 3, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- A Nilsson, J.; Cleveland, J.L. Myc pathways provoking cell suicide and cancer. Oncogene 2003, 22, 9007–9021. [Google Scholar] [CrossRef] [PubMed]
- Proud, C.G. mTORC1 regulates the efficiency and cellular capacity for protein synthesis. Biochem. Soc. Trans. 2013, 41, 923–926. [Google Scholar] [CrossRef]
- Radtke, F.; Raj, K. The role of Notch in tumorigenesis: Oncogene or tumour suppressor? Nat. Rev. Cancer 2003, 3, 756–767. [Google Scholar] [CrossRef]
- Reya, T.; Clevers, H. Wnt signalling in stem cells and cancer. Nature 2005, 434, 843–850. [Google Scholar] [CrossRef]
- Sporn, M.B.; Liby, K.T. NRF2 and cancer: The good, the bad and the importance of context. Nat. Rev. Cancer 2012, 12, 564–571. [Google Scholar] [CrossRef]
- Wade, M.; Li, Y.-C.; Wahl, G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 2013, 13, 83–96. [Google Scholar] [CrossRef]
- Frampton, G.M.; Fichtenholtz, A.; Otto, G.A.; Wang, K.; Downing, S.R.; He, J.; Schnall-Levin, M.; White, J.; Sanford, E.M.; An, P.; et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef]
- Büttner, R.; Longshore, J.W.; López-Ríos, F.; Merkelbach-Bruse, S.; Normanno, N.; Rouleau, E.; Penault-Llorca, F. Implementing TMB measurement in clinical practice: Considerations on assay requirements. ESMO Open 2019, 4, e000442. [Google Scholar] [CrossRef] [PubMed]
- Petti, M.; Farina, L. Network medicine for patients’ stratification: From single-layer to multi-omics. WIREs Mech. Dis. 2023, 15, e1623. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Mezlini, A.M.; Demir, F.; Fiume, M.; Tu, Z.; Brudno, M.; Haibe-Kains, B.; Goldenberg, A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 2014, 11, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Rahiminejad, S.; Maurya, M.R.; Subramaniam, S. Topological and functional comparison of community detection algorithms in biological networks. BMC Bioinform. 2019, 20, 212. [Google Scholar] [CrossRef]
- Cortellini, A.; Giusti, R.; Filetti, M.; Citarella, F.; Adamo, V.; Santini, D.; Buti, S.; Nigro, O.; Cantini, L.; Di Maio, M.; et al. High familial burden of cancer correlates with improved outcome from immunotherapy in patients with NSCLC independent of somatic DNA damage response gene status. J. Hematol. Oncol. 2022, 15, 9. [Google Scholar] [CrossRef]
- Jiang, M.; Jia, K.; Wang, L.; Li, W.; Chen, B.; Liu, Y.; Wang, H.; Zhao, S.; He, Y.; Zhou, C. Alterations of DNA damage repair in cancer: From mechanisms to applications. Ann. Transl. Med. 2020, 8, 1685. [Google Scholar] [CrossRef]
- Hill, W.; Lim, E.L.; Weeden, C.E.; Lee, C.; Augustine, M.; Chen, K.; Kuan, F.-C.; Marongiu, F.; Evans, E.J.; Moore, D.A.; et al. Lung adenocarcinoma promotion by air pollutants. Nature 2023, 616, 159–167. [Google Scholar] [CrossRef]
- Musacchio, L.; Cicala, C.M.; Camarda, F.; Ghizzoni, V.; Giudice, E.; Carbone, M.V.; Ricci, C.; Perri, M.T.; Tronconi, F.; Gentile, M.; et al. Combining PARP inhibition and immune checkpoint blockade in ovarian cancer patients: A new perspective on the horizon? ESMO Open 2022, 7, 100536. [Google Scholar] [CrossRef]
- Wanderley, C.W.S.; Correa, T.S.; Scaranti, M.; Cunha, F.Q.; Barroso-Sousa, R. Targeting PARP1 to enhance anticancer checkpoint immunotherapy response: Rationale and clinical implications. Front. Immunol. 2022, 13, 816642. [Google Scholar] [CrossRef]
- Ngoi, N.Y.L.; Peng, G.; Yap, T.A. A tale of two checkpoints: ATR inhibition and PD-(L)1 blockade. Annu. Rev. Med. 2022, 73, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef] [PubMed]
| Clinical Characteristic | S-pts N = 111 | NS-pts N = 31 | p-Value (S vs. NS) |
|---|---|---|---|
| Age, median (range), years | 65 (42–82) | 62 (38–84) | 0.069 |
| Sex, n (%) | |||
| Male | 73 (65.8) | 15 (48.4) | 0.095 |
| Female | 38 (34.2) | 16 (51.6) | |
| Histologic profile, n (%) | |||
| Squamous | 16 (14.4) | 3 (9.7) | 0.765 |
| Nonsquamous | 95 (85.6) | 28 (90.3) | |
| ECOG performance status, n (%) | |||
| 0–1 | 103 (92.8) | 29 (93.5) | 1 |
| ≥2 | 7 (6.2) | 2 (6.5) | |
| Not assessed | 1 (1) | 0 | |
| TMB, median (range), Mut/Mb | 8 (0–55) | 4 (0–39.09) | 0.129 |
| Pathway | S-pts; N = 111 n (%) | NS-pts; N = 31 n (%) |
|---|---|---|
| Cell Cycle | 50 (45) | 14 (45.2) |
| Hippo | 1 (1) | 1 (3.2) |
| Myc | 9 (8.1) | 3 (9.7) |
| Notch | 13 (11.7) | 8 (25.8) |
| Oxidative Stress/Nrf2 | 16 (14.4) | 7 (22.6) |
| PI3K | 42 (37.8) | 7 (22.6) |
| RTK/RAS/MAP | 86 (77.5) | 26 (83.9) |
| TGF-β | 2 (1.8) | 1 (3.2) |
| p53 | 85 (76.6) | 23 (74.2) |
| β-catenin/Wnt | 7 (6.3) | 5 (16.1) |
| DDR * | 21 (18.9) | 14 (45.2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filetti, M.; Occhipinti, M.; Cirillo, A.; Scirocchi, F.; Ugolini, A.; Giusti, R.; Lombardi, P.; Daniele, G.; Botticelli, A.; Lo Russo, G.; et al. Exploring Genomic Biomarkers for Pembrolizumab Response: A Real-World Approach and Patient Similarity Network Analysis Reveal DNA Response and Repair Gene Mutations as a Signature. Cancers 2024, 16, 3955. https://doi.org/10.3390/cancers16233955
Filetti M, Occhipinti M, Cirillo A, Scirocchi F, Ugolini A, Giusti R, Lombardi P, Daniele G, Botticelli A, Lo Russo G, et al. Exploring Genomic Biomarkers for Pembrolizumab Response: A Real-World Approach and Patient Similarity Network Analysis Reveal DNA Response and Repair Gene Mutations as a Signature. Cancers. 2024; 16(23):3955. https://doi.org/10.3390/cancers16233955
Chicago/Turabian StyleFiletti, Marco, Mario Occhipinti, Alessio Cirillo, Fabio Scirocchi, Alessio Ugolini, Raffaele Giusti, Pasquale Lombardi, Gennaro Daniele, Andrea Botticelli, Giuseppe Lo Russo, and et al. 2024. "Exploring Genomic Biomarkers for Pembrolizumab Response: A Real-World Approach and Patient Similarity Network Analysis Reveal DNA Response and Repair Gene Mutations as a Signature" Cancers 16, no. 23: 3955. https://doi.org/10.3390/cancers16233955
APA StyleFiletti, M., Occhipinti, M., Cirillo, A., Scirocchi, F., Ugolini, A., Giusti, R., Lombardi, P., Daniele, G., Botticelli, A., Lo Russo, G., De Braud, F., Marchetti, P., Nuti, M., Ferretti, E., Farina, L., Rughetti, A., & Petti, M. (2024). Exploring Genomic Biomarkers for Pembrolizumab Response: A Real-World Approach and Patient Similarity Network Analysis Reveal DNA Response and Repair Gene Mutations as a Signature. Cancers, 16(23), 3955. https://doi.org/10.3390/cancers16233955








