TFE3-Rearranged Tumors of the Kidney: An Emerging Conundrum
Abstract
Simple Summary
Abstract
1. Introduction
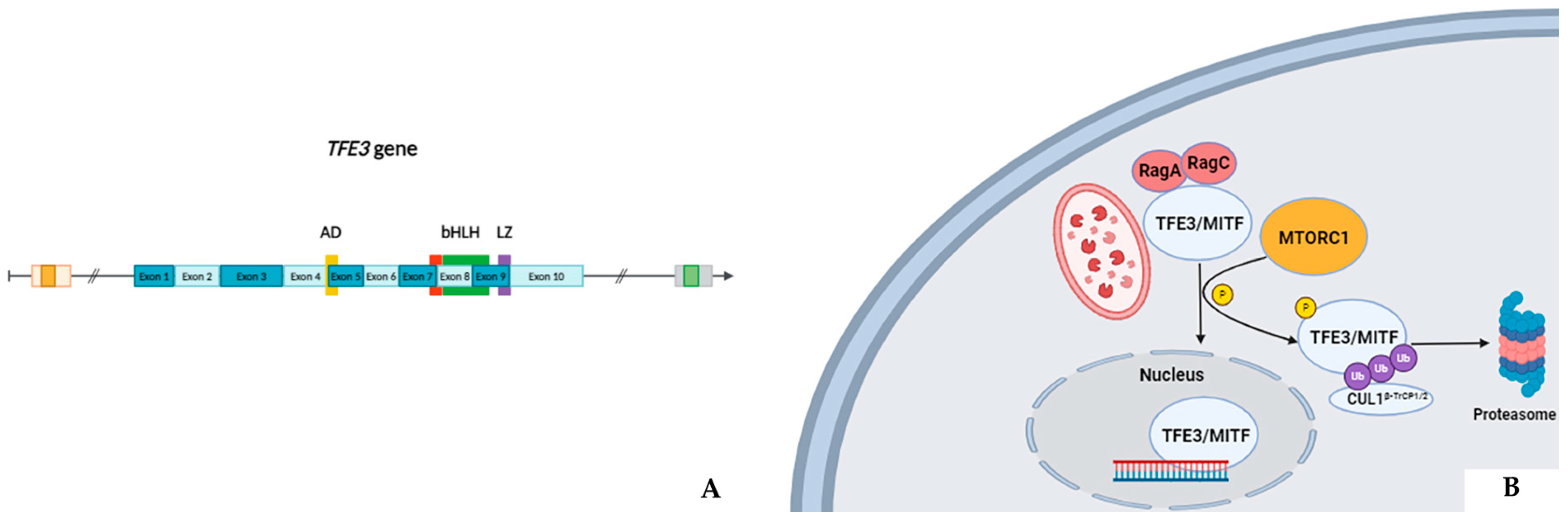
2. TFE3 Gene
- Autophagy/cellular senescence: The ASPL/ASPSCR1 gene, mapped at 17q25.3, encodes a protein containing a UBX domain which physiologically interacts with glucose transporter type 4 (GLUT4). This protein is a tether, which sequesters the GLUT4 in intracellular vesicles in muscle and fat cells in the absence of insulin and redistributes the transporter to the plasma membrane under insulin stimulation [17]. Experimental models have demonstrated that the aberrant ASPL/ASPSCR1-TFE3 protein induces p21 transcription in a p53-independent manner. This latter further up-regulates pro-inflammatory cytokines associated with autophagy [18] and a senescence-associated secretory phenotype (SASP) [19] which can, ultimately, promote a tumorigenic microenvironment. The PRCC gene instead, located at 1q23.1, codifies for a protein playing a role in pre-mRNA splicing. Similarly, the PRCC-TFE3 fusion products have been shown to stimulate the macroautophagy elimination of superfluous, aged, or damaged mitochondria, a process also called “mitophagy”. In detail, the aberrant nuclear PRCC-TFE3 proteins constitutively activate the E3 ubiquitin ligase PRKN which allows cell survival and proliferation under mitochondrial oxidative damage through a reduction in mitochondrial ROS formation [20].
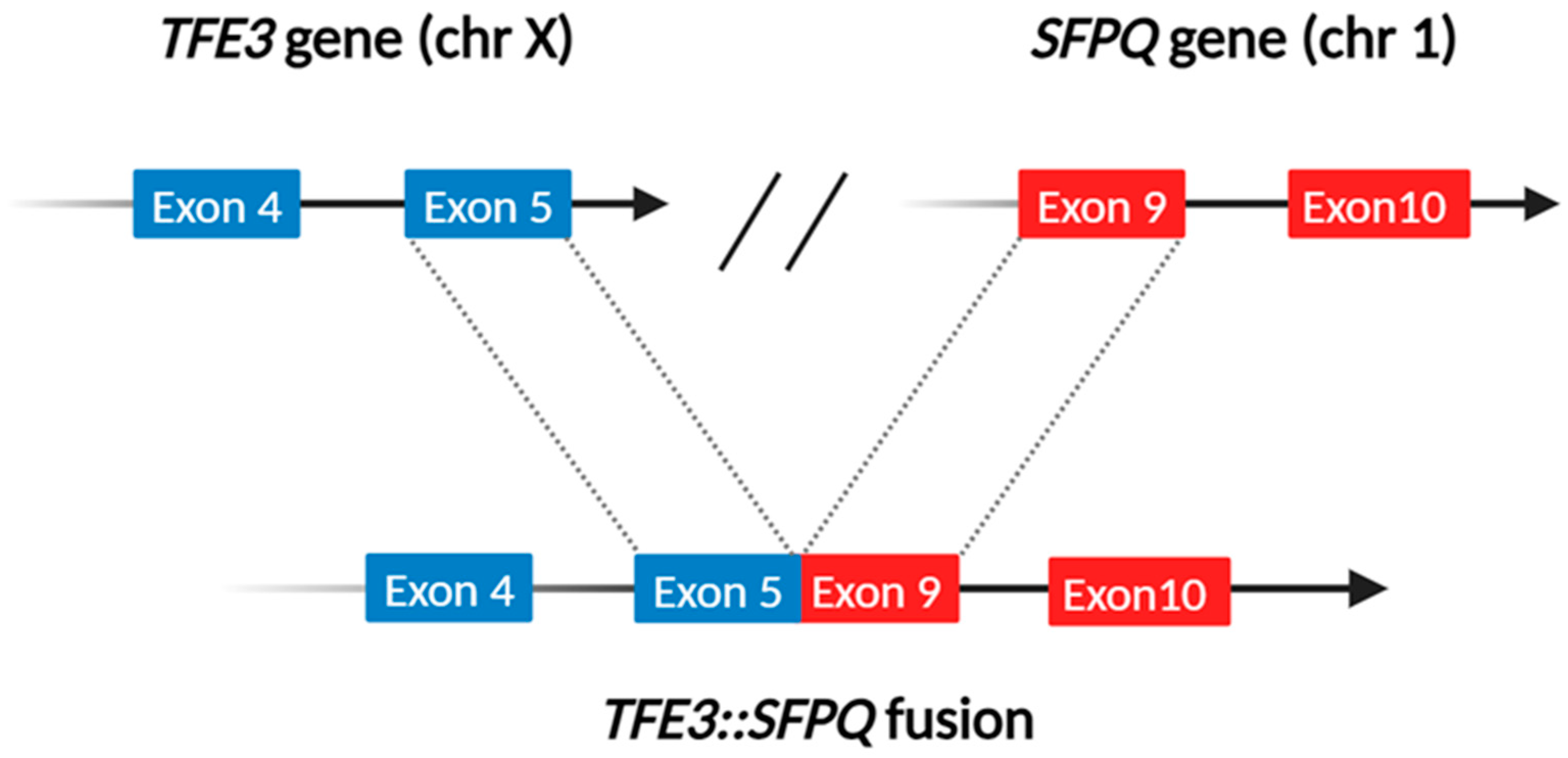
- Speckle–paraspeckles structures: The SFPQ/PSF and NONO genes, mapped at 1p34 and Xq13.1, respectively, belong to the “Drosophila behavior human splicing” family. They encode for non-coding RNAs playing an essential role in the formation and integrity of the so-called speckle–paraspeckles structures [21]. These membranelles’ nuclear bodies carry out specific regulating functions on the transcriptional process and, overall, gene expression [22] and innate immune response [23]. The proteins they codify work as (hetero)dimers and interact with the NEAT1 long non-coding RNA in constructing the structural backbone of paraspeckles. Notably, plenty of other genes whose fusions are mainly linked to TFEB-rearranged renal cell carcinomas, such as NEAT1 and MALAT1, are involved in forming the speckle–paraspeckles complex. In this view, it is indeed worth mentioning that SFPQ/PFS::TFE3-rearranged renal cell carcinoma may morphologically mimic TFEB-rearranged renal cell carcinoma. Such a finding is due to the presence of clusters of small cells or “pseudorosettes” giving various examples of these neoplasms a distinctive biphasic growth pattern [24], more commonly described in TFEB-rearranged renal cell carcinoma [25].
- Lipid accumulation: The MED15 gene is located at 22q11.21 and its derived protein works as a transcriptional coactivator in RNA polymerase II transcription. Concerning renal cell carcinoma, preclinical models have shown that HIF2α-dependent MED15 over-expression may promote lipid deposition and tumor progression through sterol regulatory element binding protein (SREBP) coactivation via the AKT pathway [26].
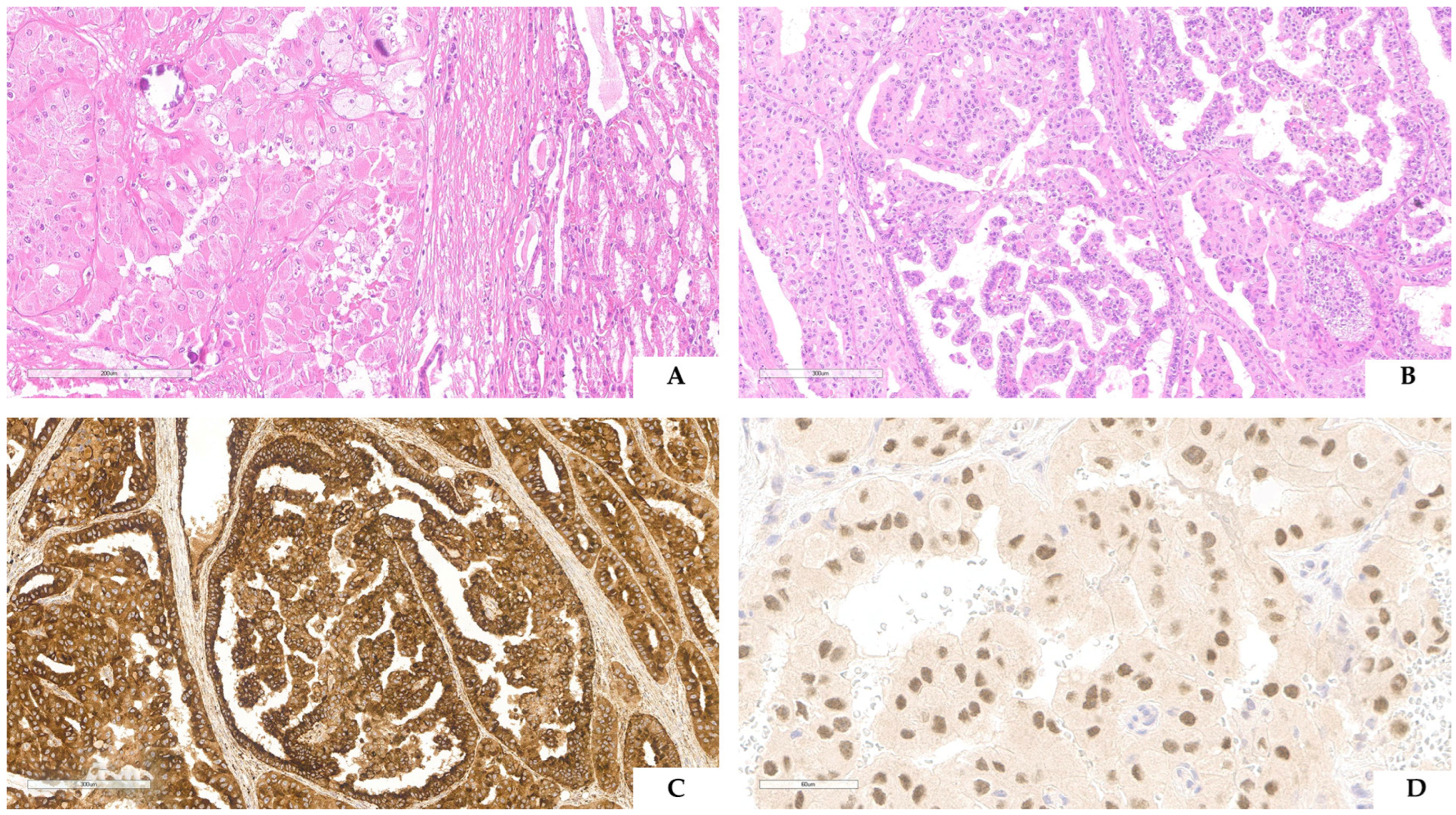
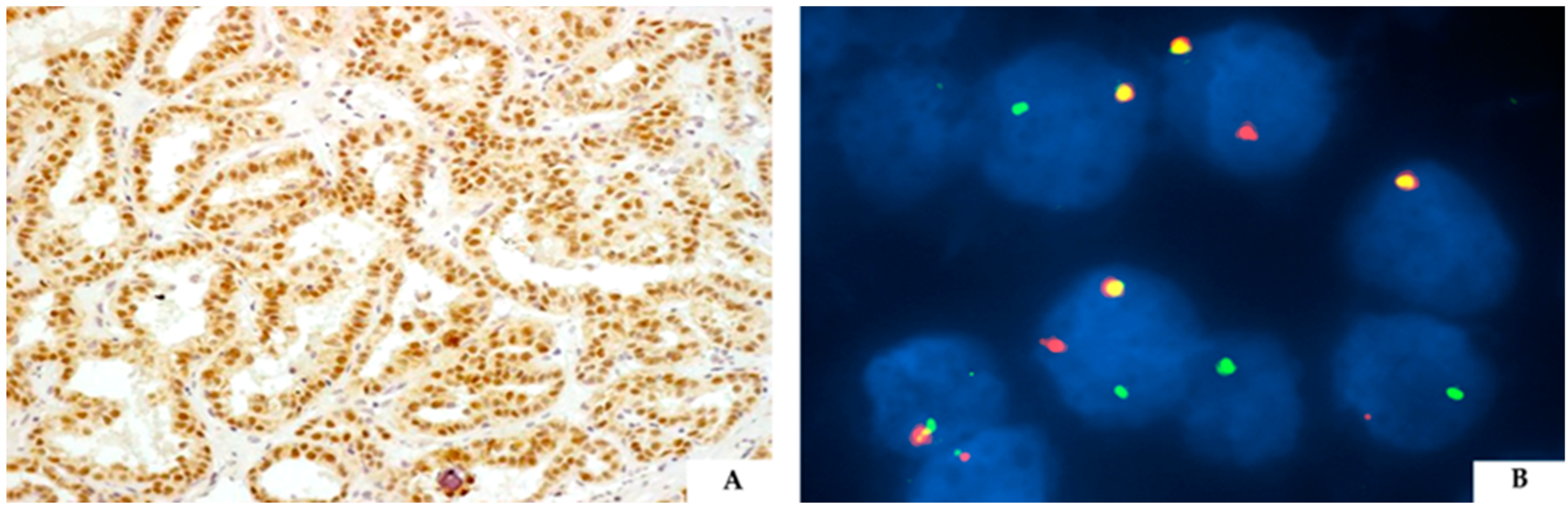
3. TFE3-Rearranged Renal Cell Carcinoma
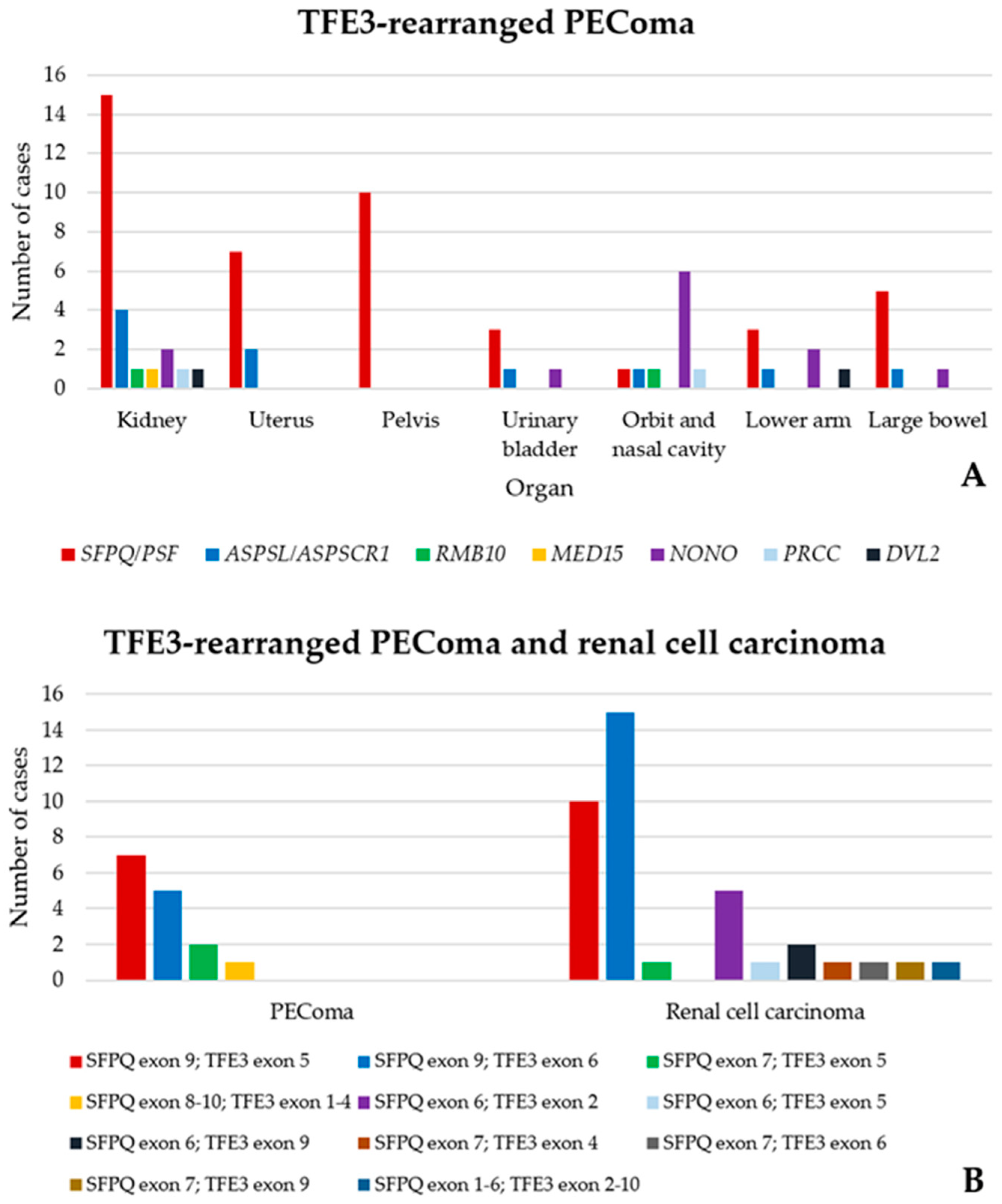
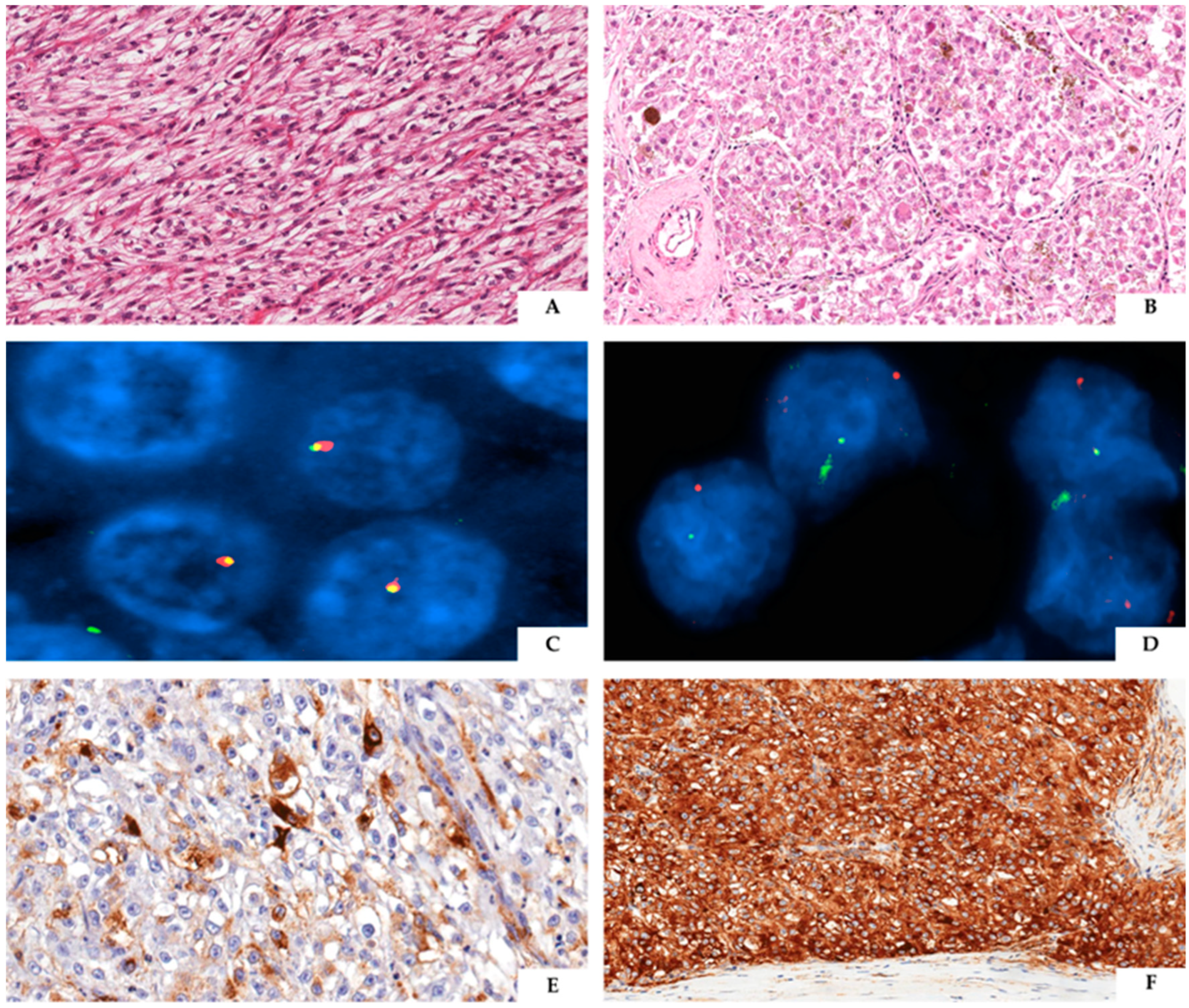
4. TFE3-Rearranged Renal PEComa
5. Emerging Conundrum
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hemesath, T.J.; Steingrímsson, E.; McGill, G.; Hansen, M.J.; Vaught, J.; Hodgkinson, C.A.; Arnheiter, H.; Copeland, N.G.; Jenkins, N.A.; Fisher, D.E. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994, 8, 2770–2780. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.M.; Di Malta, C.; Ballabio, A. MiT/TFE Family of Transcription Factors, Lysosomes, and Cancer. Annu. Rev. Cancer Biol. 2019, 3, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Pinto, K.; Chetty, R. Gene of the month: TFE 3. J. Clin. Pathol. 2020, 73, 691–694. [Google Scholar] [CrossRef] [PubMed]
- Steingrimsson, E.; Tessarollo, L.; Pathak, B.; Hou, L.; Arnheiter, H.; Copeland, N.G.; Jenkins, N.A. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc. Natl. Acad. Sci. USA 2002, 99, 4477–4482. [Google Scholar] [CrossRef] [PubMed]
- Yagil, Z.; Hadad Erlich, T.; Ofir-Birin, Y.; Tshori, S.; Kay, G.; Yekhtin, Z.; Fisher, D.E.; Cheng, C.; Wong, W.S.F.; Hartmann, K.; et al. Transcription factor E3, a major regulator of mast cell-mediated allergic response. J. Allergy Clin. Immunol. 2012, 129, 1357–1366.e5. [Google Scholar] [CrossRef]
- Huan, C.; Kelly, M.L.; Steele, R.; Shapira, I.; Gottesman, S.R.S.; Roman, C.A.J. Transcription factors TFE3 and TFEB are critical for CD40 ligand expression and thymus-dependent humoral immunity. Nat. Immunol. 2006, 7, 1082–1091. [Google Scholar] [CrossRef]
- Nardone, C.; Palanski, B.A.; Scott, D.C.; Timms, R.T.; Barber, K.W.; Gu, X.; Mao, A.; Leng, Y.; Watson, E.V.; Schulman, B.A.; et al. A central role for regulated protein stability in the control of TFE3 and MITF by nutrients. Mol. Cell 2023, 83, 57–73.e9. [Google Scholar] [CrossRef]
- Martina, J.A.; Diab, H.I.; Brady, O.A.; Puertollano, R. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 2016, 35, 479–495. [Google Scholar] [CrossRef]
- Diaz, J.; Berger, S.; Leon, E. TFE3-associated neurodevelopmental disorder: A distinct recognizable syndrome. Am. J. Med. Genet. A 2020, 182, 584–590. [Google Scholar] [CrossRef]
- Lehalle, D.; Vabres, P.; Sorlin, A.; Bierhals, T.; Avila, M.; Carmignac, V.; Chevarin, M.; Torti, E.; Abe, Y.; Bartolomaeus, T.; et al. De novo mutations in the X-linked TFE3 gene cause intellectual disability with pigmentary mosaicism and storage disorder-like features. J. Med. Genet. 2020, 57, 808–819. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board (Ed.) WHO Classification of Tumours, Urinary and Male Genital Tumours, 5th ed.; IARC Press: Lyon, France, 2022. [Google Scholar]
- Hodge, J.C.; Pearce, K.E.; Wang, X.; Wiktor, A.E.; Oliveira, A.M.; Greipp, P.T. Molecular cytogenetic analysis for TFE3 rearrangement in Xp11.2 renal cell carcinoma and alveolar soft part sarcoma: Validation and clinical experience with 75 cases. Mod. Pathol. 2014, 27, 113–127. [Google Scholar] [CrossRef]
- Agaimy, A.; Stoehr, R.; Michal, M.; Christopoulos, P.; Winter, H.; Zhang, L.; Stenzinger, A.; Michal, M.; Mechtersheimer, G.; Antonescu, C.R. Recurrent YAP1-TFE3 Gene Fusions in Clear Cell Stromal Tumor of the Lung. Am. J. Surg. Pathol. 2021, 45, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- Antonescu, C.R.; Le Loarer, F.; Mosquera, J.-M.; Sboner, A.; Zhang, L.; Chen, C.-L.; Chen, H.-W.; Pathan, N.; Krausz, T.; Dickson, B.C.; et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer 2013, 52, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Suurmeijer, A.J.H.; Song, W.; Sung, Y.-S.; Zhang, L.; Swanson, D.; Fletcher, C.D.M.; Dickson, B.C.; Antonescu, C.R. Novel recurrent PHF1-TFE3 fusions in ossifying fibromyxoid tumors. Genes Chromosomes Cancer 2019, 58, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Martignoni, G.; Bonetti, F.; Chilosi, M.; Brunelli, M.; Segala, D.; Amin, M.B.; Argani, P.; Eble, J.N.; Gobbo, S.; Pea, M. Cathepsin K expression in the spectrum of perivascular epithelioid cell (PEC) lesions of the kidney. Mod. Pathol. 2012, 25, 100–111. [Google Scholar] [CrossRef]
- Bogan, J.S.; Hendon, N.; McKee, A.E.; Tsao, T.-S.; Lodish, H.F. Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature 2003, 425, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Wang, X.; Xia, Q.; Zhao, M.; Zhang, H.; Wang, X.; Ye, S.; Cheng, K.; Liang, Y.; Cheng, Y.; et al. Nuclear translocation of ASPL-TFE3 fusion protein creates favorable metabolism by mediating autophagy in translocation renal cell carcinoma. Oncogene 2021, 40, 3303–3317. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, N.; Yoshida, H. ASPL-TFE3 Oncoprotein Regulates Cell Cycle Progression and Induces Cellular Senescence by Up-Regulating p21. Neoplasia 2016, 18, 626–635. [Google Scholar] [CrossRef]
- Wang, B.; Yin, X.; Gan, W.; Pan, F.; Li, S.; Xiang, Z.; Han, X.; Li, D. PRCC-TFE3 fusion-mediated PRKN/parkin-dependent mitophagy promotes cell survival and proliferation in PRCC-TFE3 translocation renal cell carcinoma. Autophagy 2021, 17, 2475–2493. [Google Scholar] [CrossRef]
- Lee, P.W.; Marshall, A.C.; Knott, G.J.; Kobelke, S.; Martelotto, L.; Cho, E.; McMillan, P.J.; Lee, M.; Bond, C.S.; Fox, A.H. Paraspeckle subnuclear bodies depend on dynamic heterodimerisation of DBHS RNA-binding proteins via their structured domains. J. Biol. Chem. 2022, 298, 102563. [Google Scholar] [CrossRef]
- Fox, A.H.; Lamond, A.I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010, 2, a000687. [Google Scholar] [CrossRef] [PubMed]
- Morchikh, M.; Cribier, A.; Raffel, R.; Amraoui, S.; Cau, J.; Severac, D.; Dubois, E.; Schwartz, O.; Bennasser, Y.; Benkirane, M. HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response. Mol. Cell 2017, 67, 387–399.e5. [Google Scholar] [CrossRef] [PubMed]
- Tretiakova, M.S. Chameleon TFE3-translocation RCC and How Gene Partners Can Change Morphology: Accurate Diagnosis Using Contemporary Modalities. Adv. Anat. Pathol. 2022, 29, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Caliò, A.; Marletta, S.; Brunelli, M.; Martignoni, G. WHO 2022 Classification of Kidney Tumors: What is relevant? An update and future novelties for the pathologist. Pathologica 2022, 115, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Ge, S.; Zhang, L.; Jiang, Q.; Chen, J.; Xiao, H.; Liang, C. MED15 is upregulated by HIF-2α and promotes proliferation and metastasis in clear cell renal cell carcinoma via activation of SREBP-dependent fatty acid synthesis. Cell Death Discov. 2024, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, G.E.; Nisen, P.D.; Timmons, C.F.; Schneider, N.R. Cytogenetics of a renal cell carcinoma in a 17-month-old child. Evidence for Xp11.2 as a recurring breakpoint. Cancer Genet. Cytogenet. 1991, 57, 11–17. [Google Scholar] [CrossRef]
- Meloni, A.M.; Dobbs, R.M.; Pontes, J.E.; Sandberg, A.A. Translocation (X;1) in papillary renal cell carcinoma. A new cytogenetic subtype. Cancer Genet. Cytogenet. 1993, 65, 1–6. [Google Scholar] [CrossRef]
- Argani, P.; Antonescu, C.R.; Illei, P.B.; Lui, M.Y.; Timmons, C.F.; Newbury, R.; Reuter, V.E.; Garvin, A.J.; Perez-Atayde, A.R.; Fletcher, J.A.; et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: A distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am. J. Pathol. 2001, 159, 179–192. [Google Scholar] [CrossRef]
- Ladanyi, M.; Lui, M.Y.; Antonescu, C.R.; Krause-Boehm, A.; Meindl, A.; Argani, P.; Healey, J.H.; Ueda, T.; Yoshikawa, H.; Meloni-Ehrig, A.; et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene 2001, 20, 48–57. [Google Scholar] [CrossRef]
- Weterman, M.A.; Wilbrink, M.; Geurts van Kessel, A. Fusion of the transcription factor TFE3 gene to a novel gene, PRCC, in t(X;1)(p11;q21)-positive papillary renal cell carcinomas. Proc. Natl. Acad. Sci. USA 1996, 93, 15294–15298. [Google Scholar] [CrossRef]
- Clark, J.; Lu, Y.J.; Sidhar, S.K.; Parker, C.; Gill, S.; Smedley, D.; Hamoudi, R.; Linehan, W.M.; Shipley, J.; Cooper, C.S. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 1997, 15, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, M.; Ichikawa, H.; Arai, E.; Chiku, S.; Sakamoto, H.; Fujimoto, H.; Hiramoto, M.; Nammo, T.; Yasuda, K.; Yoshida, T.; et al. Comprehensive exploration of novel chimeric transcripts in clear cell renal cell carcinomas using whole transcriptome analysis. Genes Chromosomes Cancer 2014, 53, 1018–1032. [Google Scholar] [CrossRef] [PubMed]
- Classe, M.; Malouf, G.G.; Su, X.; Yao, H.; Thompson, E.J.; Doss, D.J.; Grégoire, V.; Lenobin, J.; Fantoni, J.-C.; Sudour-Bonnange, H.; et al. Incidence, clinicopathological features and fusion transcript landscape of translocation renal cell carcinomas. Histopathology 2017, 70, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Camparo, P.; Vasiliu, V.; Molinie, V.; Couturier, J.; Dykema, K.J.; Petillo, D.; Furge, K.A.; Comperat, E.M.; Lae, M.; Bouvier, R.; et al. Renal translocation carcinomas: Clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of the literature. Am. J. Surg. Pathol. 2008, 32, 656–670. [Google Scholar] [CrossRef]
- Argani, P. MiT family translocation renal cell carcinoma. Semin. Diagn. Pathol. 2015, 32, 103–113. [Google Scholar] [CrossRef]
- Achom, M.; Sadagopan, A.; Bao, C.; McBride, F.; Li, J.; Konda, P.; Tourdot, R.W.; Xu, Q.; Nakhoul, M.; Gallant, D.S.; et al. A genetic basis for sex differences in Xp11 translocation renal cell carcinoma. Cell 2024, 181, 5735–5752.e25. [Google Scholar] [CrossRef]
- Caliò, A.; Brunelli, M.; Segala, D.; Pedron, S.; Remo, A.; Ammendola, S.; Munari, E.; Pierconti, F.; Mosca, A.; Bollito, E.; et al. Comprehensive analysis of 34 MiT family translocation renal cell carcinomas and review of the literature: Investigating prognostic markers and therapy targets. Pathology 2020, 52, 297–309. [Google Scholar] [CrossRef]
- Wang, X.-T.; Xia, Q.-Y.; Ye, S.-B.; Wang, X.; Li, R.; Fang, R.; Shi, S.-S.; Zhang, R.-S.; Tan, X.; Chen, J.-Y.; et al. RNA sequencing of Xp11 translocation-associated cancers reveals novel gene fusions and distinctive clinicopathologic correlations. Mod. Pathol. 2018, 31, 1346–1360. [Google Scholar] [CrossRef]
- Hayes, M.; Peckova, K.; Martinek, P.; Hora, M.; Kalusova, K.; Straka, L.; Daum, O.; Kokoskova, B.; Rotterova, P.; Pivovarčikova, K.; et al. Molecular-genetic analysis is essential for accurate classification of renal carcinoma resembling Xp11.2 translocation carcinoma. Virchows Arch. 2015, 466, 313–322. [Google Scholar] [CrossRef]
- Inamura, K. Translocation Renal Cell Carcinoma: An Update on Clinicopathological and Molecular Features. Cancers 2017, 9, 111. [Google Scholar] [CrossRef]
- Argani, P.; Zhong, M.; Reuter, V.E.; Fallon, J.T.; Epstein, J.I.; Netto, G.J.; Antonescu, C.R. TFE3-Fusion Variant Analysis Defines Specific Clinicopathologic Associations Among Xp11 Translocation Cancers. Am. J. Surg. Pathol. 2016, 40, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yin, X.; Xia, Q.; Zheng, L.; Yao, J.; Zeng, H.; Nie, L.; Gong, J.; Zhou, Q.; Chen, N. Xp11 translocation renal cell carcinoma with morphological features mimicking multilocular cystic renal neoplasm of low malignant potential: A series of six cases with molecular analysis. J. Clin. Pathol. 2021, 74, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Martignoni, G.; Pea, M.; Gobbo, S.; Brunelli, M.; Bonetti, F.; Segala, D.; Pan, C.-C.; Netto, G.; Doglioni, C.; Hes, O.; et al. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod. Pathol. 2009, 22, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Reuter, V.E.; Argani, P.; Zhou, M.; Delahunt, B. Best practices recommendations in the application of immunohistochemistry in the kidney tumors: Report from the International Society of Urologic Pathology consensus conference. Am. J. Surg. Pathol. 2014, 38, e35–e49. [Google Scholar] [CrossRef] [PubMed]
- Caliò, A.; Marletta, S.; Brunelli, M.; Pedron, S.; Portillo, S.C.; Segala, D.; Bariani, E.; Gobbo, S.; Netto, G.; Martignoni, G. TFE3 and TFEB-rearranged renal cell carcinomas: An immunohistochemical panel to differentiate from common renal cell neoplasms. Virchows Arch. 2022, 481, 877–891. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-M.; Zhang, Y.; Mannan, R.; Skala, S.L.; Rangaswamy, R.; Chinnaiyan, A.; Su, F.; Cao, X.; Zelenka-Wang, S.; McMurry, L.; et al. TRIM63 is a sensitive and specific biomarker for MiT family aberration-associated renal cell carcinoma. Mod. Pathol. 2021, 34, 1596–1607. [Google Scholar] [CrossRef]
- Baba, M.; Furuya, M.; Motoshima, T.; Lang, M.; Funasaki, S.; Ma, W.; Sun, H.-W.; Hasumi, H.; Huang, Y.; Kato, I.; et al. TFE3 Xp11.2 Translocation Renal Cell Carcinoma Mouse Model Reveals Novel Therapeutic Targets and Identifies GPNMB as a Diagnostic Marker for Human Disease. Mol. Cancer Res. 2019, 17, 1613–1626. [Google Scholar] [CrossRef]
- Li, H.; Argani, P.; Halper-Stromberg, E.; Lotan, T.L.; Merino, M.J.; Reuter, V.E.; Matoso, A. Positive GPNMB Immunostaining Differentiates Renal Cell Carcinoma With Fibromyomatous Stroma Associated With TSC1/2/MTOR Alterations From Others. Am. J. Surg. Pathol. 2023, 47, 1267–1273. [Google Scholar] [CrossRef]
- Salles, D.C.; Asrani, K.; Woo, J.; Vidotto, T.; Liu, H.B.; Vidal, I.; Matoso, A.; Netto, G.J.; Argani, P.; Lotan, T.L. GPNMB expression identifies TSC1/2/mTOR-associated and MiT family translocation-driven renal neoplasms. J. Pathol. 2022, 257, 158–171. [Google Scholar] [CrossRef]
- Argani, P.; Lal, P.; Hutchinson, B.; Lui, M.Y.; Reuter, V.E.; Ladanyi, M. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: A sensitive and specific immunohistochemical assay. Am. J. Surg. Pathol. 2003, 27, 750–761. [Google Scholar] [CrossRef]
- Sharain, R.F.; Gown, A.M.; Greipp, P.T.; Folpe, A.L. Immunohistochemistry for TFE3 lacks specificity and sensitivity in the diagnosis of TFE3-rearranged neoplasms: A comparative, 2-laboratory study. Hum. Pathol. 2019, 87, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Choi, Y.; Jeong, H.Y.; Lee, K.; Chae, J.Y.; Moon, K.C. Usefulness of a break-apart FISH assay in the diagnosis of Xp11.2 translocation renal cell carcinoma. Virchows Arch. 2011, 459, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Rao, Q.; Williamson, S.R.; Zhang, S.; Eble, J.N.; Grignon, D.J.; Wang, M.; Zhou, X.-J.; Huang, W.; Tan, P.-H.; Maclennan, G.T.; et al. TFE3 break-apart FISH has a higher sensitivity for Xp11.2 translocation-associated renal cell carcinoma compared with TFE3 or cathepsin K immunohistochemical staining alone: Expanding the morphologic spectrum. Am. J. Surg. Pathol. 2013, 37, 804–815. [Google Scholar] [CrossRef] [PubMed]
- Green, W.M.; Yonescu, R.; Morsberger, L.; Morris, K.; Netto, G.J.; Epstein, J.I.; Illei, P.B.; Allaf, M.; Ladanyi, M.; Griffin, C.A.; et al. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am. J. Surg. Pathol. 2013, 37, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, D.; Roy, S.; Quiroga-Garza, G.; Cieply, K.; Mahaffey, A.L.; Bastacky, S.; Dhir, R.; Parwani, A. V Validation and utilization of a TFE3 break-apart FISH assay for Xp11.2 translocation renal cell carcinoma and alveolar soft part sarcoma. Diagn. Pathol. 2015, 10, 179. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Zhang, L.; Reuter, V.E.; Tickoo, S.K.; Antonescu, C.R. RBM10-TFE3 Renal Cell Carcinoma: A Potential Diagnostic Pitfall Due to Cryptic Intrachromosomal Xp11.2 Inversion Resulting in False-negative TFE3 FISH. Am. J. Surg. Pathol. 2017, 41, 655–662. [Google Scholar] [CrossRef]
- Kato, I.; Furuya, M.; Baba, M.; Kameda, Y.; Yasuda, M.; Nishimoto, K.; Oyama, M.; Yamasaki, T.; Ogawa, O.; Niino, H.; et al. RBM10-TFE3 renal cell carcinoma characterised by paracentric inversion with consistent closely split signals in break-apart fluorescence in-situ hybridisation: Study of 10 cases and a literature review. Histopathology 2019, 75, 254–265. [Google Scholar] [CrossRef]
- Xia, Q.-Y.; Wang, Z.; Chen, N.; Gan, H.-L.; Teng, X.-D.; Shi, S.-S.; Wang, X.; Wei, X.; Ye, S.-B.; Li, R.; et al. Xp11.2 translocation renal cell carcinoma with NONO-TFE3 gene fusion: Morphology, prognosis, and potential pitfall in detecting TFE3 gene rearrangement. Mod. Pathol. 2017, 30, 416–426. [Google Scholar] [CrossRef]
- Harada, S.; Caliò, A.; Janowski, K.M.; Morlote, D.; Rodriguez Pena, M.D.; Canete-Portillo, S.; Harbi, D.; DeFrank, G.; Magi-Galluzzi, C.; Netto, G.J.; et al. Diagnostic utility of one-stop fusion gene panel to detect TFE3/TFEB gene rearrangement and amplification in renal cell carcinomas. Mod. Pathol. 2021, 34, 2055–2063. [Google Scholar] [CrossRef]
- Tretiakova, M.S.; Wang, W.; Wu, Y.; Tykodi, S.S.; True, L.; Liu, Y.J. Gene fusion analysis in renal cell carcinoma by FusionPlex RNA-sequencing and correlations of molecular findings with clinicopathological features. Genes Chromosomes Cancer 2020, 59, 40–49. [Google Scholar] [CrossRef]
- Martignoni, G.; Pea, M.; Reghellin, D.; Zamboni, G.; Bonetti, F. PEComas: The past, the present and the future. Virchows Arch. 2008, 452, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Bonetti, F.; Pea, M.; Martignoni, G.; Doglioni, C.; Zamboni, G.; Capelli, P.; Rimondi, P.; Andrion, A. Clear cell (“sugar”) tumor of the lung is a lesion strictly related to angiomyolipoma--the concept of a family of lesions characterized by the presence of the perivascular epithelioid cells (PEC). Pathology 1994, 26, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Gaffey, M.J.; Mills, S.E.; Zarbo, R.J.; Weiss, L.M.; Gown, A.M. Clear cell tumor of the lung. Immunohistochemical and ultrastructural evidence of melanogenesis. Am. J. Surg. Pathol. 1991, 15, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Stone, C.H.; Lee, M.W.; Amin, M.B.; Yaziji, H.; Gown, A.M.; Ro, J.Y.; Têtu, B.; Paraf, F.; Zarbo, R.J. Renal angiomyolipoma: Further immunophenotypic characterization of an expanding morphologic spectrum. Arch. Pathol. Lab. Med. 2001, 125, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Bonetti, F.; Chiodera, P.L.; Pea, M.; Martignoni, G.; Bosi, F.; Zamboni, G.; Mariuzzi, G.M. Transbronchial biopsy in lymphangiomyomatosis of the lung. HMB45 for diagnosis. Am. J. Surg. Pathol. 1993, 17, 1092–1102. [Google Scholar] [CrossRef]
- Tsui, W.M.; Ng, I.O.; Colombari, R.; Pea, M. Hepatic angiomyolipomas. Histopathology 1993, 22, 602–603. [Google Scholar] [CrossRef]
- Green, A.J.; Sepp, T.; Yates, J.R. Clonality of tuberous sclerosis harmatomas shown by non-random X-chromosome inactivation. Hum. Genet. 1996, 97, 240–243. [Google Scholar] [CrossRef]
- Carbonara, C.; Longa, L.; Grosso, E.; Mazzucco, G.; Borrone, C.; Garrè, M.L.; Brisigotti, M.; Filippi, G.; Scabar, A.; Giannotti, A.; et al. Apparent preferential loss of heterozygosity at TSC2 over TSC1 chromosomal region in tuberous sclerosis hamartomas. Genes Chromosomes Cancer 1996, 15, 18–25. [Google Scholar] [CrossRef]
- Qin, W.; Bajaj, V.; Malinowska, I.; Lu, X.; MacConaill, L.; Wu, C.-L.; Kwiatkowski, D.J. Angiomyolipoma have common mutations in TSC2 but no other common genetic events. PLoS ONE 2011, 6, e24919. [Google Scholar] [CrossRef]
- Giannikou, K.; Malinowska, I.A.; Pugh, T.J.; Yan, R.; Tseng, Y.-Y.; Oh, C.; Kim, J.; Tyburczy, M.E.; Chekaluk, Y.; Liu, Y.; et al. Whole Exome Sequencing Identifies TSC1/TSC2 Biallelic Loss as the Primary and Sufficient Driver Event for Renal Angiomyolipoma Development. PLoS Genet. 2016, 12, e1006242. [Google Scholar] [CrossRef]
- Alesi, N.; Akl, E.W.; Khabibullin, D.; Liu, H.-J.; Nidhiry, A.S.; Garner, E.R.; Filippakis, H.; Lam, H.C.; Shi, W.; Viswanathan, S.R.; et al. TSC2 regulates lysosome biogenesis via a non-canonical RAGC and TFEB-dependent mechanism. Nat. Commun. 2021, 12, 4245. [Google Scholar] [CrossRef] [PubMed]
- Alesi, N.; Khabibullin, D.; Rosenthal, D.M.; Akl, E.W.; Cory, P.M.; Alchoueiry, M.; Salem, S.; Daou, M.; Gibbons, W.F.; Chen, J.A.; et al. TFEB drives mTORC1 hyperactivation and kidney disease in Tuberous Sclerosis Complex. Nat. Commun. 2024, 15, 406. [Google Scholar] [CrossRef] [PubMed]
- Pastore, N.; Ballabio, A.; Brunetti-Pierri, N. Autophagy master regulator TFEB induces clearance of toxic SERPINA1/α-1-antitrypsin polymers. Autophagy 2013, 9, 1094–1096. [Google Scholar] [CrossRef] [PubMed]
- Caliò, A.; Brunelli, M.; Gobbo, S.; Argani, P.; Munari, E.; Netto, G.; Martignoni, G. Cathepsin K: A Novel Diagnostic and Predictive Biomarker for Renal Tumors. Cancers 2021, 13, 2441. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Aulmann, S.; Karanjawala, Z.; Fraser, R.B.; Ladanyi, M.; Rodriguez, M.M. Melanotic Xp11 translocation renal cancers: A distinctive neoplasm with overlapping features of PEComa, carcinoma, and melanoma. Am. J. Surg. Pathol. 2009, 33, 609–619. [Google Scholar] [CrossRef]
- Argani, P.; Aulmann, S.; Illei, P.B.; Netto, G.J.; Ro, J.; Cho, H.; Dogan, S.; Ladanyi, M.; Martignoni, G.; Goldblum, J.R.; et al. A distinctive subset of PEComas harbors TFE3 gene fusions. Am. J. Surg. Pathol. 2010, 34, 1395–1406. [Google Scholar] [CrossRef]
- Argani, P.; Gross, J.M.; Baraban, E.; Rooper, L.M.; Chen, S.; Lin, M.-T.; Gocke, C.; Agaimy, A.; Lotan, T.; Suurmeijer, A.J.H.; et al. TFE3-Rearranged PEComa/PEComa-like Neoplasms: Report of 25 New Cases Expanding the Clinicopathologic Spectrum and Highlighting its Association With Prior Exposure to Chemotherapy. Am. J. Surg. Pathol. 2024, 48, 777–789. [Google Scholar] [CrossRef]
- Rao, Q.; Shen, Q.; Xia, Q.; Wang, Z.; Liu, B.; Shi, S.; Shi, Q.; Yin, H.; Wu, B.; Ye, S.; et al. PSF/SFPQ is a very common gene fusion partner in TFE3 rearrangement-associated perivascular epithelioid cell tumors (PEComas) and melanotic Xp11 translocation renal cancers: Clinicopathologic, immunohistochemical, and molecular characteristics suggesting. Am. J. Surg. Pathol. 2015, 39, 1181–1196. [Google Scholar] [CrossRef]
- Schmiester, M.; Dolnik, A.; Kornak, U.; Pfitzner, B.; Hummel, M.; Treue, D.; Hartmann, A.; Agaimy, A.; Weyerer, V.; Lekaj, A.; et al. TFE3 activation in a TSC1-altered malignant PEComa: Challenging the dichotomy of the underlying pathogenic mechanisms. J. Pathol. Clin. Res. 2021, 7, 3–9. [Google Scholar] [CrossRef]
- Ohe, C.; Kuroda, N.; Hes, O.; Michal, M.; Vanecek, T.; Grossmann, P.; Tanaka, Y.; Tanaka, M.; Inui, H.; Komai, Y.; et al. A renal epithelioid angiomyolipoma/perivascular epithelioid cell tumor with TFE3 gene break visualized by FISH. Med. Mol. Morphol. 2012, 45, 234–237. [Google Scholar] [CrossRef]
- Shen, Q.; Rao, Q.; Xia, Q.-Y.; Yu, B.; Shi, Q.-L.; Zhang, R.-S.; Zhou, X.-J. Perivascular epithelioid cell tumor (PEComa) with TFE3 gene rearrangement: Clinicopathological, immunohistochemical, and molecular features. Virchows Arch. 2014, 465, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Zhang, L.; Sung, Y.-S.; White, M.J.; Miller, K.; Hopkins, M.; Small, D.; Pratilas, C.A.; Swanson, D.; Dickson, B.; et al. A novel RBMX-TFE3 gene fusion in a highly aggressive pediatric renal perivascular epithelioid cell tumor. Genes Chromosomes Cancer 2020, 59, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-T.; Fang, R.; Zhang, R.-S.; Ye, S.-B.; Li, R.; Wang, X.; Pan, R.; Liu, C.; Chen, J.-Y.; Zhao, M.; et al. Malignant melanotic Xp11 neoplasms exhibit a clinicopathologic spectrum and gene expression profiling akin to alveolar soft part sarcoma: A proposal for reclassification. J. Pathol. 2020, 251, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-T.; Xia, Q.-Y.; Ni, H.; Ye, S.-B.; Li, R.; Wang, X.; Shi, S.-S.; Zhou, X.-J.; Rao, Q. SFPQ/PSF-TFE3 renal cell carcinoma: A clinicopathologic study emphasizing extended morphology and reviewing the differences between SFPQ-TFE3 RCC and the corresponding mesenchymal neoplasm despite an identical gene fusion. Hum. Pathol. 2017, 63, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Wobker, S.E.; Gross, J.M.; Matoso, A.; Fletcher, C.D.M.; Antonescu, C.R. PEComa-like Neoplasms Characterized by ASPSCR1-TFE3 Fusion: Another Face of TFE3-related Mesenchymal Neoplasia. Am. J. Surg. Pathol. 2022, 46, 1153–1159. [Google Scholar] [CrossRef]
- Sun, G.; Chen, J.; Liang, J.; Yin, X.; Zhang, M.; Yao, J.; He, N.; Armstrong, C.M.; Zheng, L.; Zhang, X.; et al. Integrated exome and RNA sequencing of TFE3-translocation renal cell carcinoma. Nat. Commun. 2021, 12, 5262. [Google Scholar] [CrossRef]
- Yu, D.; Huang, C.-J.; Tucker, H.O. Established and Evolving Roles of the Multifunctional Non-POU Domain-Containing Octamer-Binding Protein (NonO) and Splicing Factor Proline- and Glutamine-Rich (SFPQ). J. Dev. Biol. 2024, 12, 3. [Google Scholar] [CrossRef]
- Zhang, S.; Cooper, J.A.; Chong, Y.S.; Naveed, A.; Mayoh, C.; Jayatilleke, N.; Liu, T.; Amos, S.; Kobelke, S.; Marshall, A.C.; et al. NONO enhances mRNA processing of super-enhancer-associated GATA2 and HAND2 genes in neuroblastoma. EMBO Rep. 2023, 24, e54977. [Google Scholar] [CrossRef]
- Bi, O.; Anene, C.A.; Nsengimana, J.; Shelton, M.; Roberts, W.; Newton-Bishop, J.; Boyne, J.R. SFPQ promotes an oncogenic transcriptomic state in melanoma. Oncogene 2021, 40, 5192–5203. [Google Scholar] [CrossRef]
- de Silva, H.C.; Lin, M.Z.; Phillips, L.; Martin, J.L.; Baxter, R.C. IGFBP-3 interacts with NONO and SFPQ in PARP-dependent DNA damage repair in triple-negative breast cancer. Cell. Mol. Life Sci. 2019, 76, 2015–2030. [Google Scholar] [CrossRef]
- Lu, Y.; Lin, Y.; Zhang, X.; Yan, J.; Kong, Z.; Zhang, L.; Wang, C.; Huang, Y.; Zhao, S.; Li, Y. LncRNA-AP006284.1 promotes prostate cancer cell growth and motility by forming RNA-DNA triplexes and recruiting GNL3/SFPQ complex to facilitate RASSF7 transcription. Genes. Dis. 2023, 10, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhong, W.; Lin, Q.; Ye, Y.; Li, S.; Chen, H.; Liu, H.; Xu, L.; Zhuang, W.; Chen, S.; et al. SFPQ promotes the proliferation, migration and invasion of hepatocellular carcinoma cells and is associated with poor prognosis. Am. J. Cancer Res. 2023, 13, 2269–2284. [Google Scholar] [PubMed]
- Ji, Q.; Zhang, L.; Liu, X.; Zhou, L.; Wang, W.; Han, Z.; Sui, H.; Tang, Y.; Wang, Y.; Liu, N.; et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br. J. Cancer 2014, 111, 736–748. [Google Scholar] [CrossRef]
- Cheng, M.; Jiang, Y.; Xu, J.; Mentis, A.-F.A.; Wang, S.; Zheng, H.; Sahu, S.K.; Liu, L.; Xu, X. Spatially resolved transcriptomics: A comprehensive review of their technological advances, applications, and challenges. J. Genet. Genom. 2023, 50, 625–640. [Google Scholar] [CrossRef] [PubMed]
- Beksac, A.T.; Paulucci, D.J.; Blum, K.A.; Yadav, S.S.; Sfakianos, J.P.; Badani, K.K. Heterogeneity in renal cell carcinoma. Urol. Oncol. 2017, 35, 507–515. [Google Scholar] [CrossRef]
- Vahdat, L.T.; Schmid, P.; Forero-Torres, A.; Blackwell, K.; Telli, M.L.; Melisko, M.; Möbus, V.; Cortes, J.; Montero, A.J.; Ma, C.; et al. Glembatumumab vedotin for patients with metastatic, gpNMB overexpressing, triple-negative breast cancer (“METRIC”): A randomized multicenter study. NPJ Breast Cancer 2021, 7, 57. [Google Scholar] [CrossRef]
- Ott, P.A.; Pavlick, A.C.; Johnson, D.B.; Hart, L.L.; Infante, J.R.; Luke, J.J.; Lutzky, J.; Rothschild, N.E.; Spitler, L.E.; Cowey, C.L.; et al. A phase 2 study of glembatumumab vedotin, an antibody-drug conjugate targeting glycoprotein NMB, in patients with advanced melanoma. Cancer 2019, 125, 1113–1123. [Google Scholar] [CrossRef]
- Kopp, L.M.; Malempati, S.; Krailo, M.; Gao, Y.; Buxton, A.; Weigel, B.J.; Hawthorne, T.; Crowley, E.; Moscow, J.A.; Reid, J.M.; et al. Phase II trial of the glycoprotein non-metastatic B-targeted antibody-drug conjugate, glembatumumab vedotin (CDX-011), in recurrent osteosarcoma AOST1521: A report from the Children’s Oncology Group. Eur. J. Cancer 2019, 121, 177–183. [Google Scholar] [CrossRef]
- Pal, S.K.; Tangen, C.; Thompson, I.M.J.; Balzer-Haas, N.; George, D.J.; Heng, D.Y.C.; Shuch, B.; Stein, M.; Tretiakova, M.; Humphrey, P.; et al. A comparison of sunitinib with cabozantinib, crizotinib, and savolitinib for treatment of advanced papillary renal cell carcinoma: A randomised, open-label, phase 2 trial. Lancet 2021, 397, 695–703. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caliò, A.; Marletta, S.; Brunelli, M.; Antonini, P.; Martelli, F.M.; Marcolini, L.; Stefanizzi, L.; Martignoni, G. TFE3-Rearranged Tumors of the Kidney: An Emerging Conundrum. Cancers 2024, 16, 3396. https://doi.org/10.3390/cancers16193396
Caliò A, Marletta S, Brunelli M, Antonini P, Martelli FM, Marcolini L, Stefanizzi L, Martignoni G. TFE3-Rearranged Tumors of the Kidney: An Emerging Conundrum. Cancers. 2024; 16(19):3396. https://doi.org/10.3390/cancers16193396
Chicago/Turabian StyleCaliò, Anna, Stefano Marletta, Matteo Brunelli, Pietro Antonini, Filippo Maria Martelli, Lisa Marcolini, Lavinia Stefanizzi, and Guido Martignoni. 2024. "TFE3-Rearranged Tumors of the Kidney: An Emerging Conundrum" Cancers 16, no. 19: 3396. https://doi.org/10.3390/cancers16193396
APA StyleCaliò, A., Marletta, S., Brunelli, M., Antonini, P., Martelli, F. M., Marcolini, L., Stefanizzi, L., & Martignoni, G. (2024). TFE3-Rearranged Tumors of the Kidney: An Emerging Conundrum. Cancers, 16(19), 3396. https://doi.org/10.3390/cancers16193396







