Potential Use of Exosomal Non-Coding MicroRNAs in Leukemia Therapy: A Systematic Review
Simple Summary
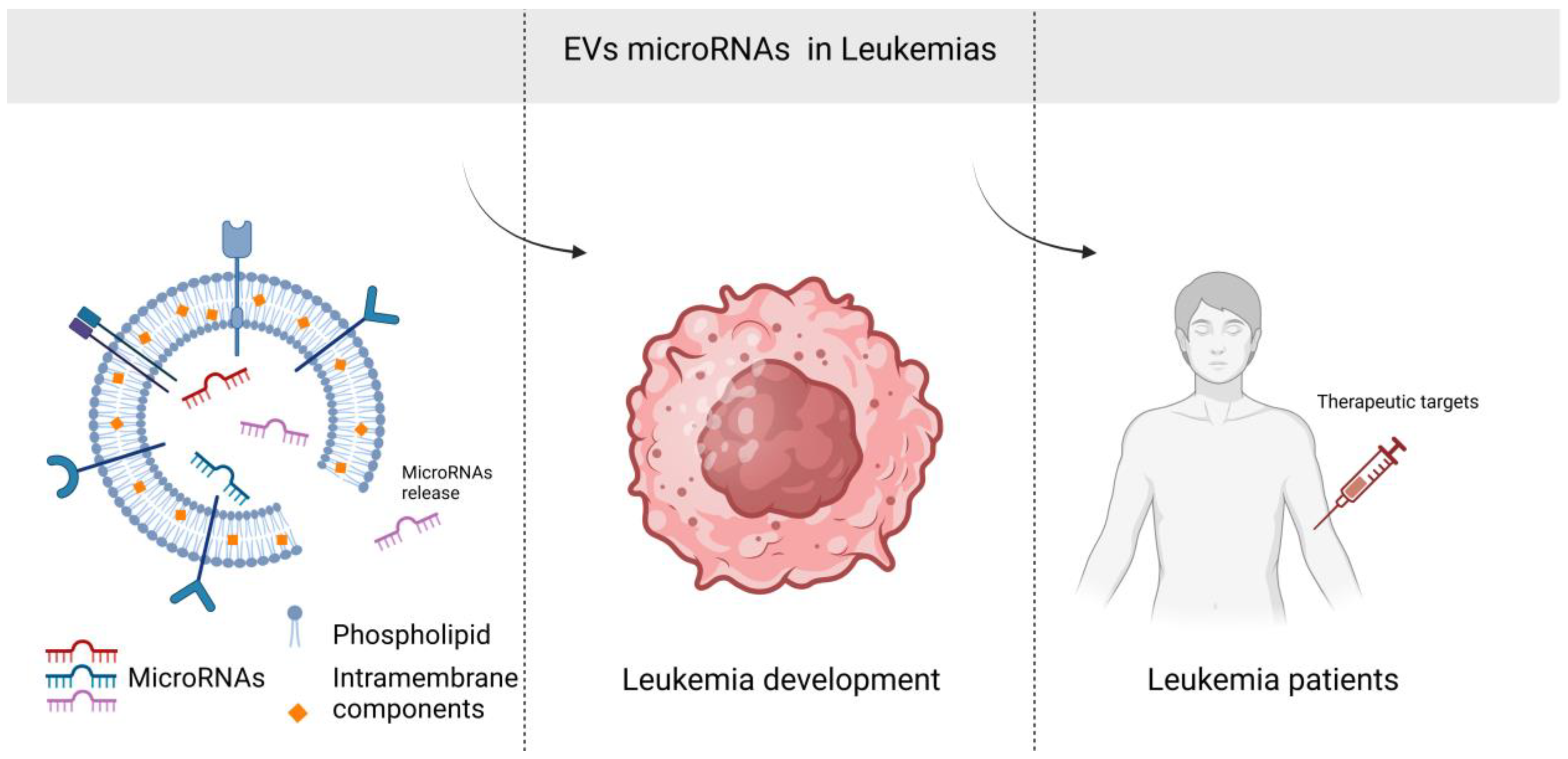
Abstract
1. Introduction
2. Methods
2.1. Protocol
2.2. Data Sources and Search Strategy
2.3. Eligibility Criteria
2.4. Data Collection and Selection Process
2.5. Data Items and Effect Measures
2.6. Study Risk of Bias Assessment
3. Results
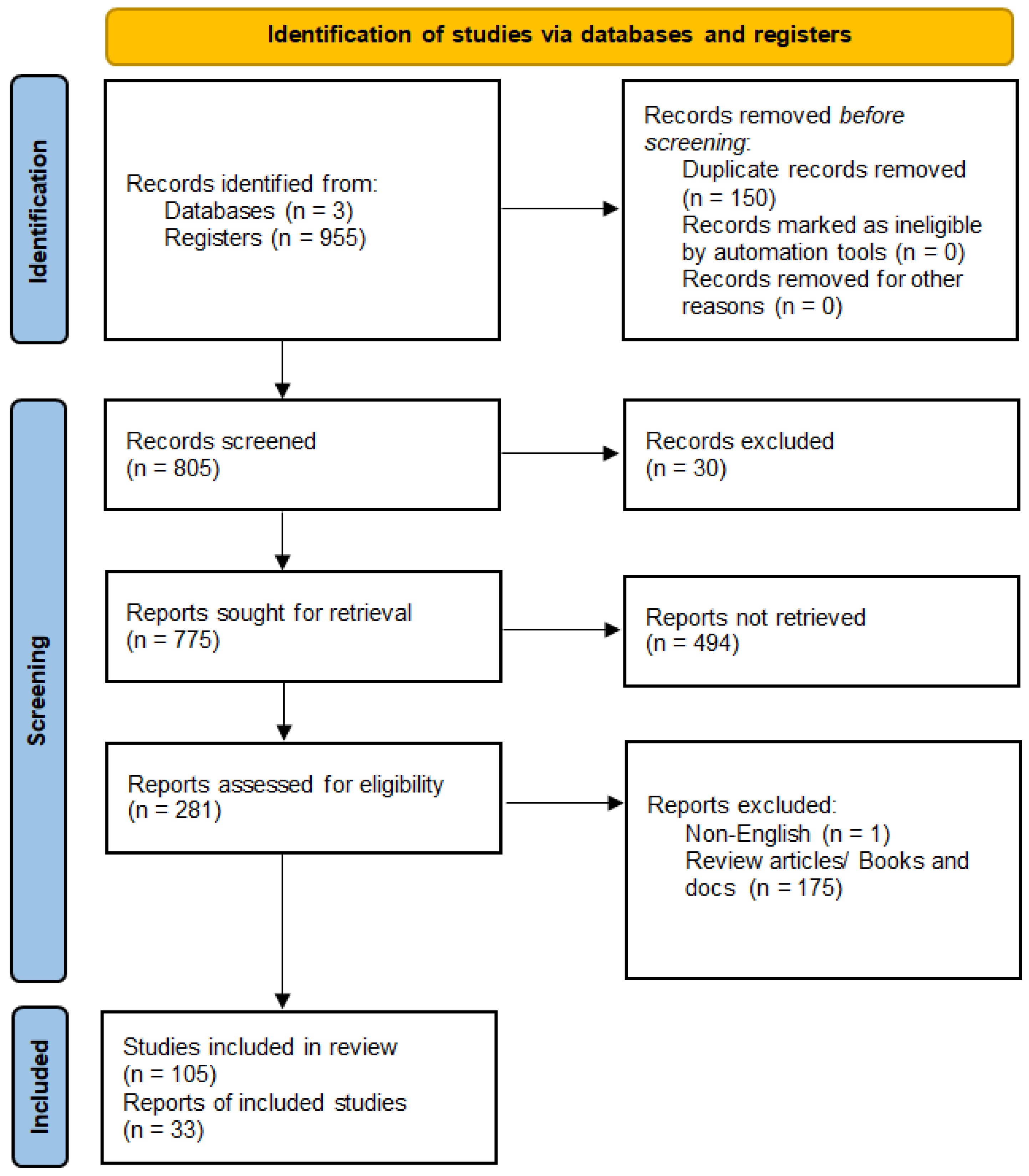
3.1. Study Selection
3.2. Study Characteristics
3.3. Risk of Bias in Studies
| Author, Date, Reference | Material | Type of RNA Evaluated | miRNA Target | Impact of the Evaluated RNA on the Patient/Cells |
|---|---|---|---|---|
| CLL | ||||
| Stamatopoulos, 2015 [28] | Plasma and B lymphocytes of patients | miR-150 | – | miR-150 level is associated with tumor aggressiveness |
| Yeh, 2015 [29] | Patients’ B lymphocytes | miR-150 | – | miR-150 level is associated with tumor aggressiveness [23] |
| miR-155 | – | Influence on disease aggression and weakening of response to chemotherapy | ||
| Farahani, 2015 [30] | B lymphocytes from patients, MEC1 and HS5 cell lines | miR-202-3p | Sufu | Suppressive effect on tumor development |
| Smallwood 2016 [31] | Peripheral blood of patients, CD19+ mononuclear cells | miR-363 | CD69 | Critical role in the regulation of T-cell motility and immune synapse signaling function |
| Paggetti 2015 [32] | Bone marrow stem cells from patients | miR-146a, miR-155 | – | Increased stromal cell proliferation, migration, and inflammatory cytokine secretion |
| CML | ||||
| Taverna, 2014 [33] | LAMA84 cell lines | miR-126 | CXCL12 i VCAM1 | Reduction in LAMA84 cell migration and adhesion |
| Chen, 2022 [34] | Bone marrow from patients, K562 cell lines, NOD-SCID mice | miR-145a-5p | USP6 | Increases imatinib-induced K562 apoptosis |
| Taverna, 2015 [35] | K562 and LAMA84 cell lines, SCID mice | miR-21 | PTEN | Increases VEGF secretion, formation of larger cell colonies |
| miR-196b | – | Decreased Bcr-Abl protein level | ||
| Gao, 2019 [36] | Bone marrow and peripheral blood of patients, K562 and LAMA84 cell lines, SCL-tTa X TER-BCR/ABL mice | miR-320 | BCR/ABL | Inhibition of K562 proliferation |
| Chai, 2023 [37] | Bone marrow and peripheral blood of patients, cell lines HEK293T, HL60, K562, BALL-1, and Jurkat cells | miR-130a/b | Cx43 | Increases the immunosuppressive properties of cells, supports immune escape in tumors |
| Ohyashiki 2016 [38] | Peripheral blood from patients | miR-215 | – | Imatinib therapy causes miR-215 expression levels to decrease, maintaining undetectable minimal residual disease (UMRD) |
| Taverna 2016 [39] | HUVEC cell lines | miR-21 | PTEN | Decreased expression of anti-apoptotic Bcl-2 and decreased expression of WT1, growth of leukemic cells by decreased expression of PTEN |
| miR-15a i miR-16 | Bcl-2, WT1 | |||
| ALL | ||||
| Huang, 2022 [40] | Mouse cell lines L1210 and p388 and DBA/2 mice | shRNA—RNA synthetic | PD-L1 | Increasing immunological properties, increasing the lifespan of mice |
| Yan, 2021 [41] | Patients’ peripheral blood, BALL-1 cell line | miR-181b-5p | – | Increased proliferation and migration and decreased apoptosis of ALL cells |
| Chai, 2023 [37] | Bone marrow and peripheral blood of patients, cell lines HEK293T, HL60, K562, BALL-1 and Jurkat cells | miR-130a/b | Cx43 | Increases the immunosuppressive properties of cells, supports immune escape in tumors |
| Saffari 2024 [42] | Peripheral blood of patients, CD10 − /CD34 − cell lines, RN95, Nalm6 cell lines | miR-326 | – | Cancer cell viability was dramatically suppressed in an exosomal miRNA dose-dependent manner |
| Rzepiel 2023 [43] | Platelet-free plasma (PFP) | miR-128-3p | – | Reduced expression positively correlates with minimal residual disease (MRD) in bone marrow flow cytometry at day 15 of treatment (potential therapeutic marker) |
| Habiel 2021 [44] | SCID/Bg mice | miR-101-3p, miR-193b-3p, miR-21-5p, miR-34a-5p | MMR, BRCA1 | Marked reduction in the expression of components of the mismatch repair (MMR) pathway and BRCA1 (divergence of leukemic cells located in their microenvironment and the generation of therapy resistance) |
| Haque 2020 [45] | Cell lines M1, SUP-B15, NALM-6, REH, NALM-16 | miR-181 | – | Role in chemoresistance in relapsed leukemia |
| Colangelo 2022 [46] | CUTLL1 cell lines | miR-223-3p | NOTCH1 | Increased population of resistant T-lymphocytic leukemia cells in response to conventional therapies [47] |
| AML | ||||
| Cheng, 2021 [48] | Blood from patients, cell lines Kasumi-1, HL-60, THP-1, HMSC, and bone marrow cells | miR-23b-5p | TRIM14 | Increased apoptosis of THP-1 cells |
| Jiang, 2022 [49] | Patient plasma, cell lines HL60, THP1, U937, KG-1, MOLM13, MV4-11, GM12878, B-NDG mice | miR-7-5p | OSBPL11 | Limiting proliferation and stimulating cell apoptosis |
| Otmani, 2023 [50] | Peripheral blood from patients | miR-24-3p | DENN/MADD | Increased T-cell apoptosis |
| Zhao, 2019 [51] | Cord blood from healthy women, cell lines HL-60, Molm-14, OCI-AML3, ML-2 | miR-4532 | LDOC1 | Inhibition of hematopoiesis |
| Xu, 2020 [52] | KG-1a cell line | hsa-miR-124-5p | SMC4 | Decreased proliferation and inhibition of the KG-1a cell cycle and increased apoptosis of KG-1a |
| Taniguchi, 2022 [53] | HL-60 and HL-60/ADR cell lines | miR-484 | – | Increased cell proliferation |
| Hu, 2020 [24] | Bone marrow from patients, cell lines THP-1, KG1a, KASUMI-1 | miR-34a | DHAC2 | Reduction in cell proliferation, increase in apoptosis of leukemia cells, prolongs survival time [54] |
| Jiang 2018 [55] | Peripheral blood from patients | miR-125b | – | Higher risks of relapse and overall death |
| Yoshida 2019 [56] | Bone marrow stem cells, HTS-5 cells | miR-7977 | signaling pathway Hippo-YAP | Inhibits the Hippo-YAP signaling pathway in bone marrow stem cells, spreading functionally impaired MSCs |
| Li 2022 [57] | Peripheral blood of patients | miR92a | PTEN, signaling pathway Wnt/β-catenin | Reduction in PTEN expression promotes cytarabine resistance in cells by activating the Wnt/β-catenin pathway |
| Yuan 2023 [58] | Bone marrow from patients | miRNA-222-3p | IRF2/INPP4B | Increased Th1/Th2 ratio and promotes apoptosis |
| Barrera-Ramirez 2017 [59] | Bone marrow from patients | miR-26a-5p i miR-101-3p | GSK3β, EZH2 | Phosphorylation of GSK-3β in AML may activate the Akt pathway and is associated with poorer overall survival; genomic loss of EZH2 may lead to epigenetic changes and overexpression of HOX genes |
| miR-23b-5p, miR-339-3p i miR-425-5p | APOBEC3A | - | ||
| Li 2022 [60] | U937 cell line | miR-3064-3p, miR-339-5p | p62 | Increased expression of p62 may promote the maturation of AML cells into granulocytes, depending on NF-κB activation, predicting poor prognosis in AML |
| ATL (adult T-cell leukemia/lymphoma) | ||||
| El-Saghir 2016 [61] | Peripheral blood of deceased patients (frozen mononuclear cells), leukemia cell lines (Molt-4, C81, and HuT-102) | miR-21, miR-155 | signaling pathway NF-κB | Changes in cellular morphology, increased proliferation, and induction of gene expression of migration and angiogenic markers |
| Studies | Criteria from the Mixed Methods Appraisal Tool | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.1 | 1.2 | 1.3 | 1.4 | 1.5 | 2.1 | 2.2 | 2.3 | 2.4 | 2.5 | 3.1 | 3.2 | 3.3 | 3.4 | 3.5 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 5.1 | 5.2 | 5.3 | 5.4 | 5.5 | |
| Type of Study | Qualitative | Quantitative Randomized Controlled Trials | Quantitative Non Randomized | Quantitative Descriptive | Mixed Methods | ||||||||||||||||||||
| Taverna, 2014 [33] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| Yan, 2021 [41] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| Jiang, 2022 [49] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| Taverna, 2015 [35] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| Zhao, 2019 [51] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| Farahani, 2015 [30] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| Gao, 2019 [36] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| Chai, 2023 [37] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| Huang, 2022 [40] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Stamatopoulos, 2015 [28] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Yeh, 2015 [29] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Cheng, 2021 [48] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| El-Saghir, 2016 [61] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Otmani, 2023 [50] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Jiang, 2018 [55] | 1 | 1 | 0 | 1 | 1 | ||||||||||||||||||||
| Ohyashiki, 2016 [38] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Chen, 2022 [34] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Xu, 2020 [52] | 0 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Taniguchi, 2022 [53] | 0 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Hu, 2020 [24] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Yuan, 2023 [58] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Taverna, 2016 [39] | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||
| Colangelo, 2022 [46] | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||
| Saffari, 2024 [42] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | |||||||||||||||
| Barrera-Ramirez, 2017 [59] | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | |||||||||||||||
| Smallwood, 2016 [31] | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | |||||||||||||||
| Habiel, 2016 [44] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | |||||||||||||||
| Yoshida 2019 [56] | 1 | 0 | 1 | 0 | 1 | ||||||||||||||||||||
| Rzepiel, 2023 [43] | 1 | 0 | 1 | 0 | 1 | ||||||||||||||||||||
| Li, 2022 [20] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Li, 2022 [57] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Haque, 2020 [45] | 1 | 1 | 1 | 0 | 1 | ||||||||||||||||||||
| Paggetti, 2015 [32] | 1 | 0 | 1 | 0 | 1 | ||||||||||||||||||||
4. Discussion
4.1. Therapeutical Potential of Exosomal miRNA in Leukemia
4.2. Exosomal miRNA in Leukemia: Pathogenesis and Therapy
4.2.1. Acute Myeloid Leukemia
4.2.2. Chronic Lymphocytic Leukemia
4.2.3. Acute Lymphoblastic Leukemia
4.2.4. Chronic Myeloid Leukemia
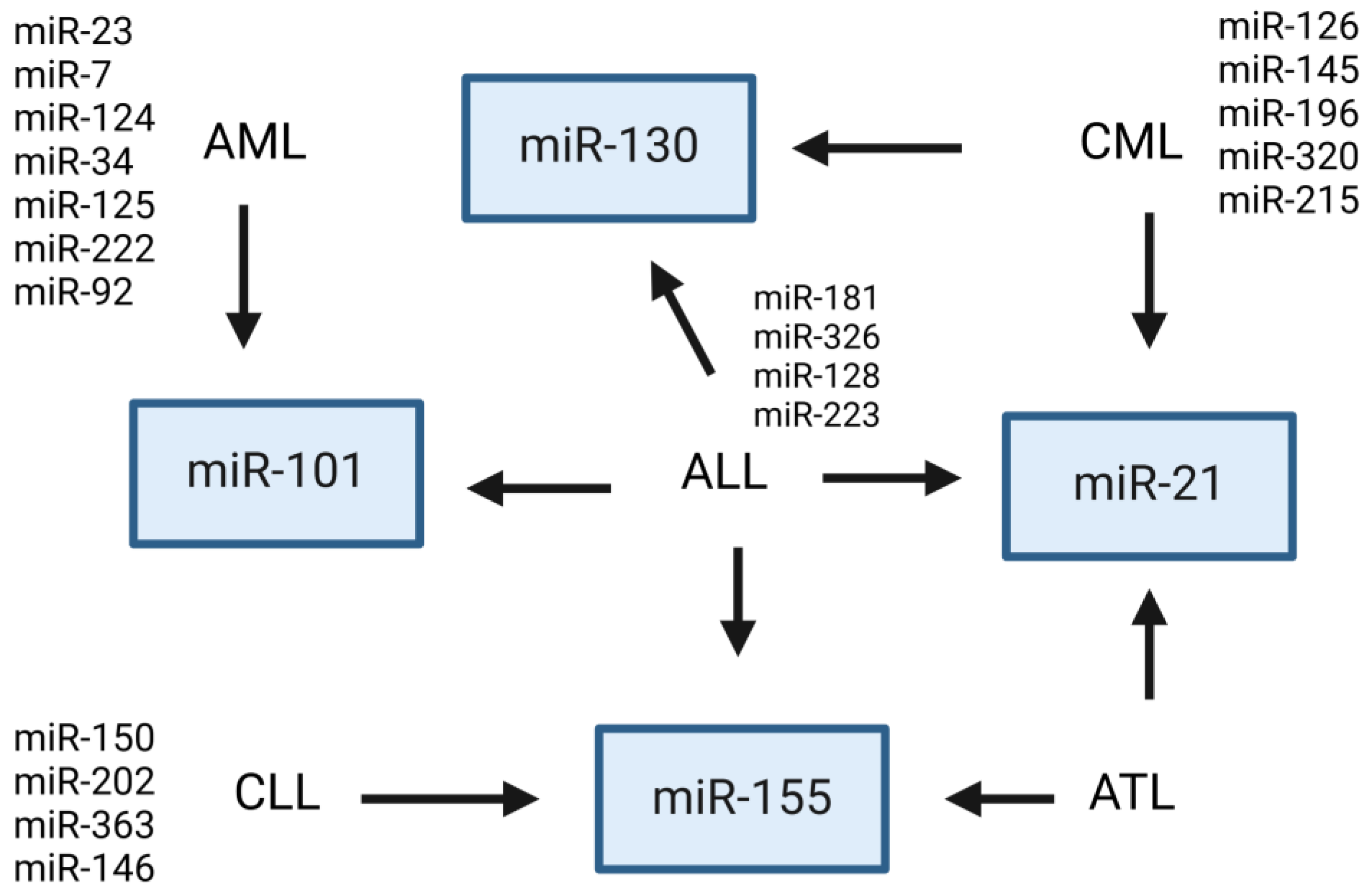
4.3. Most Frequent Exosomal miRNAs Showing Dysregulated Expression in Leukemia
4.4. Circular RNA in Leukemia: Pathogenesis and Therapy
4.5. Potential Therapeutic Applications of Exosomal-Derived miRNAs
4.6. Exosome Engineering
4.7. Clinical Trials
5. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today (Version 1.1); International Agency for Research on Cancer: Lyon, France, 2024; Available online: https://gco.iarc.who.int/today (accessed on 16 August 2024).
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Bartoszewska, E.; Tota, M.; Kisielewska, M.; Skowron, I.; Sebastianka, K.; Stefaniak, O.; Molik, K.; Rubin, J.; Kraska, K.; Choromańska, A. Overcoming Antigen Escape and T-Cell Exhaustion in CAR-T Therapy for Leukemia. Cells 2024, 13, 1596. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.; Singh, A. Leukemias in Children. Indian J. Pediatr. 2015, 82, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Salama, M.M.; Aborehab, N.M.; El Mahdy, N.M.; Zayed, A.; Ezzat, S.M. Nanotechnology in leukemia: Diagnosis, efficient-targeted drug delivery, and clinical trials. Eur. J. Med. Res. 2023, 28, 566. [Google Scholar] [CrossRef]
- Campo, E.; Harris, N.L.; Jaffe, E.S. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; Swerdlow, S.H., Ed.; International Agency for Research on Cancer: Lyon, France, 2008. [Google Scholar]
- Jagadev, P.; Virani, H.G. Detection of leukemia and its types using image processing and machine learning. In Proceedings of the 2017 International Conference on Trends in Electronics and Informatics (ICEI), Tirunelveli, India, 11–12 May 2017; pp. 522–526. [Google Scholar]
- Inaba, H.; Greaves, M.; Mullighan, C.G. Acute lymphoblastic leukaemia. Lancet 2013, 381, 1943–1955. [Google Scholar] [CrossRef]
- Srinivasan Rajsri, K.; Roy, N.; Chakraborty, S. Acute Myeloid Leukemia Stem Cells in Minimal/Measurable Residual Disease Detection. Cancers 2023, 15, 2866. [Google Scholar] [CrossRef]
- Antar, A.I.; Otrock, Z.K.; Jabbour, E.; Mohty, M.; Bazarbachi, A. FLT3 inhibitors in acute myeloid leukemia: Ten frequently asked questions. Leukemia 2020, 34, 682–696. [Google Scholar] [CrossRef]
- Giebel, S.; Czyz, A.; Ottmann, O.; Baron, F.; Brissot, E.; Ciceri, F.; Cornelissen, J.J.; Esteve, J.; Gorin, N.C.; Savani, B.; et al. Use of tyrosine kinase inhibitors to prevent relapse after allogeneic hematopoietic stem cell transplantation for patients with Philadelphia chromosome-positive acute lymphoblastic leukemia: A position statement of the Acute Leukemia Working Party of the European Society for Blood and Marrow Transplantation. Cancer 2016, 122, 2941–2951. [Google Scholar] [CrossRef]
- Ross, D.D. Novel mechanisms of drug resistance in leukemia. Leukemia 2000, 14, 467–473. [Google Scholar] [CrossRef]
- Litwińska, Z.; Łuczkowska, K.; Machaliński, B. Extracellular vesicles in hematological malignancies. Leuk. Lymphoma 2019, 60, 29–36. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Kakarla, R.; Hur, J.; Kim, Y.J.; Kim, J.; Chwae, Y.J. Apoptotic cell-derived exosomes: Messages from dying cells. Exp. Mol. Med. 2020, 52, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Garcia, V.; Rodriguez, M.; Compte, M.; Cisneros, E.; Veguillas, P.; Garcia, J.M.; Dominguez, G.; Campos-Martin, Y.; Cuevas, J.; et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer 2012, 51, 409–418. [Google Scholar] [CrossRef]
- Jahromi, F.N.A.; Dowran, R.; Jafari, R. Recent advances in the roles of exosomal microRNAs (exomiRs) in hematologic neoplasms: Pathogenesis, diagnosis, and treatment. Cell Commun. Signal. CCS 2023, 21, 88. [Google Scholar] [CrossRef]
- Li, C.; Ni, Y.Q.; Xu, H.; Xiang, Q.Y.; Zhao, Y.; Zhan, J.K.; He, J.Y.; Li, S.; Liu, Y.S. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target. Ther. 2021, 6, 383. [Google Scholar] [CrossRef] [PubMed]
- Aheget, H.; Mazini, L.; Martin, F.; Belqat, B.; Marchal, J.A.; Benabdellah, K. Exosomes: Their Role in Pathogenesis, Diagnosis and Treatment of Diseases. Cancers 2020, 13, 84. [Google Scholar] [CrossRef]
- Li, C.; Zhou, T.; Chen, J.; Li, R.; Chen, H.; Luo, S.; Chen, D.; Cai, C.; Li, W. The role of Exosomal miRNAs in cancer. J. Transl. Med. 2022, 20, 6. [Google Scholar] [CrossRef]
- Turk, A.; Calin, G.A.; Kunej, T. MicroRNAs in Leukemias: A Clinically Annotated Compendium. Int. J. Mol. Sci. 2022, 23, 3469. [Google Scholar] [CrossRef]
- Lv, M.; Zhu, S.; Peng, H.; Cheng, Z.; Zhang, G.; Wang, Z. B-cell acute lymphoblastic leukemia-related microRNAs: Uncovering their diverse and special roles. Am. J. Cancer Res. 2021, 11, 1104–1120. [Google Scholar]
- Anelli, L.; Zagaria, A.; Specchia, G.; Musto, P.; Albano, F. Dysregulation of miRNA in Leukemia: Exploiting miRNA Expression Profiles as Biomarkers. Int. J. Mol. Sci. 2021, 22, 7156. [Google Scholar] [CrossRef]
- Hu, Y.; Ma, X.; Wu, Z.; Nong, Q.; Liu, F.; Wang, Y.; Dong, M. MicroRNA-34a-mediated death of acute myeloid leukemia stem cells through apoptosis induction and exosome shedding inhibition via histone deacetylase 2 targeting. IUBMB Life 2020, 72, 1481–1490. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xie, Y.; Liu, D.; Liu, P.; Li, F.; Zhang, Z.; Zhang, M.; Wang, X.; Zhang, Y.; Sun, X.; et al. Downregulation of miR-142a Contributes to the Enhanced Anti-Apoptotic Ability of Murine Chronic Myelogenous Leukemia Cells. Front. Oncol. 2021, 11, 718731. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. Available online: https://www.prisma-statement.org/prisma-2020-statement (accessed on 30 September 2024). [CrossRef]
- Hong, Q.N.; Pluye, P.; Fàbregues, S.; Bartlett, G.; Boardman, F.; Cargo, M.; Dagenais, P.; Gagnon, M.P.; Griffiths, F.; Nicolau, B.; et al. Mixed Methods Appraisal Tool (MMAT), Version 2018; Registration of Copyright (#1148552); Canadian Intellectual Property Office, Industry Canada: Gatineau, QC, Canada, 2018. [Google Scholar]
- Stamatopoulos, B.; Van Damme, M.; Crompot, E.; Dessars, B.; Housni, H.E.; Mineur, P.; Meuleman, N.; Bron, D.; Lagneaux, L. Opposite Prognostic Significance of Cellular and Serum Circulating MicroRNA-150 in Patients with Chronic Lymphocytic Leukemia. Mol. Med. 2015, 21, 123–133. [Google Scholar] [CrossRef]
- Yeh, Y.Y.; Ozer, H.G.; Lehman, A.M.; Maddocks, K.; Yu, L.; Johnson, A.J.; Byrd, J.C. Characterization of CLL exosomes reveals a distinct microRNA signature and enhanced secretion by activation of BCR signaling. Blood 2015, 125, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Farahani, M.; Rubbi, C.; Liu, L.; Slupsky, J.R.; Kalakonda, N. CLL Exosomes Modulate the Transcriptome and Behaviour of Recipient Stromal Cells and Are Selectively Enriched in miR-202-3p. PLoS ONE 2015, 10, e0141429. [Google Scholar] [CrossRef]
- Smallwood, D.T.; Apollonio, B.; Willimott, S.; Lezina, L.; Alharthi, A.; Ambrose, A.R.; De Rossi, G.; Ramsay, A.G.; Wagner, S.D. Extracellular vesicles released by CD40/IL-4-stimulated CLL cells confer altered functional properties to CD4+ T cells. Blood 2016, 128, 542–552. [Google Scholar] [CrossRef]
- Paggetti, J.; Haderk, F.; Seiffert, M.; Janji, B.; Distler, U.; Ammerlaan, W.; Kim, Y.J.; Adam, J.; Lichter, P.; Solary, E.; et al. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood 2015, 126, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Taverna, S.; Amodeo, V.; Saieva, L.; Russo, A.; Giallombardo, M.; De Leo, G.; Alessandro, R. Exosomal shuttling of miR-126 in endothelial cells modulates adhesive and migratory abilities of chronic myelogenous leukemia cells. Mol. Cancer 2014, 13, 169. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Y.; Zhang, M.; Cheng, H.; Mai, H.; Yi, M.; Xu, H.; Yuan, X.; Liu, S.; Wen, F. HucMSC exosomes promoted imatinib-induced apoptosis in K562-R cells via a miR-145a-5p/USP6/GLS1 axis. Cell Death Dis. 2022, 13, 92. [Google Scholar] [CrossRef]
- Taverna, S.; Giallombardo, M.; Pucci, M.; Flugy, A.; Manno, M.; Raccosta, S.; Rolfo, C.; De Leo, G.; Alessandro, R. Curcumin inhibits in vitro and in vivo chronic myelogenous leukemia cells growth: A possible role for exosomal disposal of miR-21. Oncotarget 2015, 6, 21918–21933. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wan, Z.; Wei, M.; Dong, Y.; Zhao, Y.; Chen, X.; Li, Z.; Qin, W.; Yang, G.; Liu, L. Chronic myelogenous leukemia cells remodel the bone marrow niche via exosome-mediated transfer of miR-320. Theranostics 2019, 9, 5642–5656. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.; Sui, K.; Tang, J.; Yu, H.; Yang, C.; Zhang, H.; Li, S.C.; Zhong, J.F.; Wang, Z.; Zhang, X. BCR-ABL1-driven exosome-miR130b-3p-mediated gap-junction Cx43 MSC intercellular communications imply therapies of leukemic subclonal evolution. Theranostics 2023, 13, 3943–3963. [Google Scholar] [CrossRef]
- Ohyashiki, K.; Umezu, T.; Katagiri, S.; Kobayashi, C.; Azuma, K.; Tauchi, T.; Okabe, S.; Fukuoka, Y.; Ohyashiki, J.H. Downregulation of Plasma miR-215 in Chronic Myeloid Leukemia Patients with Successful Discontinuation of Imatinib. Int. J. Mol. Sci. 2016, 17, 570. [Google Scholar] [CrossRef] [PubMed]
- Taverna, S.; Fontana, S.; Monteleone, F.; Pucci, M.; Saieva, L.; De Caro, V.; Cardinale, V.G.; Giallombardo, M.; Vicario, E.; Rolfo, C.; et al. Curcumin modulates chronic myelogenous leukemia exosomes composition and affects angiogenic phenotype via exosomal miR-21. Oncotarget 2016, 7, 30420–30439. [Google Scholar] [CrossRef]
- Huang, F.; Li, Z.; Zhang, W.; Li, J.; Hao, S. Enhancing the anti-leukemia immunity of acute lymphocytic leukemia-derived exosome-based vaccine by downregulation of PD-L1 expression. Cancer Immunol. Immunother. CII 2022, 71, 2197–2212. [Google Scholar] [CrossRef]
- Yan, W.; Song, L.; Wang, H.; Yang, W.; Hu, L.; Yang, Y. Extracellular vesicles carrying miRNA-181b-5p affects the malignant progression of acute lymphoblastic leukemia. J. Transl. Med. 2021, 19, 511. [Google Scholar] [CrossRef]
- Saffari, N.; Rahgozar, S.; Faraji, E.; Sahin, F. Plasma-derived exosomal miR-326, a prognostic biomarker and novel candidate for treatment of drug resistant pediatric acute lymphoblastic leukemia. Sci. Rep. 2024, 14, 691. [Google Scholar] [CrossRef]
- Rzepiel, A.; Horváth, A.; Kutszegi, N.; Gézsi, A.; Sági, J.C.; Almási, L.; Egyed, B.; Lőrincz, P.; Visnovitz, T.; Kovács, G.T.; et al. MiR-128-3p as blood based liquid biopsy biomarker in childhood acute lymphoblastic leukemia. Mol. Cell. Probes 2023, 67, 101893. [Google Scholar] [CrossRef]
- Habiel, D.M.; Krepostman, N.; Lilly, M.; Cavassani, K.; Coelho, A.L.; Shibata, T.; Elenitoba-Johnson, K.; Hogaboam, C.M. Senescent stromal cell-induced divergence and therapeutic resistance in T cell acute lymphoblastic leukemia/lymphoma. Oncotarget 2016, 7, 83514–83529. [Google Scholar] [CrossRef]
- Haque, S.; Vaiselbuh, S.R. Vincristine and prednisone regulates cellular and exosomal miR-181a expression differently within the first time diagnosed and the relapsed leukemia B cells. Leuk. Res. Rep. 2020, 14, 100221. [Google Scholar] [CrossRef] [PubMed]
- Colangelo, T.; Panelli, P.; Mazzarelli, F.; Tamiro, F.; Melocchi, V.; De Santis, E.; Cuttano, R.; Palumbo, O.; Rossi, G.; Bianchi, F.; et al. Extracellular vesicle microRNAs contribute to Notch signaling pathway in T-cell acute lymphoblastic leukemia. Mol. Cancer 2022, 21, 226. [Google Scholar] [CrossRef] [PubMed]
- Litzow, M.R.; Ferrando, A.A. How I treat T-cell acute lymphoblastic leukemia in adults. Blood 2015, 126, 833–841. [Google Scholar] [CrossRef]
- Cheng, H.; Ding, J.; Tang, G.; Huang, A.; Gao, L.; Yang, J.; Chen, L. Human mesenchymal stem cells derived exosomes inhibit the growth of acute myeloid leukemia cells via regulating miR-23b-5p/TRIM14 pathway. Mol. Med. 2021, 27, 128. [Google Scholar] [CrossRef]
- Jiang, D.; Wu, X.; Sun, X.; Tan, W.; Dai, X.; Xie, Y.; Du, A.; Zhao, Q. Bone mesenchymal stem cell-derived exosomal microRNA-7-5p inhibits progression of acute myeloid leukemia by targeting OSBPL11. J. Nanobiotechnol. 2022, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Otmani, K.; Rouas, R.; Lagneaux, L.; Krayem, M.; Duvillier, H.; Berehab, M.; Lewalle, P. Acute myeloid leukemia-derived exosomes deliver miR-24-3p to hinder the T-cell immune response through DENN/MADD targeting in the NF-κB signaling pathways. Cell Commun. Signal. CCS 2023, 21, 253. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Du, F.; Zhao, Y.; Wang, S.; Qi, L. Acute myeloid leukemia cells secrete microRNA-4532-containing exosomes to mediate normal hematopoiesis in hematopoietic stem cells by activating the LDOC1-dependent STAT3 signaling pathway. Stem Cell Res. Ther. 2019, 10, 384. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.C.; Lin, Y.S.; Zhang, L.; Lu, Y.; Sun, Y.L.; Fang, Z.G.; Li, Z.Y.; Fan, R.F. MicroRNAs of bone marrow mesenchymal stem cell-derived exosomes regulate acute myeloid leukemia cell proliferation and apoptosis. Chin. Med. J. 2020, 133, 2829–2839. [Google Scholar] [CrossRef]
- Taniguchi, M.; Nagaya, S.; Yuyama, K.; Kotani, A.; Igarashi, Y.; Okazaki, T. Ceramide Metabolism Regulated by Sphingomyelin Synthase 2 Is Associated with Acquisition of Chemoresistance via Exosomes in Human Leukemia Cells. Int. J. Mol. Sci. 2022, 23, 10648. [Google Scholar] [CrossRef]
- Amiri, B.S.; Sabernia, N.; Abouali, B.; Amini, P.; Rezaeeyan, H. Evaluation of MicroRNA as Minimal Residual Disease in Leukemia: Diagnostic and Prognostic Approach: A Review. Iran. J. Public Health 2023, 52, 2541–2553. [Google Scholar] [CrossRef]
- Jiang, L.; Deng, T.; Wang, D.; Xiao, Y. Elevated Serum Exosomal miR-125b Level as a Potential Marker for Poor Prognosis in Intermediate-Risk Acute Myeloid Leukemia. Acta Haematol. 2018, 140, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Horiguchi, H.; Kikuchi, S.; Iyama, S.; Ikeda, H.; Goto, A.; Kawano, Y.; Murase, K.; Takada, K.; Miyanishi, K.; et al. miR-7977 inhibits the Hippo-YAP signaling pathway in bone marrow mesenchymal stromal cells. PLoS ONE 2019, 14, e0213220. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xie, C.; Lu, Y.; Chang, K.; Guan, F.; Li, X. Exosomal miR92a Promotes Cytarabine Resistance in Myelodysplastic Syndromes by Activating Wnt/β-catenin Signal Pathway. Biomolecules 2022, 12, 1448. [Google Scholar] [CrossRef]
- Yuan, Y.; Tan, S.; Wang, H.; Zhu, J.; Li, J.; Zhang, P.; Wang, M.; Zhang, F. Mesenchymal Stem Cell-Derived Exosomal miRNA-222-3p Increases Th1/Th2 Ratio and Promotes Apoptosis of Acute Myeloid Leukemia Cells. Anal. Cell. Pathol. 2023, 2023, 4024887. [Google Scholar] [CrossRef]
- Barrera-Ramirez, J.; Lavoie, J.R.; Maganti, H.B.; Stanford, W.L.; Ito, C.; Sabloff, M.; Brand, M.; Rosu-Myles, M.; Le, Y.; Allan, D.S. Micro-RNA Profiling of Exosomes from Marrow-Derived Mesenchymal Stromal Cells in Patients with Acute Myeloid Leukemia: Implications in Leukemogenesis. Stem Cell Rev. Rep. 2017, 13, 817–825. [Google Scholar] [CrossRef]
- Li, C.; Long, X.; Liang, P.; Liu, Z.; Wang, C.; Hu, R. Analysis of microRNA expression profiles in exosomes derived from acute myeloid leukemia by p62 knockdown and effect on angiogenesis. PeerJ 2022, 10, e13498. [Google Scholar] [CrossRef] [PubMed]
- El-Saghir, J.; Nassar, F.; Tawil, N.; El-Sabban, M. ATL-derived exosomes modulate mesenchymal stem cells: Potential role in leukemia progression. Retrovirology 2016, 13, 73. [Google Scholar] [CrossRef]
- Mezher, M.; Abdallah, S.; Ashekyan, O.; Shoukari, A.A.; Choubassy, H.; Kurdi, A.; Temraz, S.; Nasr, R. Insights on the Biomarker Potential of Exosomal Non-Coding RNAs in Colorectal Cancer: An In Silico Characterization of Related Exosomal lncRNA/circRNA-miRNA-Target Axis. Cells 2023, 12, 1081. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, X.; Kong, Q.; He, H.; Sun, J.; Qiu, W.; Zhang, L.; Yang, M. Extracellular Vesicle Preparation and Analysis: A State-of-the-Art Review. Adv. Sci. 2024, 11, e2401069. [Google Scholar] [CrossRef]
- Wang, J.; Yue, B.L.; Huang, Y.Z.; Lan, X.Y.; Liu, W.J.; Chen, H. Exosomal RNAs: Novel Potential Biomarkers for Diseases-A Review. Int. J. Mol. Sci. 2022, 23, 2461. [Google Scholar] [CrossRef]
- Yang, C.; Yang, H.; Liu, J.; Zhu, L.; Yu, S.; Zhang, X.; Gao, L. Focus on exosomes: Novel pathogenic components of leukemia. Am. J. Cancer Res. 2019, 9, 1815–1829. [Google Scholar] [PubMed]
- Salehi, A. A novel therapeutic strategy: The significance of exosomal miRNAs in acute myeloid leukemia. Med. Oncol. 2024, 41, 62. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.H.; Sharifi, L.M.A.; Kakhharov, A.J.; Opulencia, M.J.C.; Alsaikhan, F.; Bokov, D.O.; Majdi, H.S.; Jawad, M.A.; Hammid, A.T.; Shalaby, M.N.; et al. Role of Acute Myeloid Leukemia (AML)-Derived exosomes in tumor progression and survival. Biomed. Pharmacother. = Biomed. Pharmacother. 2022, 150, 113009. [Google Scholar] [CrossRef] [PubMed]
- Cariello, M.; Squilla, A.; Piacente, M.; Venutolo, G.; Fasano, A. Drug Resistance: The Role of Exosomal miRNA in the Microenvironment of Hematopoietic Tumors. Molecules 2022, 28, 116. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, X.; Li, C.; Chen, T.; Yang, Q. Exosomal non-coding RNAs: Emerging roles in bilateral communication between cancer cells and macrophages. Mol. Ther. J. Am. Soc. Gene Ther. 2022, 30, 1036–1053. [Google Scholar] [CrossRef]
- Liu, Q.W.; He, Y.; Xu, W.W. Molecular functions and therapeutic applications of exosomal noncoding RNAs in cancer. Exp. Mol. Med. 2022, 54, 216–225. [Google Scholar] [CrossRef]
- Tang, B.J.; Sun, B.; Chen, L.; Xiao, J.; Huang, S.T.; Xu, P. The Landscape of Exosome-Derived Non-Coding RNA in Leukemia. Front. Pharmacol. 2022, 13, 912303. [Google Scholar] [CrossRef]
- Knaus, H.A.; Berglund, S.; Hackl, H.; Blackford, A.L.; Zeidner, J.F.; Montiel-Esparza, R.; Mukhopadhyay, R.; Vanura, K.; Blazar, B.R.; Karp, J.E.; et al. Signatures of CD8+ T cell dysfunction in AML patients and their reversibility with response to chemotherapy. JCI Insight 2018, 3, e120974. [Google Scholar] [CrossRef]
- Otmani, K.; Rouas, R.; Lewalle, P. OncomiRs as noncoding RNAs having functions in cancer: Their role in immune suppression and clinical implications. Front. Immunol. 2022, 13, 913951. [Google Scholar] [CrossRef]
- Batista, I.A.; Machado, J.C.; Melo, S.A. Advances in exosomes utilization for clinical applications in cancer. Trends Cancer 2024, 10, 947–968. [Google Scholar] [CrossRef]
- Bernardi, S.; Farina, M.; Bosio, K.; Di Lucanardo, A.; Leoni, A.; Re, F.; Polverelli, N.; Turra, A.; Morello, E.; Accorsi Buttini, E.; et al. Feasibility of Leukemia-Derived Exosome Enrichment and Co-isolated dsDNA Sequencing in Acute Myeloid Leukemia Patients: A Proof of Concept for New Leukemia Biomarkers Detection. Cancers 2022, 14, 4504. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.A.; Chan, L.K.W.; Hung, L.C.; Phoebe, L.K.W.; Park, Y.; Yi, K.-H. Clinical Applications of Exosomes: A Critical Review. Int. J. Mol. Sci. 2024, 25, 7794. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhou, L.; Yu, F.; Zhang, Y.; Li, P.; Wang, K. The functional roles of exosomal long non-coding RNAs in cancer. Cell. Mol. Life Sci. CMLS 2019, 76, 2059–2076. [Google Scholar] [CrossRef] [PubMed]
- Marima, R.; Basera, A.; Miya, T.; Damane, B.P.; Kandhavelu, J.; Mirza, S.; Penny, C.; Dlamini, Z. Exosomal long non-coding RNAs in cancer: Interplay, modulation, and therapeutic avenues. Non-Coding RNA Res. 2024, 9, 887–900. [Google Scholar] [CrossRef]
- Xiao, Q.; Lin, C.; Peng, M.; Ren, J.; Jing, Y.; Lei, L.; Tao, Y.; Huang, J.; Yang, J.; Sun, M.; et al. Circulating plasma exosomal long non-coding RNAs LINC00265, LINC00467, UCA1, and SNHG1 as biomarkers for diagnosis and treatment monitoring of acute myeloid leukemia. Front. Oncol. 2022, 12, 1033143. [Google Scholar] [CrossRef]
- Deng, W.; Chao, R.; Zhu, S. Emerging roles of circRNAs in leukemia and the clinical prospects: An update. Immun. Inflamm. Dis. 2023, 11, e725. [Google Scholar] [CrossRef]
- Gharib, E.; Nasrabadi, P.N.; Robichaud, G.A. Circular RNA Expression Signatures Provide Promising Diagnostic and Therapeutic Biomarkers for Chronic Lymphocytic Leukemia. Cancers 2023, 15, 1554. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz, E.; Sole, C.; Manterola, L.; Iparraguirre, L.; Otaegui, D.; Lawrie, C.H. CircRNAs and cancer: Biomarkers and master regulators. Semin. Cancer Biol. 2019, 58, 90–99. [Google Scholar] [CrossRef]
- Cui, X.; Wang, J.; Guo, Z.; Li, M.; Li, M.; Liu, S.; Liu, H.; Li, W.; Yin, X.; Tao, J.; et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol. Cancer 2018, 17, 123. [Google Scholar] [CrossRef]
- Li, Q.; Luan, Q.; Zhu, H.; Zhao, Y.; Ji, J.; Wu, F.; Yan, J. Circular RNA circ_0005774 contributes to proliferation and suppresses apoptosis of acute myeloid leukemia cells via circ_0005774/miR-192-5p/ULK1 ceRNA pathway. Biochem. Biophys. Res. Commun. 2021, 551, 78–85. [Google Scholar] [CrossRef]
- Li, Q.; Ren, X.; Wang, Y.; Xin, X. CircRNA: A rising star in leukemia. PeerJ 2023, 11, e15577. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, J.; Fan, Y.; Xu, X.; Xiong, M.; Qi, Y.; Wu, W.; Zhao, Y. circZNF91 Promotes the Malignant Phenotype of Chronic Lymphocytic Leukemia Cells by Targeting the miR-1283/WEE1 Axis. BioMed Res. Int. 2022, 2022, 2855394. [Google Scholar] [CrossRef] [PubMed]
- Zimta, A.A.; Tomuleasa, C.; Sahnoune, I.; Calin, G.A.; Berindan-Neagoe, I. Long Non-coding RNAs in Myeloid Malignancies. Front. Oncol. 2019, 9, 1048. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.A.; Younes, S.N.; Raza, S.S.; Zarif, L.; Nisar, S.; Ahmed, I.; Mir, R.; Kumar, S.; Sharawat, S.K.; Hashem, S.; et al. Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance. Mol. Cancer 2020, 19, 57. [Google Scholar] [CrossRef]
- Xu, T.; Wang, M.; Jiang, L.; Ma, L.; Wan, L.; Chen, Q.; Wei, C.; Wang, Z. CircRNAs in anticancer drug resistance: Recent advances and future potential. Mol. Cancer 2020, 19, 127. [Google Scholar] [CrossRef]
- Ma, S.; Kong, S.; Wang, F.; Ju, S. CircRNAs: Biogenesis, functions, and role in drug-resistant Tumours. Mol. Cancer 2020, 19, 119. [Google Scholar] [CrossRef]
- Ji, T.; Chen, Q.; Tao, S.; Shi, Y.; Chen, Y.; Shen, L.; Wang, C.; Yu, L. The research progress of circular RNAs in hematological malignancies. Hematology 2019, 24, 727–731. [Google Scholar] [CrossRef]
- Zhong, A.N.; Yin, Y.; Tang, B.J.; Chen, L.; Shen, H.W.; Tan, Z.P.; Li, W.Q.; He, Q.; Sun, B.; Zhu, Y.; et al. CircRNA Microarray Profiling Reveals hsa_circ_0058493 as a Novel Biomarker for Imatinib-Resistant CML. Front. Pharmacol. 2021, 12, 728916. [Google Scholar] [CrossRef]
- Ding, J.; Zhang, X.; Xue, J.; Fang, L.; Ban, C.; Song, B.; Wu, L. CircNPM1 strengthens Adriamycin resistance in acute myeloid leukemia by mediating the miR-345-5p/FZD5 pathway. Cent.-Eur. J. Immunol. 2021, 46, 162–182. [Google Scholar] [CrossRef]
- Rezaie, J.; Feghhi, M.; Etemadi, T. A review on exosomes application in clinical trials: Perspective, questions, and challenges. Cell Commun. Signal. CCS 2022, 20, 145. [Google Scholar] [CrossRef]
- Wang, X.; Xia, J.; Yang, L.; Dai, J.; He, L. Recent progress in exosome research: Isolation, characterization and clinical applications. Cancer Gene Ther. 2023, 30, 1051–1065. [Google Scholar] [CrossRef] [PubMed]
- Dilsiz, N. A comprehensive review on recent advances in exosome isolation and characterization: Toward clinical applications. Transl. Oncol. 2024, 50, 102121. [Google Scholar] [CrossRef] [PubMed]
- Coughlan, C.; Bruce, K.D.; Burgy, O.; Boyd, T.D.; Michel, C.R.; Garcia-Perez, J.E.; Adame, V.; Anton, P.; Bettcher, B.M.; Chial, H.J.; et al. Exosome Isolation by Ultracentrifugation and Precipitation and Techniques for Downstream Analyses. Curr. Protoc. Cell Biol. 2020, 88, e110. [Google Scholar] [CrossRef] [PubMed]
- Karttunen, J.; Heiskanen, M.; Navarro-Ferrandis, V.; Das Gupta, S.; Lipponen, A.; Puhakka, N.; Rilla, K.; Koistinen, A.; Pitkänen, A. Precipitation-based extracellular vesicle isolation from rat plasma co-precipitate vesicle-free microRNAs. J. Extracell. Vesicles 2018, 8, 1555410. [Google Scholar] [CrossRef]
- Araujo-Abad, S.; Berna, J.M.; Lloret-Lopez, E.; López-Cortés, A.; Saceda, M.; de Juan Romero, C. Exosomes: From basic research to clinical diagnostic and therapeutic applications in cancer. Cell. Oncol. 2024; Advance online publication. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef]
- Singh, S.; Dansby, C.; Agarwal, D.; Bhat, P.D.; Dubey, P.K.; Krishnamurthy, P. Exosomes: Methods for Isolation and Characterization in Biological Samples. Methods Mol. Biol. 2024, 2835, 181–213. [Google Scholar] [CrossRef]
- Bachurski, D.; Schuldner, M.; Nguyen, P.H.; Malz, A.; Reiners, K.S.; Grenzi, P.C.; Babatz, F.; Schauss, A.C.; Hansen, H.P.; Hallek, M.; et al. Extracellular vesicle measurements with nanoparticle tracking analysis—An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles 2019, 8, 1596016. [Google Scholar] [CrossRef] [PubMed]
- Dixson, A.C.; Dawson, T.R.; Di Vizio, D.; Weaver, A.M. Context-specific regulation of extracellular vesicle biogenesis and cargo selection. Nature reviews. Mol. Cell Biol. 2023, 24, 454–476. [Google Scholar] [CrossRef]
- Dutta, A. Exosomes-based cell-free cancer therapy: A novel strategy for targeted therapy. Immunol. Med. 2021, 44, 116–123. [Google Scholar] [CrossRef]
- Tan, F.; Li, X.; Wang, Z.; Li, J.; Shahzad, K.; Zheng, J. Clinical applications of stem cell-derived exosomes. Signal Transduct. Target. Ther. 2024, 9, 17. [Google Scholar] [CrossRef]
- Sonar, S. Clinical trial status of exosomes-based cancer theranostics. Clin. Transl. Disc. 2024, 4, e327. [Google Scholar] [CrossRef]
- Wu, M.; Wang, G.; Hu, W.; Yao, Y.; Yu, X.F. Emerging roles and therapeutic value of exosomes in cancer metastasis. Mol. Cancer 2019, 18, 53. [Google Scholar] [CrossRef] [PubMed]
- Urabe, F.; Kosaka, N.; Ito, K.; Kimura, T.; Egawa, S.; Ochiya, T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am. J. Physiol. Cell Physiol. 2020, 318, C29–C39. [Google Scholar] [CrossRef] [PubMed]
- Clinicaltrial.gov. Available online: https://clinicaltrials.gov/ (accessed on 10 November 2024).
- Chen, Y.S.; Lin, E.Y.; Chiou, T.W.; Harn, H.J. Exosomes in clinical trial and their production in compliance with good manufacturing practice. Tzu Chi Med. J. 2019, 32, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Shahraki, K.; Boroumand, P.G.; Lotfi, H.; Radnia, F.; Shahriari, H.; Sargazi, S.; Mortazavi, S.S.; Shirvaliloo, M.; Shirvalilou, S.; Sheervalilou, R. An update in the applications of exosomes in cancer theranostics: From research to clinical trials. J. Cancer Res. Clin. Oncol. 2023, 149, 8087–8116. [Google Scholar] [CrossRef]
- Garza Treviño, E.N.; Quiroz Reyes, A.G.; Delgado Gonzalez, P.; Rojas Murillo, J.A.; Islas, J.F.; Alonso, S.S.; Gonzalez Villarreal, C.A. Applications of Modified Mesenchymal Stem Cells as Targeted Systems against Tumor Cells. Int. J. Mol. Sci. 2024, 25, 7791. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil-Kulik, P.; Kluz, N.; Przywara, D.; Petniak, A.; Wasilewska, M.; Frączek-Chudzik, N.; Cieśla, M. Potential Use of Exosomal Non-Coding MicroRNAs in Leukemia Therapy: A Systematic Review. Cancers 2024, 16, 3948. https://doi.org/10.3390/cancers16233948
Gil-Kulik P, Kluz N, Przywara D, Petniak A, Wasilewska M, Frączek-Chudzik N, Cieśla M. Potential Use of Exosomal Non-Coding MicroRNAs in Leukemia Therapy: A Systematic Review. Cancers. 2024; 16(23):3948. https://doi.org/10.3390/cancers16233948
Chicago/Turabian StyleGil-Kulik, Paulina, Natalia Kluz, Dominika Przywara, Alicja Petniak, Małgorzata Wasilewska, Natalia Frączek-Chudzik, and Marek Cieśla. 2024. "Potential Use of Exosomal Non-Coding MicroRNAs in Leukemia Therapy: A Systematic Review" Cancers 16, no. 23: 3948. https://doi.org/10.3390/cancers16233948
APA StyleGil-Kulik, P., Kluz, N., Przywara, D., Petniak, A., Wasilewska, M., Frączek-Chudzik, N., & Cieśla, M. (2024). Potential Use of Exosomal Non-Coding MicroRNAs in Leukemia Therapy: A Systematic Review. Cancers, 16(23), 3948. https://doi.org/10.3390/cancers16233948







