Simple Summary
This study aimed to determine the gene expression profiles associated with chemotherapeutic responses in conventional osteosarcomas (COS) within South Africa. We observed a significant downregulation in the ATP binding cassette subfamily C members (ABCC3 and ABCB1-p-glycoprotein), excision repair cross-complimenting group 1 (ERCC 1), replication factor C subunit 1 (RFC1), and tumour protein 53 (p53) genes in the COS tumours compared to the healthy donors. Furthermore, an upregulated ERCC1 gene expression level predicted a poor chemotherapeutic response. Additionally, the predictors of COS chemotherapeutic response comprised age, chondroblastic and osteoblastic histological subtypes, and ABCC3, ERCC1, and RFC1 gene expression.
Abstract
Background: We determined the predictive gene expression profiles associated with chemo-response in conventional osteosarcomas (COS) within South Africa. Materials and methods: In 28 patients, we performed an RNA extraction, cDNA synthesis, and quantitative analysis using the RT-PCR 2−∆∆CT method to determine the fold change in gene expression alongside GAPDH (housekeeping gene). Results: We observed a significant downregulation in the mRNA expression profiles of ABCB1-p-glycoprotein (p = 0.0007), ABCC3 (p = 0.002), ERCC1 (p = 0.007), p-53 (p = 0.007), and RFC1 (p = 0.003) in the COS patients compared to the healthy donors. Furthermore, ABCB1-p-glycoprotein (p = 0.008) and ABCC3 (p = 0.020) exhibited a significant downregulation in the COS tumour tissues when compared to the healthy donors. In our univariate logistic regression, the predictors of chemotherapeutic response comprised ERCC1 [restricted cubic spline (RCS) knot: OR −0.27; CI −0.504 to −0.032; p = 0.036]; osteoblastic subtype [OR −0.36; CI −0.652 to −0.092; p = 0.026); fibroblastic subtype [OR 0.91; CI 0.569 to 1.248; p < 0.001]; and mixed subtype [OR 0.53; CI 0.232 to 0.032; p = 0.032]. In our multivariable logistic regression, the significant predictors of chemotherapeutic response comprised age [RCS knot: OR −2.5; CI −3.616 to −1.378; p = 0.022]; ABCC3 [RCS knot: OR 0.67; CI 0.407 to 0.936, p = 0.016]; ERCC1 [RCS knot: OR 0.57; CI 0.235 to 0.901; p = 0.044]; RFC1 [RCS knot: OR −1.04; CI −1.592 to −0.487; p = 0.035]; chondroblastic subtype [OR −0.83; CI −1.106 to −0.520; p = 0.012]; and osteoblastic subtype [OR −1.28; CI −1.664 to −0.901; p = 0.007]. Conclusions: In this South African cohort, we observed the unique gene expression profiles of osteosarcoma tumourigenesis and chemotherapeutic responses. These may serve as prognostication and therapeutic targets. Larger-scale research is needed on the African continent.
1. Introduction
Osteosarcomas are a group of aggressive primary malignant bone tumours of mesenchymal cancer stem cell origin that produce osteoid matrices. They most commonly occur in children and adolescents [1]. According to the WHO classification, central high-grade conventional osteosarcoma (COS) is the most common subtype, comprising 90% of all osteosarcoma variants [1,2]. Currently, the treatment of COS typically comprises neoadjuvant chemotherapy and wide surgical excision, followed by adjuvant chemotherapy [3,4,5,6]. The predictors of a poor survival prognosis include male sex, older age, advanced Enneking staging, non-extremity tumour, proximal long bone sites, metastasis, poor response to chemotherapy, no surgical treatment, and amputations [3,4,5,6]. The COS tumour’s chemotherapy response has emerged as the most important independent risk factor for long-term survival [3,4,5,6]. The Rosen protocol is widely utilised to measure the response to neoadjuvant chemotherapy during histological analysis after the final tumour surgical resection. A good response is defined as ≥90% tumour necrosis (Huvos grade III/IV) with a poor response defined as <90% tumour necrosis (Huvos grade I/II) [3,4].
Despite the advances in multidrug chemotherapy (MDR) and surgical procedures over the past decades, the 5-year survival of non-metastatic COS has plateaued at 60–70% [3,4,5,6]. MDR remains the most significant obstacle to improving long-term survival. The response to neoadjuvant chemotherapy is poor in up to 40–45% of cases [5]. Neither tailored postoperative chemotherapy (according to the histological response to preoperative chemotherapy agents) nor dose intensification have improved survival rates [5,6,7]. Heterogeneous gene expression patterns underpin the tumour microenvironment, with a host of molecular markers associated with chemotherapy response [8,9,10,11,12]. Examining gene expression profiles commonly known for tumourigenesis and chemoresistance in COS could assist with prognostication and guide treatment. Risk stratification using surrogate markers of disease burden and chemo-response offers great promise for the future treatment of osteosarcoma [8,9,10,11,12]. Gene therapy targeting specific genes involved in MDR is being widely researched globally but not in Africa. [8,9,10,11,12]
For instance, the ATP Binding Cassette Subfamily C Members (ABCC3 and ABCB1 genes), the latter formerly known as the MDR1 gene or P-glycoprotein (P-pg), are integral in drug uptake and transport mechanisms, thereby influencing the intracellular concentration of chemotherapy agents [9,12,13]. Contrary to drug efflux, the Replication Factor C Subunit 1 (RFC1 gene), situated on the tumour cell membrane, modulates the accumulation of methotrexate (MTX), a chemotherapeutic agent commonly used in osteosarcomas [9,12,14]. On the other hand, the Excision Repair Cross-Complimenting Group 1 (ERCC1) plays a pivotal role in tumour cell DNA repair mechanisms, impacting the efficacy of DNA-damaging chemotherapy agents such as cisplatin. Elevated ERCC1 expression is associated with reduced sensitivity to cisplatin-based chemotherapy regimens [9,12,15]. Meanwhile, the tumour protein 53 (p53 gene) is an oncogene that plays a multifaceted role in cancer progression and chemotherapy response. Its overexpression contributes to tumourigenesis and chemoresistance by inhibiting apoptosis, a key mechanism of chemotherapy-induced cell death [9,12,16]. Exploring the expression and functional implications of conventional chemotherapy drug sensitivity genes in African populations is paramount for developing personalised treatment strategies and improving cancer outcomes in this demographic. By elucidating the interplay between genetic variations and chemotherapy response, we can pave the way for more effective and tailored cancer treatments for individuals of African descent.
This study aimed to examine the gene expression profiles associated with multidrug chemotherapy responses in central high-grade COS within the South African context, explicitly referencing the ABCB1—p-glycoprotein, ABCC3, ERCC1, RFC1, and p53 genes. Hopefully, the results emanating from this research will inform therapeutic targets in our population.
2. Materials and Methods
This prospective study comprises 28 patients diagnosed with histologically confirmed primary high-grade COS as defined by the WHO criteria [1]. We examined 28 paired (normal muscle and tumour) tissue samples from patients < 40 years of age with chemotherapy-naïve osteosarcomas of the extremities and pelvis of patients treated in out-training hospitals in South Africa between 2021 and 2022. In addition, to compare, we sampled normal muscles from nine healthy donor (non-cancerous) patients during their routine elective orthopaedic procedures. Ethical approval was obtained from the Biomedical Research Ethics Committee, and all patients or legal guardians consented to study involvement. The study proceeded once all the necessary ethical and local regulatory approvals were obtained (BREC/00002737/2021) and per the rules of the Declaration of Helsinki on human studies. The response to chemotherapy in each case was determined after neoadjuvant chemotherapy and wide surgical resection per Rosen protocol with Huvos criterion as defined previously [3,4]. We compared the gene expression profile of the primary tumours before chemotherapy induction in non-responders (NR) to neoadjuvant chemotherapy to responders (R) [8,11]. Exclusion criteria were non-conventional osteosarcoma subtypes and osteosarcoma presenting as a secondary malignancy. The clinical data of patients’ characteristics and management is provided in Table 1. Tumour volume was calculated using a previously described technique, using the formula for an ellipsoidal mass (width × height × diameter × 0.52) [17].

Table 1.
Clinical characteristics of individual patients.
Immediately after CT-guided percutaneous or incisional biopsy, tumour specimens were snap-frozen in liquid nitrogen and stored at –80° C until RNA extraction. For diagnostic purposes, all samples used for RNA extraction were coded using a tissue ID corresponding to the histological section. Our previous study informed the selection of five candidate genes, which was a systematic review of the heterogeneous gene expression patterns associated with chemoresistance in COS in genome-wide sequencing studies [12]. Patients were classified according to the Huvos criteria as responders (R) if tumours exhibited ≥90% tumour necrosis and non-responders (NR) if <90% necrosis, following preoperative chemotherapy, consisting of methotrexate 8–12 g/m2, cisplatin 100 mg/m2, and doxorubicin, 25 mg/m2—MAP [4] [Figure 1 and Table 1].
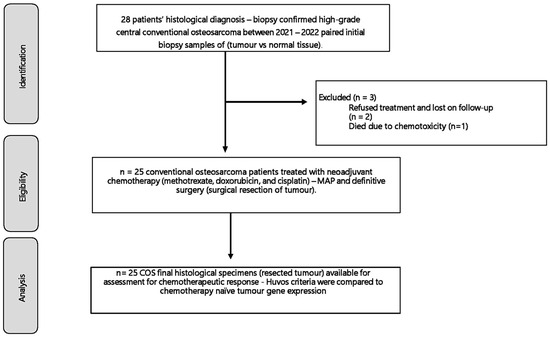
Figure 1.
The 28 patients included had a histological biopsy-confirmed high-grade conventional osteosarcoma of the appendicular skeleton. Between 2021 and 2022, after ethical approval, clinical and histological data, and a biopsy of the tumour and normal muscle tissue were collected for a gene expression analysis that was compared to their chemotherapy response.
2.1. RNA Extraction and cDNA Synthesis
- (a)
- Washing: All specimens (tumour and normal tissue) contained in an Eppendorf tube were washed using 300 μL of phosphate-buffered saline (PBS). Five millimetres of tissues were placed into a PBS tube and vortexed for 60 s. The supernatant was then discarded. Then, 250 μL of PBS was added to the Eppendorf tube, and the steps were repeated. The tissues were then cut into small pieces and placed in 100 μL PBS.
- (b)
- Extraction process: We transferred the emulsified tissue and PBS into a clean 1.5 mL Eppendorf tube and added 100 μL of QuickExtract RNA Extraction Kit (LGC Biosearch Technologies, Oxford, UK). The mixture was vortexed for 2 min. The tube was then centrifuged for 2 min at 14,000 rpm. The supernatant was removed and transferred to a clean 96-well plate. We used an ND8000 nanodrop (Thermofisher, Waltham, MA, USA) to determine the concentration of RNA within our extracted samples. Thereafter, RNA was standardised to 50 ng/uL.
- (c)
- cDNA synthesis: We used the Vilo superscriPT cDNA synthesis kit (Thermofisher, USA) to synthesise cDNA. Briefly, a single reaction was made up using 4 μL of 5× VILOTM Reaction Mix (Thermofisher Scientific, Carlsbad, CA, USA), 2 μL of 10× SuperScriptTM Enzyme Mix, and 8 μL of standardised RNA. Thereafter, the reaction mix was incubated at 25 °C for 10 min, followed by 42 °C for 60 min using the SimpliAmp Thermal Cycler (Thermofisher Scientific, Carlsbad, CA, USA). The reaction was terminated at 85 °C for 5 min. Diluted cDNA (1:400) was stored at −20 °C (Table 2).
 Table 2. A cDNA synthesis reaction is set up for a single reaction.
Table 2. A cDNA synthesis reaction is set up for a single reaction.
2.2. RT-PCR
RT-PCR was used to determine the expression of ABCC3, ABCB1, ERCC1, RFC1, and p53. Briefly, a single reaction was made up using 6 μL of 5× Powerup SYBR green (Thermofisher Scientific, Carlsbad, CA, USA), 1 μL of forward primer (10 μM), 1 μL of reverse primer (10 μM), and 5 μL of cDNA (Table 3). All samples were run in triplicate per gene. The sequences for the primers used were obtained from PrimerBank (Table 3). The quality of assays was assessed by calculating the ratio of 3′:5′ features for the reference gene, GAPDH, which remained stable and when the ratio was greater than 2, the experimental results were nullified. Samples were amplified using the Quantstudio 5 (Thermofisher, Waltham, MA, USA) with the following cycling conditions: 95 °C for 2 min, 40 cycles of 95 °C for 30 s, 60 °C for 1 min, and 72 °C for 30 s (Table 3).

Table 3.
Forward and reverse primer sequences used in RT-PCR reaction.
Relative gene expression was determined using the Livak and Schmittgen quantitation method. Gene expression of the target gene was normalised against the reference gene, GAPDH and it was reported as fold change using a 2-delta-delta CT method (2−∆∆CT) [18].
2.3. Outcome Measures
The primary outcome of interest was candidate gene expression levels in chemotherapy-naïve tumour specimens compared to normal muscle tissues (paired samples). Gene expression was determined by measuring the fold change in the messenger RNA (mRNA) level from a specific gene.
The secondary outcomes were the associations of mRNA expression of ERCC1, FRC, p53, ABCB1, and ABCC3 of tumour samples with (a) the chemotherapy response as per the Huvos grading system and (b) the patient clinical factors.
2.4. Statistical Analysis
The statistical analysis used GraphPad Prism version 8.0.2 (263) and R (R Foundation for Statistical Computing, Vienna, Austria). Continuous variables were reported as mean (standard deviation [SD]) or median (with interquartile range [IQR]), and categorical variables as numbers and percentages unless otherwise stated. The average expression levels across individuals were compared, and the fold change for each gene was calculated. The significance level was determined using an unpaired, nonparametric, Mann–Whitney, two-tailed experimental design with a significant difference set at p < 0.05. We examined the relationship between gene expression and tissue histology using univariate logistic regression with gene concentrations and age modelled using a three-knot restricted cubic spline. Multivariate logistic regression was performed, and the most parsimonious model was chosen using stepwise selection by minimising the Akaike Information Criteria (AIC).
3. Results
3.1. Clinical Characteristics
The median age was 16.0 years (IQR 11.3–20.3), and there was a predilection for male sex of 64% (n = 18). The most common tumour locations were around the knee, involving the distal femur in 57% (n = 16) and the proximal tibia in 18% (n = 5) of cases. Other locations (n = 7) included the proximal femur 11% (n = 3), pelvis 7% (n = 2), proximal humerus 4% (n = 1), and distal radius 4% (n = 1). At the time of diagnosis, the median MRI tumour volume was 780 cm3 (IQR = 511–1133; CI = 664–1092), median alkaline phosphatase (ALP) was 237.0 U/L (IQR = 141.0–431; CI = 218.4–384.1) and lactate dehydrogenase (LDH) was 624 U/L (IQR = 229.0–663.0; CI = 386.1–801.2). Metastasis was present at diagnosis in 46.4% (n = 13) of the cases. The histological subtypes comprised osteoblastic (46%; n = 13), chondroblastic (32%; n = 9), mixed (11%; n = 3), and fibroblastic (11%; n = 3). Eighty-nine percent (n = 25) of the patients were treated with neoadjuvant chemotherapy drugs and subsequently received definitive surgery. Two patients refused treatment and sought alternative traditional medicine while one developed chemotoxicity and did not complete the treatment. All three of these patients died during follow-up. Of the remaining 25 patients, 84% (n = 21) did not show an adequate response to chemotherapy (Huvos criteria < 90% necrosis). The median study follow-up period was 12.7 months (IQR 9.0–17.0) [Table 4].

Table 4.
The descriptive statistics results of the cohort.
3.2. mRNA Expression of Candidate Genes
Using a Mann–Whitney test, we observed a significant downregulation in the mRNA expression levels of ABCC3, ABCB1 (p-glycoprotein), ERCC1, p53, and RFC1 of COS patients compared to the healthy donors as measured by RT-PCR (p = 0.002; p = 0.0007; p = 0.007; p = 0.007; and p = 0.003), respectively—Figure 2. Furthermore, the mRNA expression levels of ABCC3 (p = 0.020) and ABCB1, also known as p-glycoprotein (p = 0.008), exhibited a significant downregulation in the tumour tissues of COS patients tumour when compared to the healthy donors. This is in contrast to the clear but non-significant upregulation in ERCC1 (p = 0.080) and RFC1 (p = 0.075) between the tumours and normal muscle tissue of the COS patients. Meanwhile, ABCB1, ABCC3, and p53 remained non-significant (p = 0.205; p = 0.841; p = 0.111, respectively).
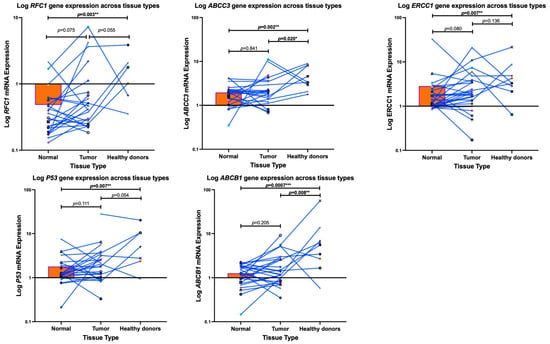
Figure 2.
A Mann–Whitney Test was employed to measure the gene expression profiles of conventional osteosarcoma patients (tumours vs. normal muscle tissue) compared to healthy donors (normal muscle) within South Africa. Each dot, shape, and colour represents each patient. Notably, a significant downregulation in the mRNA expressions of genes ABCB1 (p-glycoprotein), ABCC1, ERCC1, p-53, and RFC1 occurred in the conventional osteosarcoma patients compared to the healthy individuals. Furthermore, the tumour tissues of the conventional osteosarcoma patients exhibited a significant downregulation in the mRNA expression of ABCB1 (p-glycoprotein) and ABCC3, compared to healthy patients’ muscles, as measured using real-time PCR. The * p < 0.05, ** p < 0.01, *** p < 0.001 denotes significance.
3.3. Chemotherapy Response
3.3.1. Histology
With univariate logistic regression, the osteoblastic subtype was negatively associated with the chemotherapy response (odds ratio [OR] −0.36, CI 0.652 to −0.092; p = 0.026), while the fibroblastic subtype (OR 0.91, CI 0.569 to 1.248; p = 0.001) and mixed subtype (OR 0.53, CI 0.232 to 0.032; p = 0.032) were positively associated. With the addition of age and sex in a multivariate analysis, the osteoblastic histology (OR −0.45, CI −0.810 to −0.092; p = 0.024) remained negatively associated with the chemotherapy response, while the fibroblastic histology (OR 0.9, CI 0.523 to 1.290; p = 0.001) and mixed histology (OR 0.56, 0.076 to 1.046; p = 0.035), remained positively associated. The chondroblastic histology was not significant in the univariate (OR −0.29, CI −0.623 to 0.0351; p = 0.093) nor multivariate analysis (OR −0.41, −0.816 to −0.0135; p = 0.057).
3.3.2. Gene Candidates
Using an unpaired Mann–Whitney two-tailed analysis, only the ERCC1 gene was significantly associated with the chemotherapy response (p = 0.033) when comparing the non-responders against responders (Figure 3). The non-responders showed a mean 4.2-fold increase (95% CI −7.862 to –0.517) in ERCC1 gene expression levels. In contrast, we found no significant differences in the mRNA expression levels between the responders and non-responders for ABCC3 (p = 0.495), ABCB1 (p = 0.231), p53 (p = 0.199), and RFC1 (p = 0.658).
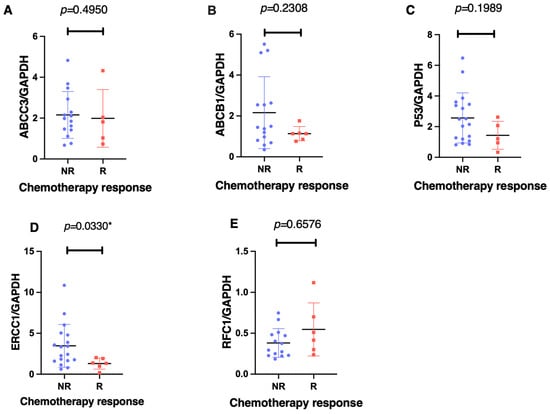
Figure 3.
The gene expression associations of ABCC3 (A), ABCB1 (B), p53 (C), ERCC1 (D), and RFC1 (E), with the effect of the chemotherapy response, as measured using an unpaired, nonparametric, Mann–Whitney, two-tailed experimental design with a significant difference set at p < 0.005. Each patient is represented by either a different colour or shape. Notably, there was a significant association between the ERCC1 expression levels and the chemotherapeutic response in the non-responders (N < 90% tumour necrosis—Huvos criteria) as compared to the responders (N > 90% tumour necrosis) post-neoadjuvant chemotherapy. The * p < 0.05 denotes significance.
Our univariate logistic regression found that ERCC1 was negatively associated with a chemotherapeutic response [restricted cubic spline (RCS) knot 1: OR −0.27, CI −0.504 to −0.032; p = 0.036; knot 2: OR = 0.39, CI −0.043 to 0.831; p = 0.036], as shown in Figure 4, while none of the other gene candidates exhibited a significant association. With multivariable logistic regression, including age and sex, this association was no longer significant for ERCC1 (RCS knot 1: OR −0.26, CI −0.508 to −0.003; p = 0.062; knot 2: OR 0.37, CI −0.103 to 0.840; p = 0.142), nor were any of the other gene candidates significant.
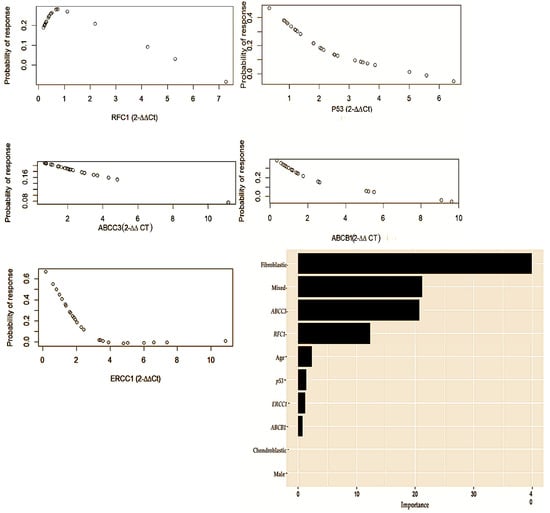
Figure 4.
Th gene concentration levels measured using the delta-delta (2−∆∆CT) method comprising RFC1, p53, ABCC3, ABCB1, and ERCC1. This multivariate logistic regression represents both the clinical and genetic factors that are predictive of chemotherapeutic responses in osteosarcoma. In accordance with the level of importance, the following variables are presented: age [Restricted cubic spline (RCS) knot: OR −2.5; CI −3.616 to −1.378; p = 0.022]; ABCC3 [RCS knot: OR 0.67; CI 0.407 to 0.936, p = 0.016]; ERCC1 [RCS knot: OR 0.57; CI 0.235 to 0.901; p = 0.044]; RFC1 [RCS knot: OR −1.04; CI −1.592 to −0.487; p = 0.035]; chondroblastic [OR −0.83; CI −1.106 to −0.520; p = 0.012]; and osteoblastic [OR −1.28; CI −1.664 to −0.91; p = 0.007].
3.3.3. Prediction of Multiple Factors in Predicting Chemotherapeutic Response
Our multivariable logistic regression, which included the age, sex, histological sub-type, and gene candidates found that age, ABCC3, ERCC1, RFC1, and chondroblastic and osteoblastic histology contributed significantly to predicting a response to chemotherapy. The age and gene candidates were modelled as three-knot restricted cubic splines (Table 5).

Table 5.
Multivariate logistic regression predicting chemotherapeutic response.
4. Discussion
This is the first prospective African study interrogating the gene expression profiles associated with the chemo-response of conventional osteosarcoma (COS). We analysed relative gene expression data using RT–PCR and the 2−∆∆CT method [18]. We observed a unique pattern of significant downregulation in the mRNA expressions of genes ABCB1 (p-glycoprotein), ABCC1, ERCC1, p-53, and RFC1 of the COS patients compared to the phenotypically healthy donor muscles. Furthermore, the tumour tissues with histologically confirmed COS exhibited a significant downregulation in their mRNA expression profiles for ABCB1 (p-glycoprotein) and ABCC3 compared to the healthy donor muscles, as measured using real-time PCR. Although our cohort had variability in their mRNA gene expression, we caution about interpreting our results as the sample size was small, aside from the intrinsic and extrinsic factors that may influence gene expression. Nonetheless, in this cohort, the downregulation in the mRNA expression of ABCB1 (p-glycoprotein), ABCC1, ERCC1, p-53, and RFC1 in the COS patients suggests the involvement of unknown biological risk factors of genomic instability such as cancer cell mutations, tumour microenvironment (cell types, signalling molecules, and extracellular matrix), epigenetic changes (DNA methylation and histone modification), dysregulation signalling pathways, immune response, and metabolic changes. In 2023, a systematic review described the gene expression patterns of COS as highly variable and heterogeneous. Furthermore, 473 differentially expressed genes (DEGs) were associated with chemotherapy response, and 57 genes were associated with MDR [12]. Unfortunately, not all 57 chemoresistance genes previously identified by the authors could be explored in this series amid our limited resources. The slight increase in COS among Africans suggests unknown risk factors and genetic alterations [19]. The genetic and epigenetic pattern needs to be studied in greater detail amongst individuals of African ethnicity. Our study indicates there are differential expression patterns amongst healthy individuals and COS patients. Novel gene expression profiles may underpin the tumourigenesis of osteosarcoma in African ethnicities; hence, the aggressive nature with an advanced Enneking stage, early metastasis, and impoverished survivals [19,20,21]. Therefore, wide genomic exploratory studies are needed to expand our understanding of this disease.
The ERCC1 gene is representative of the nucleotide excision repair genes, which assist osteosarcoma tumour cells in repairing the DNA damage caused by chemotherapy drugs, including cisplatin and cyclophosphamide [22]. In our study, the ERCC1 gene was downregulated in the COS patients, which may suggest it has undergone biological alterations. The dysregulated ERCC1 gene in the amputated osteosarcoma tumours suggests its participation in the tumourigenesis, aggressive phenotype, and poor chemo-response [23]. In our univariate and multivariate analyses of osteosarcomas, the ERCC1 gene in the COS tumour specimens was negatively associated with the chemo-response. Contrasting evidence co-exists regarding the associations between ERCC1 gene expression studies, polymorphisms, and chemo-response in COS [22,23,24,25,26,27]. In 2023, Trujillo-Paolillo et al. and colleagues concurred with our findings that ERCC1 gene expression was negatively associated with chemo-response [23]. However, Nathrath et al. (2012) found no correlation between the ERCC1 gene and chemo-response in 45 osteosarcoma patients [24]. Meanwhile, Hao et al. (2012) and Zhang et al. (2015) implicated the ERCC1 single nucleotide polymorphism (SNP) rs11615 in patients with a good chemo-response and prognosis [15,25]. Similarly, a meta-analysis by Liu et al. (2017) concurred with the previous authors and further suggested that the ERCC1 rs11615 SNP could be a useful genetic marker for predicting osteosarcoma prognosis [25]. These findings contradict Yang et al. (2012) and Li et al. (2014), who did not find a significant association between the ERCC1 polymorphism and chemo-response [26,27]. Fanelli et al. (2020) used the in vitro validation of candidate DNA repair-related therapeutic targets and drugs for tailored treatment in cisplatin-resistant osteosarcoma and found the ERCC1 gene as one of the main therapeutic targets [28]. Therefore, ERCC1 mRNA expression and polymorphisms warrant further investigations in osteosarcoma patients.
On the other hand, the ABC transporters (including the ABCC3 and ABCB1 gene products) are transmembrane ATP-dependent efflux pumps that block chemotherapy drugs from entering osteosarcoma tumour cells, leading to poor outcomes and systemic toxicity [9,12,13]. In our study, the ABCC3 and ABCB1 genes in the osteosarcoma tissues were significantly downregulated compared to the healthy donors. This may suggest that they are mutated, participate in tumour suppression, or are influenced by cancer cells’ epigenetic patterns, which can silence the gene that will normally suppress the tumour. In our multivariate logistic regression, ABCC3 was a negative predictor of chemo-response. In 2021, Ramírez-Cosmes et al. implicated the ABCC3 gene in chemotherapeutic responses in various cancers including lung, colon, breast, bladder, and gliomas [29]. Furthermore, in 2022, a systematic appraisal by Hurkmans et al., which focused on the role of genetic variants in COS, found ABCC3 and ABCB1 variants conferred advantages in chemotherapeutic responses, relapse, and event-free and overall survival [30]. Baldini et al. found that ABCB1 (P-pg) is associated with MDR of osteosarcoma, specifically referencing conventional chemotherapy drugs, including doxorubicin [31]. Furthermore, with respect to P-pg, overexpression is associated with cisplatin efficacy in osteosarcoma patients [13]. The expression of ABCB1 and ABCC3 in osteosarcoma metastatic tumour biopsies confers impoverished event-free and overall survival outcomes [23].
In contrast to drug efflux pumps, the reduced folate carrier 1 (RFC1) gene is responsible for the intracellular transportation of chemotherapy drugs, including high-dose methotrexate (HDMTX), and its decreased expression is thought to lead to a poor chemotherapeutic response [9,14,32,33]. On the other hand, in 2022, Wu et al. conducted an integrative analysis of the expression of the RFC family of genes, which exhibited an increase in sarcoma tissues [34]. In our osteosarcomas, the expression of RFC1 negatively predicted the chemotherapeutic response. HDMTX is a mainstay therapeutic agent in osteosarcoma; therefore, investigating genetic variants with the HDMTX pathway may provide important insights for future clinical practice [9,14,32,33]. The significant correlation between low RFC1 mRNA expression at diagnosis, poor histological response, and osteosarcoma recurrence warrants larger-scale investigation [32]. In addition, p53, the oncogene, is considered the prototypic tumour suppressor gene in osteosarcoma in that a complete loss of function is required before tumourigenesis [15,35]. Dysregulated or mutant P53 influences chemotherapeutic responses and impoverished COS survival [36]. In our case, the p53 gene was downregulated in the COS patients compared to the healthy donors, which may also suggest it has undergone biological alterations, as mentioned previously, which underscores the need for further elucidation.
In 2011, Kubista et al. used differentially expressed genes to classify the histological subtypes of COS and found the genes prohibitin, Annexin 1, Annexin 4, and GGH to be involved in chemoresistance [37]. Their bioinformatic analysis showed that the previously mentioned genes were expressed in osteoblastic and non-osteoblastic osteosarcoma subtypes [37]. In our series, the proportions of COS histological subtypes comprised 46% osteoblastic, 32% chondroblastic, 11% mixed lesions, and 11% fibroblastic tumours. Age, osteoblastic subtypes, and chondroblastic subtypes were negative predictors of chemo-response. In contrast, fibroblastic and mixed subtypes were positively predictive of chemo-response. Young age in osteosarcoma confers a better chemo-response, as well as event-free and overall disease survival [3,38]; this is in contrast to increased age > 22, which increases the risk of mortality in COS patients [3]. However, this remains to be seen in our prospective follow-up of this cohort. In 2002, Hauben et al. found a significant proportion of good chemo-responders in the fibroblastic subtype compared to the chondroblastic subtype [39]. Contrary to Simeland et al. (2019), who conducted the largest EURAMOS -1 osteosarcoma clinical trial and found that the histological subtype telangiectatic exhibited better survival than the unspecified conventional subtypes [5]. Good chemotherapy response in COS confers better survival [5,6,7], but unfortunately, 84% of our patients exhibited chemoresistance. This may suggest unique genetic expression in our African population.
The strengths of this study include its prospective design, and that it is the first study of this type in the African population to investigate the topic. The sampling strategy used stringent inclusion criteria for only conventional subtypes of osteosarcoma and exclusion criteria for other variants. In addition, the genes investigated underpinned COS tumourigenesis, MAP pharmacokinetics, pharmacodynamics, and MDR mechanisms. Owing to the rarity of osteosarcoma, there is a selection bias associated with the small sample size, and the lack of data regarding mRNA expression measurements in the African population, which prevented us from conducting a formal power calculation for the study. This restriction compromises the statistical power and generalizability of the findings, making it difficult to draw robust conclusions about gene expression profiles and their clinical relevance. Three patients were not available for the final analysis, of which two of those abandoned chemotherapy and surgery and went to seek alternative traditional medicine, and one demised amid chemotherapy toxicity. Furthermore, there is a selection bias when employing candidate gene selection or pre-selected target analysis rather than an accurate representation of the genome-wide expression profile. Discordances of evidence exist in the standardised experimental protocols, and the use of different technologies for data acquisition and analysis when investigating osteosarcoma gene expression, make it challenging to compare the results [8,9,10,11,12]. We used RT–PCR and the 2−∆∆CT method, which may introduce another selection bias [18].
5. Conclusions
In this South African population, we observed a significant downregulation in the mRNA expression of genes ABCB1, ABCC3, ERCC1, RFC1, and p53 in the conventional osteosarcoma (COS) patients compared to the healthy individuals. Furthermore, compared to the healthy individuals, the tumour tissues with COS exhibited a significant downregulation in the mRNA expression profiles for ABCB1 (p-glycoprotein) and ABCC3. Our univariate logistic regression analysis revealed several predictors of chemotherapeutic response, including ERCC1, along with osteoblastic, fibroblastic, and mixed tumour subtypes. The multivariable analysis further underscored the significance of age, ABCC3, ERCC1, RFC1, and specific tumour histologies in predicting treatment outcomes. These may serve as prognostication and therapeutic targets. Larger-scale research is needed on the African continent.
Author Contributions
Conceptualization, P.G.M., L.M. and V.R.; methodology, P.G.M., A.G., T.A. and V.R.; software, P.G.M.; validation, L.M., V.R. and T.A.; formal analysis, P.G.M., T.A., V.R., L.M., A.G. and R.R.; investigation, P.G.M., T.A., V.R., A.G. and L.M.; resources, P.G.M. and L.M.; data curation, P.G.M. and L.M.; writing—original draft preparation, P.G.M., T.A. and L.M.; writing—review and editing, P.G.M., L.M., V.R. and T.A.; visualization, L.M., V.R., T.A. and R.R.; supervision, L.M., V.R. and R.R.; project administration, P.G.M. and L.M.; funding acquisition, P.G.M. and L.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Discovery Foundation, grant ref. 049492, through the University of KwaZulu-Natal. The foundation is managed by Tshikululu Social Investments (www.tshikululu.org.za, accessed on 3 February 2024) and www.discovery.co.za (accessed on 3 February 2024). Email: discoveryfoundation@tshikululu.org.za. Tel.: (011)-544-0300/Fax: (011)-484-5997. Registration Number: IT 10514/05. V.R. was funded as a FLAIR Research Fellow (the Future Leader in African Independent Research (FLAIR) Fellowship Programme was a partnership between the African Academy of Sciences (AAS) and the Royal Society that was funded by the United Kingdom Government as part of the Global Challenge Research Fund (GCRF) (Grant No. FLAIR-FLR\R1\190204); supported by the South African Medical Research Council (SAMRC) with funds from the Department of Science and Technology (DST). Funding was also provided in part through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative (Grant No. DEL-15-006) by the AAS. A.G. was funded by The Poliomyelitis Research Foundation (PRF) [Grant No. PRF22/77] and CHS Funding, College of Health Science, University of KwaZulu-Natal. T.A. is funded by South African Medical Research Council Sir Grant and L’ORÉAL UNESCO Women in Science South African Young Talent fellow.
Institutional Review Board Statement
This study was conducted per the Declaration of Helsinki and approved by the University of KwaZulu-Natal Biomedical Research Ethics Committee (protocol code BREC/00002737/2021 on 31 August 2021).
Informed Consent Statement
Informed consent was obtained from all patients and/or, depending on the patient’s age, their legal guardians.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.
Acknowledgments
We thank all patients and their families, as well as the staff of the participating institutions.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Fletcher, C.D.M.; Unni, K.K.; Mertens, F. (Eds.) World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone; IARC Press: Lyon, France, 2002; pp. 1–36. [Google Scholar]
- Klein, M.J.; Siegal, G.P. Osteosarcoma: Anatomic and histologic variants. Am. J. Clin. Pathol. 2006, 125, 555–581. [Google Scholar] [CrossRef] [PubMed]
- Xin, S.; Wei, G. Prognostic factors in osteosarcoma: A study level meta-analysis and systematic review of current practice. J. Bone Oncol. 2020, 21, 100281. [Google Scholar] [CrossRef] [PubMed]
- Huvos, A.G.; Rosen, G.; Marcove, R.C. Primary osteogenic sarcoma: Pathologic aspects in 20 patients after treatment with chemotherapy en bloc resection, and prosthetic bone replacement. Arch. Pathol. Lab. Med. 1977, 101, 14–18. [Google Scholar] [PubMed]
- Smeland, S.; Bielack, S.S.; Whelan, J.; Bernstein, M.; Hogendoorn, P.; Krailo, M.D.; Gorlick, R.; Janeway, K.A.; Ingleby, F.C.; Anninga, J.; et al. survival and prognosis with osteosarcoma: Outcomes in more than 2000 patients in the EURAMOS-1 (European and American Osteosarcoma Study) cohort. Eur. J. Cancer 2019, 109, 36–50. [Google Scholar] [CrossRef]
- Marina, N.M.; Smeland, S.; Bielack, S.S.; Bernstein, M.; Jovic, G.; Krailo, M.D.; Hook, J.M.; Arndt, C.; van den Berg, H.; Brennan, B.; et al. Comparison of MAPIE versus MAP in patients with a poor response to preoperative chemotherapy for newly diagnosed high-grade osteosarcoma (EURAMOS-1): An open-label, international randomised controlled trial. Lancet Oncol. 2016, 17, 1396–1408. [Google Scholar] [CrossRef]
- Zhang, Y.; He, Z.; Duan, Y.; Wang, C.; Kamar, S.; Shi, X.; Yang, J.; Yang, J.; Zhao, N.; Han, L.; et al. Does intensified chemotherapy increase survival outcomes of osteosarcoma patients? A meta-analysis. J. Bone Oncol. 2018, 12, 54–60. [Google Scholar] [CrossRef]
- Mintz, M.B.; Sowers, R.; Brown, K.M.; Hilmer, S.C.; Mazza, B.; Huvos, A.G.; Meyers, P.A.; LaFleur, B.; McDonough, W.S.; Henry, M.M.; et al. An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res. 2005, 65, 1748–1754. [Google Scholar] [CrossRef]
- Marchandet, L.; Lallier, M.; Charrier, C.; Baud’huin, M.; Ory, B.; Lamoureux, F. Mechanisms of Resistance to Conventional Therapies for Osteosarcoma. Cancers 2021, 13, 683. [Google Scholar] [CrossRef]
- Man, T.-K.; Chintagumpala, M.; Visvanathan, J.; Shen, J.; Perlaky, L.; Hicks, J.; Johnson, M.; Davino, N.; Murray, J.; Helman, L.; et al. Expression profiles of osteosarcoma that can predict response to chemotherapy. Cancer Res. 2005, 65, 8142–8150. [Google Scholar] [CrossRef]
- Ochi, K.; Daigo, Y.; Katagiri, T.; Nagayama, S.; Tsunoda, T.; Myoui, A.; Naka, N.; Araki, N.; Kudawara, I.; Ieguchi, M.; et al. Prediction of response to neoadjuvant chemotherapy for osteosarcoma by gene-expression profiles. Int. J. Oncol. 2004, 24, 647–655. [Google Scholar] [CrossRef]
- Mthethwa, P.G.; Marais, L.C.; Ramsuran, V.; Aldous, C.M. A Systematic Review of the Heterogenous Gene Expression Patterns Associated with Multidrug Chemoresistance in Conventional Osteosarcoma. Genes 2023, 14, 832. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Sun, Z.; Hoffman, R.M.; Yang, Z.; Jiang, Y.; Wang, L.; Hao, Y. P-glycoprotein overexpression is associated with cisplatin resistance in human osteosarcoma. Anticancer Res. 2019, 39, 1711–1718. [Google Scholar] [CrossRef] [PubMed]
- Patiño-García, A.; Zalacaín, M.; Marrodán, L.; San-Julián, M.; Sierrasesúmaga, L. Methotrexate in Pediatric Osteosarcoma: Response and Toxicity in Relation to Genetic Polymorphisms and Dihydrofolate Reductase and Reduced Folate Carrier 1 Expression. J. Pediatr. 2009, 154, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lv, L.Y.; Li, B.J.; Zhang, J.; Wei, F. Investigation of ERCC1 and ERCC2 gene polymorphisms and response to chemotherapy and overall survival in osteosarcoma. Genet. Mol. Res. GMR 2015, 14, 11235–11241. [Google Scholar] [CrossRef] [PubMed]
- Kaseta, M.K.; Khaldi, L.; Gomatos, I.P.; Tzagarakis, G.P.; Alevizos, L.; Leandros, E.; Papagelopoulos, P.J.; Soucacos, P.N. Prognostic value of bax, bcl-2, and p53 staining in primary osteosarcoma. J. Surg. Oncol. 2008, 97, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.H.; Moon, S.-H.; Suh, J.-S.; Yang, W.-I. Tumor Volume Change as a Predictor of Chemotherapeutic Response in Osteosarcoma. Clin. Orthop. Relat. Res. 2000, 376, 200–208. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sadykova, L.R.; Ntekim, A.I.; Muyangwa-Semenova, M.; Rutland, C.S.; Jeyapalan, J.N.; Blatt, N.; Rizvanov, A.A. Epidemiology and Risk Factors of Osteosarcoma. Cancer Investig. 2020, 38, 259–269. [Google Scholar] [CrossRef]
- Hart, H.; Parkes, J.D. Long-term outcomes in osteosarcoma patients in the Groote Schuur Hospital patient population: A retrospective review. S. Afr. J. Oncol. 2017, 1, a17. [Google Scholar] [CrossRef]
- Mthethwa, P.G.; Marais, L.C.; Aldous, C.M. Prognostic factors for overall survival of conventional osteosarcoma of the appendicular skeleton. Bone Jt. Open 2024, 5, 210–217. [Google Scholar] [CrossRef]
- Hattinger, C.M.; Patrizio, M.P.; Luppi, S.; Magagnoli, F.; Picci, P.; Serra, M. Current understanding of pharmacogenetic implications of DNA damaging drugs used in osteosarcoma treatment. Expert Opin. Drug Metab. Toxicol. 2019, 15, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Trujillo-Paolillo, A.; Tesser-Gamba, F.; Seixas Alves, M.T.; Filho, R.J.G.; Oliveira, R.; Petrilli, A.S.; Toledo, S.R.C. Pharmacogenetics of the Primary and Metastatic Osteosarcoma: Gene Expression Profile Associated with Outcome. Int. J. Mol. Sci. 2023, 24, 5607. [Google Scholar] [CrossRef] [PubMed]
- Nathrath, M.; Kremer, M.; Letzel, H.; Remberger, K.; Höfler, H.; Ulle, T. Expression of genes of potential importance in response to chemotherapy in osteosarcoma patients. Klin. Padiatr. 2002, 214, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Feng, W.; Zhang, J.; Sun, Y.J.; Wang, G. Association of four ERCC1 and ERCC2 SNPs with survival of bone tumour patients. Asian Pac. J. Cancer Prev. 2012, 13, 3821–3824. [Google Scholar] [CrossRef]
- Liu, X.Y.; Zhang, Z.; Deng, C.B.; Tian, Y.H.; Ma, X. Meta-analysis showing that ERCC1 polymorphism is predictive of osteosarcoma prognosis. Oncotarget 2017, 8, 62769–62779. [Google Scholar] [CrossRef]
- Li, J.; Liu, S.; Wang, W.; Zhang, K.; Liu, Z.; Zhang, C.; Chen, S.; Wu, S. ERCC polymorphisms and prognosis of patients with osteosarcoma. Tumor Biol. 2014, 35, 10129–10136. [Google Scholar] [CrossRef]
- Fanelli, M.; Tavanti, E.; Patrizio, M.P.; Vella, S.; Fernandez-Ramos, A.; Magagnoli, F.; Luppi, S.; Hattinger, C.M.; Serra, M. Cisplatin Resistance in Osteosarcoma: In vitro Validation of Candidate DNA Repair-Related Therapeutic Targets and Drugs for Tailored Treatments. Front. Oncol. 2020, 10, 331. [Google Scholar] [CrossRef]
- Ramírez-Cosmes, A.; Reyes-Jiménez, E.; Zertuche-Martínez, C.; Hernández-Hernández, C.A.; García-Román, R.; Romero-Díaz, R.I.; Manuel-Martínez, A.E.; Elizarrarás-Rivas, J.; Vásquez-Garzón, V.R. The implications of ABCC3 in cancer drug resistance: Can we use it as a therapeutic target? Am. J. Cancer Res. 2021, 11, 4127–4140. [Google Scholar]
- Hurkmans, E.G.E.; Brand, A.C.A.M.; Verdonschot, J.A.J.; te Loo, D.M.W.M.; Coenen, M.J.H. Pharmacogenetics of chemotherapy treatment response and -toxicities in patients with osteosarcoma: A systematic review. BMC Cancer 2022, 22, 1326. [Google Scholar] [CrossRef]
- Baldini, N.; Scotlandi, K.; Serra, M.; Picci, P.; Bacci, G.; Sottili, S.; Campanacci, M. P-glycoprotein expression in osteosarcoma: A basis for risk-adapted adjuvant chemotherapy. J. Orthop. Res. 1999, 17, 629–632. [Google Scholar] [CrossRef]
- Ifergan, I.; Meller, I.; Issakov, J.; Assaraf, Y.G. Reduced folate carrier protein expression in osteosarcoma. Cancer 2003, 98, 1958–1966. [Google Scholar] [CrossRef]
- Flintoff, W.F.; Sadlish, H.; Gorlick, R.; Yang, R.; Williams, F.M. Functional analysis of altered reduced folate carrier sequence changes identified in osteosarcomas. Biochim. Biophys. Acta 2004, 1690, 110–117. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, G.; Zhou, J.; Zhu, X.; Tang, X.; Liu, J.; Zhou, Q.; Chen, Z.; Liu, T.; Wang, W.; Xiao, X.; et al. Integrative analysis of expression, prognostic significance and immune infiltration of RFC family genes in human sarcoma. Aging 2022, 14, 3705–3719. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Guo, J.; Zhang, K.; Guo, Y. TP53 Mutations and Survival in Osteosarcoma Patients: A Meta-Analysis of Published Data. Dis. Markers 2016, 2016, 4639575. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Shen, J.; Choy, E.; Yang, C.; Mankin, H.; Hornicek, F.; Duan, Z. p53 overexpression increases chemosensitivity in multidrug-resistant osteosarcoma cell lines. Cancer Chemother. Pharmacol. 2016, 77, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Kubista, B.; Klinglmueller, F.; Bilban, M.; Pfeiffer, M.; Lass, R.; Giurea, A.; Funovics, P.T.; Toma, C.; Dominkus, M.; Kotz, R.; et al. Microarray analysis identifies distinct gene expression profiles associated with histological subtype in human osteosarcoma. Int. Orthop. 2011, 35, 401–411. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, C.; Tang, D.; Chen, B.; Jiang, Z. Development and Validation of Nomogram-Based Prognosis Tools for Patients with Extremity Osteosarcoma: A SEER Population Study. J. Oncol. 2022, 2022, 9053663. [Google Scholar] [CrossRef]
- Hauben, E.I.; Weeden, S.; Pringle, J.; Van Marck, E.A.; Hogendoorn, P.C. Does the histological subtype of high-grade central osteosarcoma influence the response to treatment with chemotherapy and does it affect overall survival? A study on 570 patients of two consecutive trials of the European Osteosarcoma Intergroup. Eur. J. Cancer 2002, 38, 1218–1225. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).