Unveiling the Inflammatory Landscape of Recurrent Glioblastoma through Histological-Based Assessments
Abstract
Simple Summary
Abstract
1. Introduction
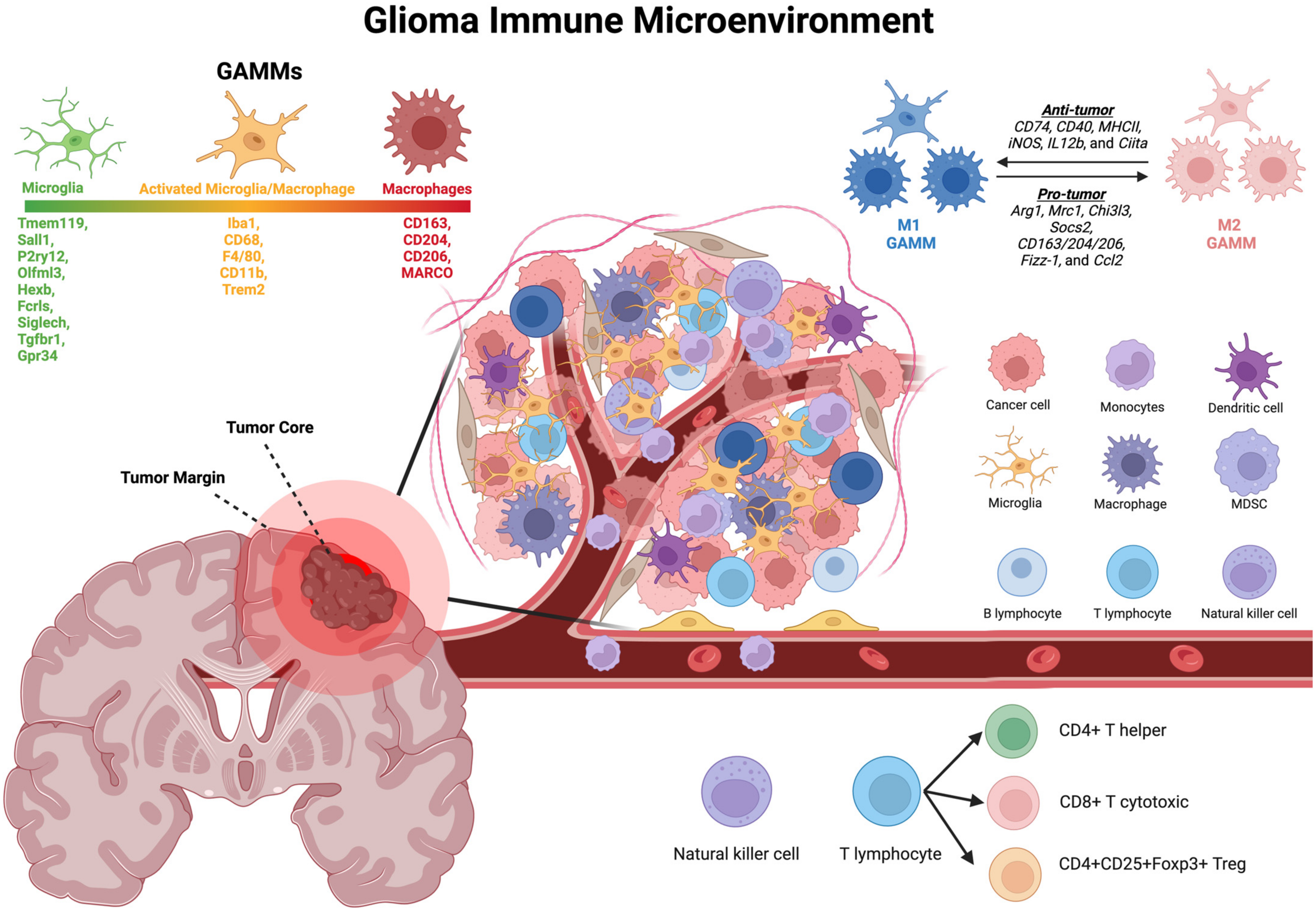
2. The Immune Microenvironment: Key Players
3. Glioma-Associated Macrophages and Microglia (GAMM)
3.1. Origins
3.2. Histological Identification and Presence in rGBM
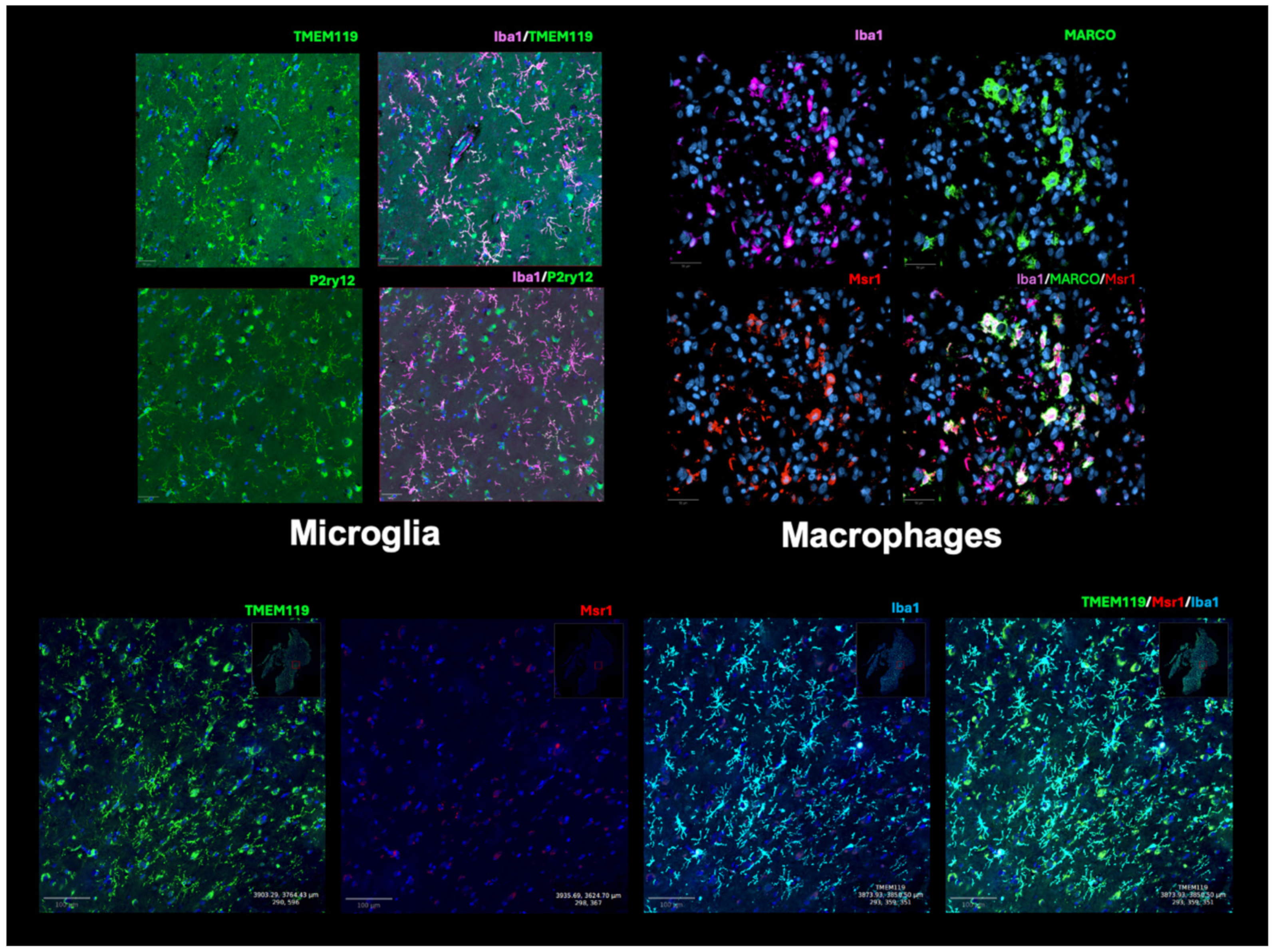
3.3. Microglia Markers
3.3.1. P2ry12 and Tmem119
3.3.2. Iba1
3.3.3. Trem2
3.4. Macrophage, MDSC, and General Monocyte Markers
3.4.1. CD68 and CD11b
3.4.2. CD163, CD204, CD206
3.4.3. MARCO
| Study | Patient Population | Setting | Markers | Analysis | Study Result | Result Implication |
|---|---|---|---|---|---|---|
| Lu-Emerson et al., 2013 [18] | 20 patients, rGBM | 12 patients received antiangiogenic treatment and chemoradiation, 8 patients received chemotherapy and/or radiotherapy (no antiangiogenic treatment) | anti-CD68, anti-CD11b, anti-CD163, anti-CD14, anti-CD45, anti-CSF1R | IHC | Increased CD68 in tumor bulk (p < 0.01) and infiltrative regions (p = 0.02). Increased CD11b+ in tumor bulk (p < 0.01) and trend increase in I filtrate region (p = 0.09). Increased CD163 in tumor bulk (p = 0.09) in therapy group. Sequencing validated by IHC. | Inflammation induces mesenchymal-like state; macrophages are enriched in the vicinity of MES-like glioblastoma cells compared with OPC-like cells. |
| Gill et al., 2014 [2] | 49 pGBM, 19rGBM | Characterized core vs. margins of p/rGBM | anti-CD44, anti-IBA1, anti-CD68, anti-SOX2 | IHC | Core samples had higher cellularity and contained a higher amount of glomeruloid-type vascular necrosis than margins (p < 0.00001). Margin biopsies contained more NeuN+ neurons than core biopsies. | Non-neoplastic cells are a major component of the non-enhancing margins of p/rGBM tumors. |
| Wang et al., 2017 [21] | 37 pGBM, 42 rGBM | Immune cell presence of p/rGBM | anti-AIF1, anti-NF1, anti-GeneTex | IHC | Increase in Iba1 (AIF1) at recurrence. | Increased inflammatory TME can drive mesenchymal-like tumor cell population. |
| Miyazaki et al., 2017 [85] | 16 patients with both pGBM and rGBM samples | Molecular expression of immune environment in p/rGBM | anti-Ki-67, anti-TP53, anti-MHC class I, anti-MHC class II, anti-IDH-1R132H, anti-CD3, anti-CD8, anti-CD20, anti-CD45RO, anti-PD-L1, anti-Granzyme B, anti-PD-1, and anti-ATRX. | IHC | CD3, CD8, and PD-1 staining scores were significantly increased in rGBM specimens compared with pGBM specimens (p ≤ 0.05). | Stimulations, including AFTV treatment, induce the recruitment of many T cell type TILs, consisting mainly of CD8+ T cells. High CD8 and PD-1 scores after the secondary surgery were significantly poor prognostic factors of survival after second resection as high PD-1 score (p < 0.05 each), while high CD3 score trended as a poor prognostic factor (p = 0.065). In tumor markers, high PD-L1 grading trended as a favorable prognostic factor of PFS second resection (p = 0.095). |
| Rahman et al., 2018 [64] | 38 pGBM, 12 rGBM | Immune markers of p/rGBM | anti-FOXP3, anti-CD70, anti-CTLA-4, anti-PD-L1, anti-PD1, anti-CD163, anti-CD68 | IHC | No significant difference was identified in any immune marker between the primary and recurrent GBM. | No significant difference was identified in any immune marker between the primary and recurrent GBM. |
| Kaffes et al., 2019 [46] | 48 pGBM, 8 rGBM | Mesenchymal vs. pro-neural GBM in pGBM and rGBM together | Anti-IBA1; anti-human FOXP3; anti-human CD8; anti-human CD3 | IHC/IF | Mesenchymal subtype of GBM showed the highest presence of TAM, CD8+, CD3+, and FOXP3+ T cells. | High expression levels of FOXP3 and CD3G were associated with improved overall survival. High AIF1 expression levels confer a worse prognosis in the PN subtype but bestow a survival benefit in MES tumors. |
| Cloughesy et al., 2019 [86] | 30 rGBM | 15 rGBM patients treated with anti-PD-1 immunotherapy with SOC, 15 rGBM patients treated with SOC | anti-CD8, anti-CD45, anti-PD-1, anti-PD-L1 | IF | Density of CD8+ T cells increased dramatically in the neoadjuvant (pembrolizumab) group. | Neoadjuvant administration of PD-1 blockade enhances the local and systemic anti-tumor immune response. |
| Liesche-Starnecker et al., 2020 [14] | 21 patients with both pGBM and rGBM samples | Intra-tumoral heterogeneity and immune environment in p/rGBM | anti-EGFR, anti-GFAP, anti-Iba1, anti-Olig2, anti-p53, anti-Mib1 | IHC/IF | Positive correlation of ALDH1A3 and Iba1 and increased levels of both in the progression (p = 0.000 and p = 0.001). | Temporal heterogeneity in GBM exists and potentially provides information important for prognosis and therapy resistance. For the recurrent tumors, a clear dominance of the mesenchymal/microglial-dominant subtype was observed. |
| Fu et al., 2020 [20] | 13 pGBM, 3 rGBM | Quantification of TAMs in p/rGBM | anti-CD45, anti-CD68, anti-TNFa, anti-IDO | IF | CD4+/CD8+ T cells secrete more IL-10, IDO, TGFβ, T-bet, and TNFβ while expressing higher levels of PD-1, LAG-3, and TIM-3 than regulatory T cells, CD4+ T cells, and CD8+ T cells in p/rGBM | Provides further understanding of the immune environment of p/rGBM. |
| Miyazaki et al., 2020 [73] | 6 paired p/rGBM | Standard radiochemotherapy and both standard and immunotherapy for assessment of preclinical models and pGBM and rGBM | anti-CD163 | IHC | Infiltration of CD163-positive cells increased 2.7× in recurrent GBM specimens from patients treated with immunotherapy, although a 1.1× increase was observed in a pair of specimens from GBM patients treated with only standard therapy. | Anti-PD-L1 antibody treatment activates infiltration of CD163-positive Mϕ, usually considered as an M2 Mϕ marker, in a TMZ-resistant murine glioma model and also pGBM/rGBM tissue. |
| Tang et al., 2021 [44] | 42 rGBM | LRRC15 characterization in the tumor environment | anti-LRRC15, anti-CD206 | IHC | LRRC15 expression was positively correlated with CD206 expression in recurrent GBM (p = 0.001). | LRRC15 expression was positively correlated with CD206 expression in recurrent GBM. High expression levels of LLRC15 promote poor prognosis of recurrent GBM patients. |
| Magri et al. 2021 [63] | 44 pGBM, 19 rGBM | pGBM (pre-treatment) and rGBM (post-treatment) with radiochemotherapy and temozolomide | anti-CD68, anti-CD8 | anti-CD68, anti-CD8 | In relapsing GBM, the presence of tumor macrophages redistributed in the two distinct areas, as the presence of BMDMs increased in the marginal area (p = 0.013) and MG decreased in the central zone (p = 0.002) Found no difference in CD68 cell density between primary and recurrent tumors; recurrent tumors demonstrated increased CD8+ cells (p = 0.015) | Increased recruitment of suppressive BMDMs in relapsing GBM. |
| Wang et al., 2022 [87] | 13 pGBM, 11 rGBM (5 of each is the same patient) | Immune environment quantification in p/rGBM | anti-CD4, anti-CD8, anti-CD68, anti-PD-1, and anti-PD-L1 | IHC/IF | Primary GBM typically had low levels of CD8+ T-cell abundance, and CD8+ T cells were sparse, isolated, and frequently confined to the perivascular space, while matched rGBM showed robust T-cell invasion of the cellular tumor. | We found that T-cell abundance was correlated with a significant increase in survival. |
| Alanio et al., 2022 [43] | 14 pGBM, 13 rGBM | Immune environment in relation to survival in p/rGBM | anti-CD3, anti-Ki-67, anti-CD8, anti-Foxp3, anti-CD68, anti-CD163, anti-EGFR, anti-p53, anti-HLA-DR | IHC | Three groups: Myeloid I (tissue-resident microglial-derived TAM), Myeloid II (type-1 myeloid dendritic cells (cDC1), and CD163hi monocyte-derived TAM), Myeloid III (CD163low monocyte-derived TAM). Higher myeloid II and III in recurrent cases. Increase in the proportion of CD80-expressing myeloid cells detected in de novo tumors (p = 0.01) Similar overall content of CD8+ T cells in perivascular regions in paired primary and recurrent but increased number of perivascular regions associated with CD8+ T cells seen in rGBM samples. Perivascular T-cell high regions in rGBM displayed a lower density of FOXP3+ Treg cells and a significantly higher ratio of CD8+ T cells to FOXP3+. | Findings identify the spatial distribution of T cells rather than their abundance as a potential key immunological determinant that is associated with the evolution and pathogenesis of GBM. Enrichment of activated T cells in perivascular regions may be a determinant of longer survival in patients with rGBM T-cell compartment, especially in the perivascular regions of the tumor, in some patients with rGBM who may be polarized toward an antitumor-activated phenotype with clinical implications. |
| Al Dalahmah et al., 2023 [3] | 45 primary and rGBM | Deconvolution of landscape in primary and recurrent GBM | anti-CD68 | IHC | Specific cell types/transcriptional states colocalize in “tissue-states” defined by Sox-2, CD68, and NeuN staining. Significant enrichment in tissue state B (high CD68) gene signatures in rGBM samples compared to pGBM. | Compared to primary GBM, rGBM has increased CD68+ staining and a mesenchymal phenotype associated with worse survival. |
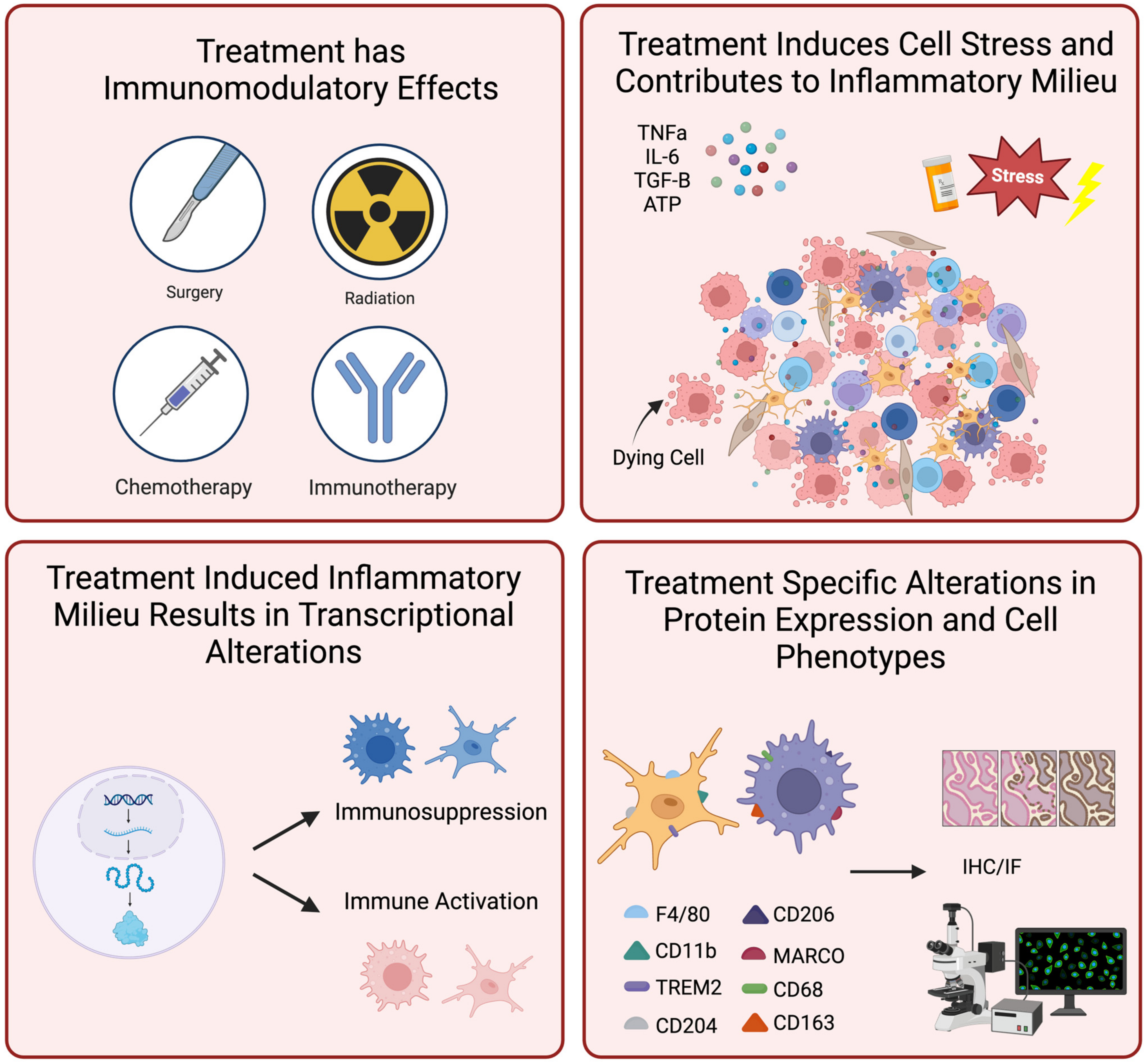
4. The Inflammatory Landscape in the Context of Standard of Care Treatment
4.1. Immune TME Baseline Can Influence Glioma Resistance to Standard of Care Treatments
4.2. TMZ Drives Inflammatory Changes in the Post-Treatment Setting
4.3. Radiation Therapy Can Drive Glioma Toward a Resistant Phenotype
4.4. The M2-Polarized GAMMs May Promote Recurrence in Post-Treatment Glioma
5. Tissue Sampling of a Heterogeneous rGBM Tumor Microenvironment
6. Harnessing the Histopathologic Inflammatory Landscape for rGBM Treatments
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Gill, B.J.; Pisapia, D.J.; Malone, H.R.; Goldstein, H.; Lei, L.; Sonabend, A.; Yun, J.; Samanamud, J.; Sims, J.S.; Banu, M.; et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc. Natl. Acad. Sci. USA 2014, 111, 12550–12555. [Google Scholar] [CrossRef]
- Al-Dalahmah, O.; Argenziano, M.G.; Kannan, A.; Mahajan, A.; Furnari, J.; Paryani, F.; Boyett, D.; Save, A.; Humala, N.; Khan, F.; et al. Re-convolving the compositional landscape of primary and recurrent glioblastoma reveals prognostic and targetable tissue states. Nat. Commun. 2023, 14, 2586. [Google Scholar] [CrossRef]
- Campos, B.; Olsen, L.R.; Urup, T.; Poulsen, H.S. A comprehensive profile of recurrent glioblastoma. Oncogene 2016, 35, 5819–5825. [Google Scholar] [CrossRef]
- Hoogstrate, Y.; Draaisma, K.; Ghisai, S.A.; van Hijfte, L.; Barin, N.; de Heer, I.; Coppieters, W.; van den Bosch, T.P.P.; Bolleboom, A.; Gao, Z.; et al. Transcriptome analysis reveals tumor microenvironment changes in glioblastoma. Cancer Cell 2023, 41, 678–692.e677. [Google Scholar] [CrossRef]
- Wang, W.; Tugaoen, J.D.; Fadda, P.; Toland, A.E.; Ma, Q.; Elder, J.B.; Giglio, P.; James Cancer Center Integrated Neuro-Oncology, T.; Otero, J.J. Glioblastoma pseudoprogression and true progression reveal spatially variable transcriptional differences. Acta Neuropathol. Commun. 2023, 11, 192. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef]
- Wang, E.J.; Chen, J.S.; Jain, S.; Morshed, R.A.; Haddad, A.F.; Gill, S.; Beniwal, A.S.; Aghi, M.K. Immunotherapy Resistance in Glioblastoma. Front. Genet. 2021, 12, 750675. [Google Scholar] [CrossRef]
- Tomaszewski, W.; Sanchez-Perez, L.; Gajewski, T.F.; Sampson, J.H. Brain Tumor Microenvironment and Host State: Implications for Immunotherapy. Clin. Cancer Res. 2019, 25, 4202–4210. [Google Scholar] [CrossRef]
- Karachi, A.; Dastmalchi, F.; Mitchell, D.A.; Rahman, M. Temozolomide for immunomodulation in the treatment of glioblastoma. Neuro-Oncology 2018, 20, 1566–1572. [Google Scholar] [CrossRef]
- Schmitt, M.J.; Company, C.; Dramaretska, Y.; Barozzi, I.; Göhrig, A.; Kertalli, S.; Großmann, M.; Naumann, H.; Sanchez-Bailon, M.P.; Hulsman, D.; et al. Phenotypic Mapping of Pathologic Cross-Talk between Glioblastoma and Innate Immune Cells by Synthetic Genetic Tracing. Cancer Discov. 2021, 11, 754–777. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.P.; Sells, B.E.; Haque, S.J.; Chakravarti, A. Tumor Heterogeneity in Glioblastomas: From Light Microscopy to Molecular Pathology. Cancers 2021, 13, 761. [Google Scholar] [CrossRef] [PubMed]
- Gromeier, M.; Brown, M.C.; Zhang, G.; Lin, X.; Chen, Y.; Wei, Z.; Beaubier, N.; Yan, H.; He, Y.; Desjardins, A.; et al. Very low mutation burden is a feature of inflamed recurrent glioblastomas responsive to cancer immunotherapy. Nat. Commun. 2021, 12, 352. [Google Scholar] [CrossRef]
- Liesche-Starnecker, F.; Mayer, K.; Kofler, F.; Baur, S.; Schmidt-Graf, F.; Kempter, J.; Prokop, G.; Pfarr, N.; Wei, W.; Gempt, J.; et al. Immunohistochemically Characterized Intratumoral Heterogeneity Is a Prognostic Marker in Human Glioblastoma. Cancers 2020, 12, 2964. [Google Scholar] [CrossRef]
- Martinez-Lage, M.; Lynch, T.M.; Bi, Y.; Cocito, C.; Way, G.P.; Pal, S.; Haller, J.; Yan, R.E.; Ziober, A.; Nguyen, A.; et al. Immune landscapes associated with different glioblastoma molecular subtypes. Acta Neuropathol. Commun. 2019, 7, 203. [Google Scholar] [CrossRef]
- Paulus, W.; Peiffer, J. Intratumoral histologic heterogeneity of gliomas. A quantitative study. Cancer 1989, 64, 442–447. [Google Scholar] [CrossRef]
- Rahimi Koshkaki, H.; Minasi, S.; Ugolini, A.; Trevisi, G.; Napoletano, C.; Zizzari, I.G.; Gessi, M.; Giangaspero, F.; Mangiola, A.; Nuti, M.; et al. Immunohistochemical Characterization of Immune Infiltrate in Tumor Microenvironment of Glioblastoma. J. Pers. Med. 2020, 10, 112. [Google Scholar] [CrossRef]
- Lu-Emerson, C.; Snuderl, M.; Kirkpatrick, N.D.; Goveia, J.; Davidson, C.; Huang, Y.; Riedemann, L.; Taylor, J.; Ivy, P.; Duda, D.G.; et al. Increase in tumor-associated macrophages after antiangiogenic therapy is associated with poor survival among patients with recurrent glioblastoma. Neuro-Oncology 2013, 15, 1079–1087. [Google Scholar] [CrossRef]
- Spinazzi, E.F.; Argenziano, M.G.; Upadhyayula, P.S.; Banu, M.A.; Neira, J.A.; Higgins, D.M.O.; Wu, P.B.; Pereira, B.; Mahajan, A.; Humala, N.; et al. Chronic convection-enhanced delivery of topotecan for patients with recurrent glioblastoma: A first-in-patient, single-centre, single-arm, phase 1b trial. Lancet Oncol. 2022, 23, 1409–1418. [Google Scholar] [CrossRef]
- Fu, W.; Wang, W.; Li, H.; Jiao, Y.; Huo, R.; Yan, Z.; Wang, J.; Wang, S.; Wang, J.; Chen, D.; et al. Single-Cell Atlas Reveals Complexity of the Immunosuppressive Microenvironment of Initial and Recurrent Glioblastoma. Front. Immunol. 2020, 11, 835. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56.E6. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Chanoch-Myers, R.; Mathewson, N.D.; Myskiw, C.; Atta, L.; Bussema, L.; Eichhorn, S.W.; Greenwald, A.C.; Kinker, G.S.; Rodman, C.; et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell 2021, 39, 779–792.E11. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Feng, X.; Herting, C.J.; Garcia, V.A.; Nie, K.; Pong, W.W.; Rasmussen, R.; Dwivedi, B.; Seby, S.; Wolf, S.A.; et al. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res. 2017, 77, 2266–2278. [Google Scholar] [CrossRef]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef]
- Carson, M.J.; Thrash, J.C.; Walter, B. The cellular response in neuroinflammation: The role of leukocytes, microglia and astrocytes in neuronal death and survival. Clin. Neurosci. Res. 2006, 6, 237–245. [Google Scholar] [CrossRef]
- Rio-Hortega, P. The microglia. Lancet 1939, 233, 1023–1026. [Google Scholar] [CrossRef]
- Juba, A. Untersuchungen über die Entwicklung der Hortegaschen Mikroglia des Menschen. Arch. Psychiatr. Nervenkrankh. 1934, 101, 577–592. [Google Scholar] [CrossRef]
- Ginhoux, F.; Prinz, M. Origin of microglia: Current concepts and past controversies. Cold Spring Harb. Perspect. Biol. 2015, 7, a020537. [Google Scholar] [CrossRef]
- De, S.; Van Deren, D.; Peden, E.; Hockin, M.; Boulet, A.; Titen, S.; Capecchi, M.R. Two distinct ontogenies confer heterogeneity to mouse brain microglia. Development 2018, 145, dev152306. [Google Scholar] [CrossRef]
- Darmanis, S.; Sloan, S.A.; Croote, D.; Mignardi, M.; Chernikova, S.; Samghababi, P.; Zhang, Y.; Neff, N.; Kowarsky, M.; Caneda, C.; et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 2017, 21, 1399–1410. [Google Scholar] [CrossRef]
- Ochocka, N.; Segit, P.; Walentynowicz, K.A.; Wojnicki, K.; Cyranowski, S.; Swatler, J.; Mieczkowski, J.; Kaminska, B. Single-cell RNA sequencing reveals functional heterogeneity of glioma-associated brain macrophages. Nat. Commun. 2021, 12, 1151. [Google Scholar] [CrossRef] [PubMed]
- Hickman, S.E.; Kingery, N.D.; Ohsumi, T.K.; Borowsky, M.L.; Wang, L.-C.; Means, T.K.; El Khoury, J. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013, 16, 1896–1905. [Google Scholar] [CrossRef] [PubMed]
- Hammond, T.R.; Dufort, C.; Dissing-Olesen, L.; Giera, S.; Young, A.; Wysoker, A.; Walker, A.J.; Gergits, F.; Segel, M.; Nemesh, J.; et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 2019, 50, 253–271.e256. [Google Scholar] [CrossRef] [PubMed]
- Woolf, Z.; Swanson, M.E.V.; Smyth, L.C.; Mee, E.W.; Schweder, P.; Heppner, P.; Kim, B.J.H.; Turner, C.; Oldfield, R.L.; Curtis, M.A.; et al. Single-cell image analysis reveals a protective role for microglia in glioblastoma. Neurooncol. Adv. 2021, 3, vdab031. [Google Scholar] [CrossRef]
- Dumas, A.A.; Borst, K.; Prinz, M. Current tools to interrogate microglial biology. Neuron 2021, 109, 2805–2819. [Google Scholar] [CrossRef]
- Deczkowska, A.; Keren-Shaul, H.; Weiner, A.; Colonna, M.; Schwartz, M.; Amit, I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018, 173, 1073–1081. [Google Scholar] [CrossRef]
- Zhu, C.; Kros, J.M.; van der Weiden, M.; Zheng, P.; Cheng, C.; Mustafa, D.A. Expression site of P2RY12 in residential microglial cells in astrocytomas correlates with M1 and M2 marker expression and tumor grade. Acta Neuropathol. Commun. 2017, 5, 4. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Biber, K. The microglial ATP-gated ion channel P2X7 as a CNS drug target. Glia 2016, 64, 1772–1787. [Google Scholar] [CrossRef]
- Swanson, M.E.V.; Murray, H.C.; Ryan, B.; Faull, R.L.M.; Dragunow, M.; Curtis, M.A. Quantitative immunohistochemical analysis of myeloid cell marker expression in human cortex captures microglia heterogeneity with anatomical context. Sci. Rep. 2020, 10, 11693. [Google Scholar] [CrossRef]
- Ito, D.; Imai, Y.; Ohsawa, K.; Nakajima, K.; Fukuuchi, Y.; Kohsaka, S. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res. Mol. Brain Res. 1998, 57, 1–9. [Google Scholar] [CrossRef]
- Kettenmann, H.; Hanisch, U.K.; Noda, M.; Verkhratsky, A. Physiology of microglia. Physiol. Rev. 2011, 91, 461–553. [Google Scholar] [CrossRef] [PubMed]
- Berger, G.; Knelson, E.H.; Jimenez-Macias, J.L.; Nowicki, M.O.; Han, S.; Panagioti, E.; Lizotte, P.H.; Adu-Berchie, K.; Stafford, A.; Dimitrakakis, N.; et al. STING activation promotes robust immune response and NK cell–mediated tumor regression in glioblastoma models. Proc. Natl. Acad. Sci. USA 2022, 119, e2111003119. [Google Scholar] [CrossRef] [PubMed]
- Alanio, C.; Binder, Z.A.; Chang, R.B.; Nasrallah, M.P.; Delman, D.; Li, J.H.; Tang, O.Y.; Zhang, L.Y.; Zhang, J.V.; Wherry, E.J.; et al. Immunologic Features in De Novo and Recurrent Glioblastoma Are Associated with Survival Outcomes. Cancer Immunol. Res. 2022, 10, 800–810. [Google Scholar] [CrossRef]
- Tang, H.; Liu, W.; Xu, Z.; Zhao, J.; Wang, W.; Yu, Z.; Wei, M. Integrated microenvironment-associated genomic profiles identify LRRC15 mediating recurrent glioblastoma-associated macrophages infiltration. J. Cell Mol. Med. 2021, 25, 5534–5546. [Google Scholar] [CrossRef]
- Pombo Antunes, A.R.; Scheyltjens, I.; Lodi, F.; Messiaen, J.; Antoranz, A.; Duerinck, J.; Kancheva, D.; Martens, L.; De Vlaminck, K.; Van Hove, H.; et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci. 2021, 24, 595–610. [Google Scholar] [CrossRef]
- Kaffes, I.; Szulzewsky, F.; Chen, Z.; Herting, C.J.; Gabanic, B.; Velázquez Vega, J.E.; Shelton, J.; Switchenko, J.M.; Ross, J.L.; McSwain, L.F.; et al. Human Mesenchymal glioblastomas are characterized by an increased immune cell presence compared to Proneural and Classical tumors. Oncoimmunology 2019, 8, e1655360. [Google Scholar] [CrossRef]
- Peshoff, M.M.; Gupta, P.; Oberai, S.; Trivedi, R.; Katayama, H.; Chakrapani, P.; Dang, M.; Migliozzi, S.; Gumin, J.; Kadri, D.B.; et al. Triggering receptor expressed on myeloid cells 2 (TREM2) regulates phagocytosis in glioblastoma. Neuro-Oncology 2024, 26, 826–839. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, L.; Zhao, S.; Zhang, W.; Chang, Y.; Bosco, D.B.; Huang, T.; Dheer, A.; Gao, S.; Xu, S.; et al. TREM2 mediates MHCII-associated CD4+ T-cell response against gliomas. Neuro-Oncology 2023, 26, 811–825. [Google Scholar] [CrossRef]
- Binnewies, M.; Pollack, J.L.; Rudolph, J.; Dash, S.; Abushawish, M.; Lee, T.; Jahchan, N.S.; Canaday, P.; Lu, E.; Norng, M.; et al. Targeting TREM2 on tumor-associated macrophages enhances immunotherapy. Cell Rep. 2021, 37, 109844. [Google Scholar] [CrossRef]
- Bingle, L.; Brown, N.J.; Lewis, C.E. The role of tumour-associated macrophages in tumour progression: Implications for new anticancer therapies. J. Pathol. 2002, 196, 254–265. [Google Scholar] [CrossRef]
- Chen, A.X.; Gartrell, R.D.; Zhao, J.; Upadhyayula, P.S.; Zhao, W.; Yuan, J.; Minns, H.E.; Dovas, A.; Bruce, J.N.; Lasorella, A.; et al. Single-cell characterization of macrophages in glioblastoma reveals MARCO as a mesenchymal pro-tumor marker. Genome Med. 2021, 13, 88. [Google Scholar] [CrossRef] [PubMed]
- Komohara, Y.; Ohnishi, K.; Kuratsu, J.; Takeya, M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 2008, 216, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Nishie, A.; Ono, M.; Shono, T.; Fukushi, J.; Otsubo, M.; Onoue, H.; Ito, Y.; Inamura, T.; Ikezaki, K.; Fukui, M.; et al. Macrophage infiltration and heme oxygenase-1 expression correlate with angiogenesis in human gliomas. Clin. Cancer Res. 1999, 5, 1107–1113. [Google Scholar]
- Rossi, M.L.; Jones, N.R.; Candy, E.; Nicoll, J.A.; Compton, J.S.; Hughes, J.T.; Esiri, M.M.; Moss, T.H.; Cruz-Sanchez, F.F.; Coakham, H.B. The mononuclear cell infiltrate compared with survival in high-grade astrocytomas. Acta Neuropathol. 1989, 78, 189–193. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Killingsworth, M.C.; Myasoedova, V.A.; Orekhov, A.N.; Bobryshev, Y.V. CD68/macrosialin: Not just a histochemical marker. Lab. Investig. 2017, 97, 4–13. [Google Scholar] [CrossRef]
- van der Kooij, M.A.; von der Mark, E.M.; Kruijt, J.K.; van Velzen, A.; van Berkel, T.J.; Morand, O.H. Human monocyte-derived macrophages express an approximately 120-kD Ox-LDL binding protein with strong identity to CD68. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 3107–3116. [Google Scholar] [CrossRef]
- Waller, R.; Baxter, L.; Fillingham, D.J.; Coelho, S.; Pozo, J.M.; Mozumder, M.; Frangi, A.F.; Ince, P.G.; Simpson, J.E.; Highley, J.R. Iba-1-/CD68+ microglia are a prominent feature of age-associated deep subcortical white matter lesions. PLoS ONE 2019, 14, e0210888. [Google Scholar] [CrossRef]
- Rolfe, N.W.; Dadario, N.B.; Canoll, P.; Bruce, J.N. A Review of Therapeutic Agents Given by Convection-Enhanced Delivery for Adult Glioblastoma. Pharmaceuticals 2024, 17, 973. [Google Scholar] [CrossRef]
- Banu, M.A.; Dovas, A.; Argenziano, M.G.; Zhao, W.; Grajal, H.C.; Higgins, D.M.O.; Sperring, C.P.; Pereira, B.; Ye, L.F.; Mahajan, A.; et al. A cell state specific metabolic vulnerability to GPX4-dependent ferroptosis in glioblastoma. bioRxiv 2023. [Google Scholar] [CrossRef]
- de Groot, J.; Penas-Prado, M.; Alfaro-Munoz, K.; Hunter, K.; Pei, B.L.; O’Brien, B.; Weathers, S.P.; Loghin, M.; Kamiya Matsouka, C.; Yung, W.K.A.; et al. Window-of-opportunity clinical trial of pembrolizumab in patients with recurrent glioblastoma reveals predominance of immune-suppressive macrophages. Neuro-Oncology 2020, 22, 539–549. [Google Scholar] [CrossRef]
- Dang, W.; Xiao, J.; Ma, Q.; Miao, J.; Cao, M.; Chen, L.; Shi, Y.; Yao, X.; Yu, S.; Liu, X.; et al. Combination of p38 MAPK inhibitor with PD-L1 antibody effectively prolongs survivals of temozolomide-resistant glioma-bearing mice via reduction of infiltrating glioma-associated macrophages and PD-L1 expression on resident glioma-associated microglia. Brain Tumor Pathol. 2021, 38, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Flores-Toro, J.A.; Luo, D.; Gopinath, A.; Sarkisian, M.R.; Campbell, J.J.; Charo, I.F.; Singh, R.; Schall, T.J.; Datta, M.; Jain, R.K.; et al. CCR2 inhibition reduces tumor myeloid cells and unmasks a checkpoint inhibitor effect to slow progression of resistant murine gliomas. Proc. Natl. Acad. Sci. USA 2020, 117, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Magri, S.; Musca, B.; Bonaudo, C.; Tushe, A.; Russo, M.G.; Masetto, E.; Zagonel, V.; Lombardi, G.; Della Puppa, A.; Mandruzzato, S. Sustained Accumulation of Blood-Derived Macrophages in the Immune Microenvironment of Patients with Recurrent Glioblastoma after Therapy. Cancers 2021, 13, 6178. [Google Scholar] [CrossRef]
- Rahman, M.; Kresak, J.; Yang, C.; Huang, J.; Hiser, W.; Kubilis, P.; Mitchell, D. Analysis of immunobiologic markers in primary and recurrent glioblastoma. J. Neurooncol. 2018, 137, 249–257. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Y. Tumor-associated macrophages: From basic research to clinical application. J. Hematol. Oncol. 2017, 10, 58. [Google Scholar] [CrossRef]
- Kaku, Y.; Imaoka, H.; Morimatsu, Y.; Komohara, Y.; Ohnishi, K.; Oda, H.; Takenaka, S.; Matsuoka, M.; Kawayama, T.; Takeya, M.; et al. Overexpression of CD163, CD204 and CD206 on alveolar macrophages in the lungs of patients with severe chronic obstructive pulmonary disease. PLoS ONE 2014, 9, e87400. [Google Scholar] [CrossRef]
- Wen, P.Y.; Chang, S.M.; Van den Bent, M.J.; Vogelbaum, M.A.; Macdonald, D.R.; Lee, E.Q. Response Assessment in Neuro-Oncology Clinical Trials. J. Clin. Oncol. 2017, 35, 2439–2449. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, C.; Maimela, N.R.; Yang, L.; Zhang, Z.; Ping, Y.; Huang, L.; Zhang, Y. Molecular and clinical characterization of CD163 expression via large-scale analysis in glioma. Oncoimmunology 2019, 8, 1601478. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhao, Q.; Zhao, S.; Zhang, P.; Zhao, H.; Li, Z.; Du, Y.; Tian, X.; Lu, J. Characterization of transcriptome profile and clinical features of a novel immunotherapy target CD204 in diffuse glioma. Cancer Med. 2019, 8, 3811–3821. [Google Scholar] [CrossRef]
- Lu, W.; Su, L.; Yu, Z.; Zhang, S.; Miao, J. The New Role of CD163 in the Differentiation of Bone Marrow Stromal Cells into Vascular Endothelial-Like Cells. Stem Cells Int. 2016, 2016, 2539781. [Google Scholar] [CrossRef]
- Zhu, C.; Kros, J.M.; Cheng, C.; Mustafa, D. The contribution of tumor-associated macrophages in glioma neo-angiogenesis and implications for anti-angiogenic strategies. Neuro-Oncology 2017, 19, 1435–1446. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Yu, H.; Zhang, Y.; Zhao, D.; Lv, P.; Zhong, Y.; Xu, Y. IL-10 secreted by M2 macrophage promoted tumorigenesis through interaction with JAK2 in glioma. Oncotarget 2016, 7, 71673–71685. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Ishikawa, E.; Matsuda, M.; Sugii, N.; Kohzuki, H.; Akutsu, H.; Sakamoto, N.; Takano, S.; Matsumura, A. Infiltration of CD163-positive macrophages in glioma tissues after treatment with anti-PD-L1 antibody and role of PI3Kγ inhibitor as a combination therapy with anti-PD-L1 antibody in in vivo model using temozolomide-resistant murine glioma-initiating cells. Brain Tumor Pathol. 2020, 37, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yu, X.; Gao, L.; Zhao, Y.; Lai, J.; Lu, D.; Bao, R.; Jia, B.; Zhong, L.; Wang, F.; et al. Noninvasive Imaging of CD206-Positive M2 Macrophages as an Early Biomarker for Post-Chemotherapy Tumor Relapse and Lymph Node Metastasis. Theranostics 2017, 7, 4276–4288. [Google Scholar] [CrossRef] [PubMed]
- Zarif, J.C.; Baena-Del Valle, J.A.; Hicks, J.L.; Heaphy, C.M.; Vidal, I.; Luo, J.; Lotan, T.L.; Hooper, J.E.; Isaacs, W.B.; Pienta, K.J.; et al. Mannose Receptor-positive Macrophage Infiltration Correlates with Prostate Cancer Onset and Metastatic Castration-resistant Disease. Eur. Urol. Oncol. 2019, 2, 429–436. [Google Scholar] [CrossRef]
- Georgieva, P.B.; Mathivet, T.; Alt, S.; Giese, W.; Riva, M.; Balcer, M.; Gerhardt, H. Long-lived tumor-associated macrophages in glioma. Neurooncol. Adv. 2020, 2, vdaa127. [Google Scholar] [CrossRef]
- Wang, B.; Li, Q.; Qin, L.; Zhao, S.; Wang, J.; Chen, X. Transition of tumor-associated macrophages from MHC class II(hi) to MHC class II(low) mediates tumor progression in mice. BMC Immunol. 2011, 12, 43. [Google Scholar] [CrossRef]
- Sørensen, M.D.; Kristensen, B.W. Tumour-associated CD204(+) microglia/macrophages accumulate in perivascular and perinecrotic niches and correlate with an interleukin-6-enriched inflammatory profile in glioblastoma. Neuropathol. Appl. Neurobiol. 2022, 48, e12772. [Google Scholar] [CrossRef]
- Leblond, M.M.; Gérault, A.N.; Corroyer-Dulmont, A.; MacKenzie, E.T.; Petit, E.; Bernaudin, M.; Valable, S. Hypoxia induces macrophage polarization and re-education toward an M2 phenotype in U87 and U251 glioblastoma models. Oncoimmunology 2016, 5, e1056442. [Google Scholar] [CrossRef]
- Buonfiglioli, A.; Hambardzumyan, D. Macrophages and microglia: The cerberus of glioblastoma. Acta Neuropathol. Commun. 2021, 9, 54. [Google Scholar] [CrossRef]
- Gjorgjevski, M.; Hannen, R.; Carl, B.; Li, Y.; Landmann, E.; Buchholz, M.; Bartsch, J.W.; Nimsky, C. Molecular profiling of the tumor microenvironment in glioblastoma patients: Correlation of microglia/macrophage polarization state with metalloprotease expression profiles and survival. Biosci. Rep. 2019, 39, BSR20182361. [Google Scholar] [CrossRef] [PubMed]
- Lisi, L.; Ciotti, G.M.; Braun, D.; Kalinin, S.; Currò, D.; Dello Russo, C.; Coli, A.; Mangiola, A.; Anile, C.; Feinstein, D.L.; et al. Expression of iNOS, CD163 and ARG-1 taken as M1 and M2 markers of microglial polarization in human glioblastoma and the surrounding normal parenchyma. Neurosci. Lett. 2017, 645, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Montemurro, N.; Pahwa, B.; Tayal, A.; Shukla, A.; De Jesus Encarnacion, M.; Ramirez, I.; Nurmukhametov, R.; Chavda, V.; De Carlo, A. Macrophages in Recurrent Glioblastoma as a Prognostic Factor in the Synergistic System of the Tumor Microenvironment. Neurol. Int. 2023, 15, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Ishikawa, E.; Matsuda, M.; Akutsu, H.; Osuka, S.; Sakamoto, N.; Takano, S.; Yamamoto, T.; Tsuboi, K.; Matsumura, A. Assessment of PD-1 positive cells on initial and secondary resected tumor specimens of newly diagnosed glioblastoma and its implications on patient outcome. J. Neurooncol. 2017, 133, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Cloughesy, T.F.; Mochizuki, A.Y.; Orpilla, J.R.; Hugo, W.; Lee, A.H.; Davidson, T.B.; Wang, A.C.; Ellingson, B.M.; Rytlewski, J.A.; Sanders, C.M.; et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat. Med. 2019, 25, 477–486. [Google Scholar] [CrossRef]
- Wang, H.; Argenziano, M.G.; Yoon, H.; Boyett, D.; Save, A.; Petridis, P.; Savage, W.; Jackson, P.; Hawkins-Daarud, A.; Tran, N.; et al. Biologically-informed deep neural networks provide quantitative assessment of intratumoral heterogeneity in post-treatment glioblastoma. bioRxiv 2022. [Google Scholar] [CrossRef]
- Di Ianni, N.; Maffezzini, M.; Eoli, M.; Pellegatta, S. Revisiting the Immunological Aspects of Temozolomide Considering the Genetic Landscape and the Immune Microenvironment Composition of Glioblastoma. Front. Oncol. 2021, 11, 747690. [Google Scholar] [CrossRef]
- Ha, W.; Sevim-Nalkiran, H.; Zaman, A.M.; Matsuda, K.; Khasraw, M.; Nowak, A.K.; Chung, L.; Baxter, R.C.; McDonald, K.L. Ibudilast sensitizes glioblastoma to temozolomide by targeting Macrophage Migration Inhibitory Factor (MIF). Sci. Rep. 2019, 9, 2905. [Google Scholar] [CrossRef]
- Kitange, G.J.; Carlson, B.L.; Schroeder, M.A.; Decker, P.A.; Morlan, B.W.; Wu, W.; Ballman, K.V.; Giannini, C.; Sarkaria, J.N. Expression of CD74 in high grade gliomas: A potential role in temozolomide resistance. J. Neurooncol. 2010, 100, 177–186. [Google Scholar] [CrossRef][Green Version]
- Alfonso, J.C.L.; Grass, G.D.; Welsh, E.; Ahmed, K.A.; Teer, J.K.; Pilon-Thomas, S.; Harrison, L.B.; Cleveland, J.L.; Mulé, J.J.; Eschrich, S.A.; et al. Tumor-immune ecosystem dynamics define an individual Radiation Immune Score to predict pan-cancer radiocurability. Neoplasia 2021, 23, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Cifarelli, C.P.; Jacques, A.; Bobko, A. Heterogeneity of radiation response in mesenchymal subtype glioblastoma: Molecular profiling and reactive oxygen species generation. J. Neurooncol. 2021, 152, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Xu, J.; Fan, Y.; Zhang, Z.; Wang, H.; Qian, M.; Zhang, P.; Deng, L.; Shen, J.; Xue, H.; et al. ARPC1B promotes mesenchymal phenotype maintenance and radiotherapy resistance by blocking TRIM21-mediated degradation of IFI16 and HuR in glioma stem cells. J. Exp. Clin. Cancer Res. 2022, 41, 323. [Google Scholar] [CrossRef]
- Hughes, R.; Qian, B.Z.; Rowan, C.; Muthana, M.; Keklikoglou, I.; Olson, O.C.; Tazzyman, S.; Danson, S.; Addison, C.; Clemons, M.; et al. Perivascular M2 Macrophages Stimulate Tumor Relapse after Chemotherapy. Cancer Res. 2015, 75, 3479–3491. [Google Scholar] [CrossRef]
- Urbantat, R.M.; Jelgersma, C.; Brandenburg, S.; Nieminen-Kelhä, M.; Kremenetskaia, I.; Zollfrank, J.; Mueller, S.; Rubarth, K.; Koch, A.; Vajkoczy, P.; et al. Tumor-Associated Microglia/Macrophages as a Predictor for Survival in Glioblastoma and Temozolomide-Induced Changes in CXCR2 Signaling with New Resistance Overcoming Strategy by Combination Therapy. Int. J. Mol. Sci. 2021, 22, 11180. [Google Scholar] [CrossRef]
- Bruyère, C.; Mijatovic, T.; Lonez, C.; Spiegl-Kreinecker, S.; Berger, W.; Kast, R.E.; Ruysschaert, J.-M.; Kiss, R.; Lefranc, F. Temozolomide-induced modification of the CXC chemokine network in experimental gliomas Corrigendum in /ijo/38/6/1767. Int. J. Oncol. 2011, 38, 1453–1464. [Google Scholar] [CrossRef]
- Hasan, T.; Caragher, S.P.; Shireman, J.M.; Park, C.H.; Atashi, F.; Baisiwala, S.; Lee, G.; Guo, D.; Wang, J.Y.; Dey, M.; et al. Interleukin-8/CXCR2 signaling regulates therapy-induced plasticity and enhances tumorigenicity in glioblastoma. Cell Death Dis. 2019, 10, 292. [Google Scholar] [CrossRef]
- Hu, J.; Zhao, Q.; Kong, L.Y.; Wang, J.; Yan, J.; Xia, X.; Jia, Z.; Heimberger, A.B.; Li, S. Regulation of tumor immune suppression and cancer cell survival by CXCL1/2 elevation in glioblastoma multiforme. Sci. Adv. 2021, 7, eabc2511. [Google Scholar] [CrossRef]
- Zeiner, P.S.; Preusse, C.; Golebiewska, A.; Zinke, J.; Iriondo, A.; Muller, A.; Kaoma, T.; Filipski, K.; Müller-Eschner, M.; Bernatz, S.; et al. Distribution and prognostic impact of microglia/macrophage subpopulations in gliomas. Brain Pathol. 2019, 29, 513–529. [Google Scholar] [CrossRef]
- Sørensen, M.D.; Dahlrot, R.H.; Boldt, H.B.; Hansen, S.; Kristensen, B.W. Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol. Appl. Neurobiol. 2018, 44, 185–206. [Google Scholar] [CrossRef]
- Li, J.; Kaneda, M.M.; Ma, J.; Li, M.; Shepard, R.M.; Patel, K.; Koga, T.; Sarver, A.; Furnari, F.; Xu, B.; et al. PI3Kγ inhibition suppresses microglia/TAM accumulation in glioblastoma microenvironment to promote exceptional temozolomide response. Proc. Natl. Acad. Sci. USA 2021, 118, e2009290118. [Google Scholar] [CrossRef] [PubMed]
- Nduom, E.K.; Wei, J.; Yaghi, N.K.; Huang, N.; Kong, L.Y.; Gabrusiewicz, K.; Ling, X.; Zhou, S.; Ivan, C.; Chen, J.Q.; et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro-Oncology 2016, 18, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Akkari, L.; Bowman, R.L.; Tessier, J.; Klemm, F.; Handgraaf, S.M.; de Groot, M.; Quail, D.F.; Tillard, L.; Gadiot, J.; Huse, J.T.; et al. Dynamic changes in glioma macrophage populations after radiotherapy reveal CSF-1R inhibition as a strategy to overcome resistance. Sci. Transl. Med. 2020, 12, eaaw7843. [Google Scholar] [CrossRef]
- Leblond, M.M.; Pérès, E.A.; Helaine, C.; Gérault, A.N.; Moulin, D.; Anfray, C.; Divoux, D.; Petit, E.; Bernaudin, M.; Valable, S. M2 macrophages are more resistant than M1 macrophages following radiation therapy in the context of glioblastoma. Oncotarget 2017, 8, 72597–72612. [Google Scholar] [CrossRef]
- Foray, C.; Valtorta, S.; Barca, C.; Winkeler, A.; Roll, W.; Müther, M.; Wagner, S.; Gardner, M.L.; Hermann, S.; Schäfers, M.; et al. Imaging temozolomide-induced changes in the myeloid glioma microenvironment. Theranostics 2021, 11, 2020–2033. [Google Scholar] [CrossRef]
- Chiang, C.S.; Fu, S.Y.; Wang, S.C.; Yu, C.F.; Chen, F.H.; Lin, C.M.; Hong, J.H. Irradiation promotes an m2 macrophage phenotype in tumor hypoxia. Front. Oncol. 2012, 2, 89. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, C.; Zhang, Z.; Han, B.; Shen, Z.; Li, L.; Liu, S.; Zhao, X.; Ye, F.; Zhang, Y. Specific clinical and immune features of CD68 in glioma via 1,024 samples. Cancer Manag. Res. 2018, 10, 6409–6419. [Google Scholar] [CrossRef]
- Jackson, R.J.; Fuller, G.N.; Abi-Said, D.; Lang, F.F.; Gokaslan, Z.L.; Shi, W.M.; Wildrick, D.M.; Sawaya, R. Limitations of stereotactic biopsy in the initial management of gliomas. Neuro-Oncology 2001, 3, 193–200. [Google Scholar] [CrossRef]
- Abbasi, A.W.; Westerlaan, H.E.; Holtman, G.A.; Aden, K.M.; van Laar, P.J.; van der Hoorn, A. Incidence of Tumour Progression and Pseudoprogression in High-Grade Gliomas: A Systematic Review and Meta-Analysis. Clin. Neuroradiol. 2018, 28, 401–411. [Google Scholar] [CrossRef]
- Liu, H.; Sun, Y.; Zhang, Q.; Jin, W.; Gordon, R.E.; Zhang, Y.; Wang, J.; Sun, C.; Wang, Z.J.; Qi, X.; et al. Pro-inflammatory and proliferative microglia drive progression of glioblastoma. Cell Rep. 2021, 36, 109718. [Google Scholar] [CrossRef]
- Tang, F.; Wang, Y.; Zeng, Y.; Xiao, A.; Tong, A.; Xu, J. Tumor-associated macrophage-related strategies for glioma immunotherapy. NPJ Precis. Oncol. 2023, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- von Roemeling, C.A.; Wang, Y.; Qie, Y.; Yuan, H.; Zhao, H.; Liu, X.; Yang, Z.; Yang, M.; Deng, W.; Bruno, K.A.; et al. Therapeutic modulation of phagocytosis in glioblastoma can activate both innate and adaptive antitumour immunity. Nat. Commun. 2020, 11, 1508. [Google Scholar] [CrossRef] [PubMed]
- Ravn-Boess, N.; Roy, N.; Hattori, T.; Bready, D.; Donaldson, H.; Lawson, C.; Lapierre, C.; Korman, A.; Rodrick, T.; Liu, E.; et al. The expression profile and tumorigenic mechanisms of CD97 (ADGRE5) in glioblastoma render it a targetable vulnerability. Cell Rep. 2023, 42, 113374. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Velagapudi, R.; Terrando, N. Neuroinflammation after surgery: From mechanisms to therapeutic targets. Nat. Immunol. 2020, 21, 1319–1326. [Google Scholar] [CrossRef]
- Iliadi, C.; Verset, L.; Bouchart, C.; Martinive, P.; Van Gestel, D.; Krayem, M. The current understanding of the immune landscape relative to radiotherapy across tumor types. Front. Immunol. 2023, 14, 1148692. [Google Scholar] [CrossRef]
- Wheeler, C.J.; Das, A.; Liu, G.; Yu, J.S.; Black, K.L. Clinical responsiveness of glioblastoma multiforme to chemotherapy after vaccination. Clin. Cancer Res. 2004, 10, 5316–5326. [Google Scholar] [CrossRef]
- Tamimi, A.F.; Juweid, M. Epidemiology and Outcome of Glioblastoma. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017. [Google Scholar]
- Yang, W.; Warrington, N.M.; Taylor, S.J.; Whitmire, P.; Carrasco, E.; Singleton, K.W.; Wu, N.; Lathia, J.D.; Berens, M.E.; Kim, A.H.; et al. Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci. Transl. Med. 2019, 11, eaao5253. [Google Scholar] [CrossRef]
- Turaga, S.M.; Silver, D.J.; Bayik, D.; Paouri, E.; Peng, S.; Lauko, A.; Alban, T.J.; Borjini, N.; Stanko, S.; Naik, U.P.; et al. JAM-A functions as a female microglial tumor suppressor in glioblastoma. Neuro-Oncology 2020, 22, 1591–1601. [Google Scholar] [CrossRef]
- Tharp, M.E.; Han, C.Z.; Balak, C.D.; Fitzpatrick, C.; O’Connor, C.; Preissl, S.; Buchanan, J.; Nott, A.; Escoubet, L.; Mavrommatis, K.; et al. The inactive X chromosome drives sex differences in microglial inflammatory activity in human glioblastoma. bioRxiv 2024. [Google Scholar] [CrossRef]
- Kfoury, N.; Sun, T.; Yu, K.; Rockwell, N.; Tinkum, K.L.; Qi, Z.; Warrington, N.M.; McDonald, P.; Roy, A.; Weir, S.J.; et al. Cooperative p16and p21 action protects female astrocytes from transformation. Acta Neuropathol. Commun. 2018, 6, 12. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dadario, N.B.; Boyett, D.M.; Teasley, D.E.; Chabot, P.J.; Winans, N.J.; Argenziano, M.G.; Sperring, C.P.; Canoll, P.; Bruce, J.N. Unveiling the Inflammatory Landscape of Recurrent Glioblastoma through Histological-Based Assessments. Cancers 2024, 16, 3283. https://doi.org/10.3390/cancers16193283
Dadario NB, Boyett DM, Teasley DE, Chabot PJ, Winans NJ, Argenziano MG, Sperring CP, Canoll P, Bruce JN. Unveiling the Inflammatory Landscape of Recurrent Glioblastoma through Histological-Based Assessments. Cancers. 2024; 16(19):3283. https://doi.org/10.3390/cancers16193283
Chicago/Turabian StyleDadario, Nicholas B., Deborah M. Boyett, Damian E. Teasley, Peter J. Chabot, Nathan J. Winans, Michael G. Argenziano, Colin P. Sperring, Peter Canoll, and Jeffrey N. Bruce. 2024. "Unveiling the Inflammatory Landscape of Recurrent Glioblastoma through Histological-Based Assessments" Cancers 16, no. 19: 3283. https://doi.org/10.3390/cancers16193283
APA StyleDadario, N. B., Boyett, D. M., Teasley, D. E., Chabot, P. J., Winans, N. J., Argenziano, M. G., Sperring, C. P., Canoll, P., & Bruce, J. N. (2024). Unveiling the Inflammatory Landscape of Recurrent Glioblastoma through Histological-Based Assessments. Cancers, 16(19), 3283. https://doi.org/10.3390/cancers16193283






