Correction: Tasci et al. RadWise: A Rank-Based Hybrid Feature Weighting and Selection Method for Proteomic Categorization of Chemoirradiation in Patients with Glioblastoma. Cancers 2023, 15, 2672
- 59.
- Zeng, S.; Li, W.; Ouyang, H.; Xie, Y.; Feng, X.; Huang, L. A Novel Prognostic Pyroptosis-Related Gene Signature Correlates to Oxidative Stress and Immune-Related Features in Gliomas. Oxid. Med. Cell. Longev. 2023, 2023, 4256116. https://doi.org/10.1155/2023/4256116.
Reference
- Tasci, E.; Jagasia, S.; Zhuge, Y.; Sproull, M.; Cooley Zgela, T.; Mackey, M.; Camphausen, K.; Krauze, A.V. RadWise: A Rank-Based Hybrid Feature Weighting and Selection Method for Proteomic Categorization of Chemoirradiation in Patients with Glioblastoma. Cancers 2023, 15, 2672. [Google Scholar] [CrossRef] [PubMed]
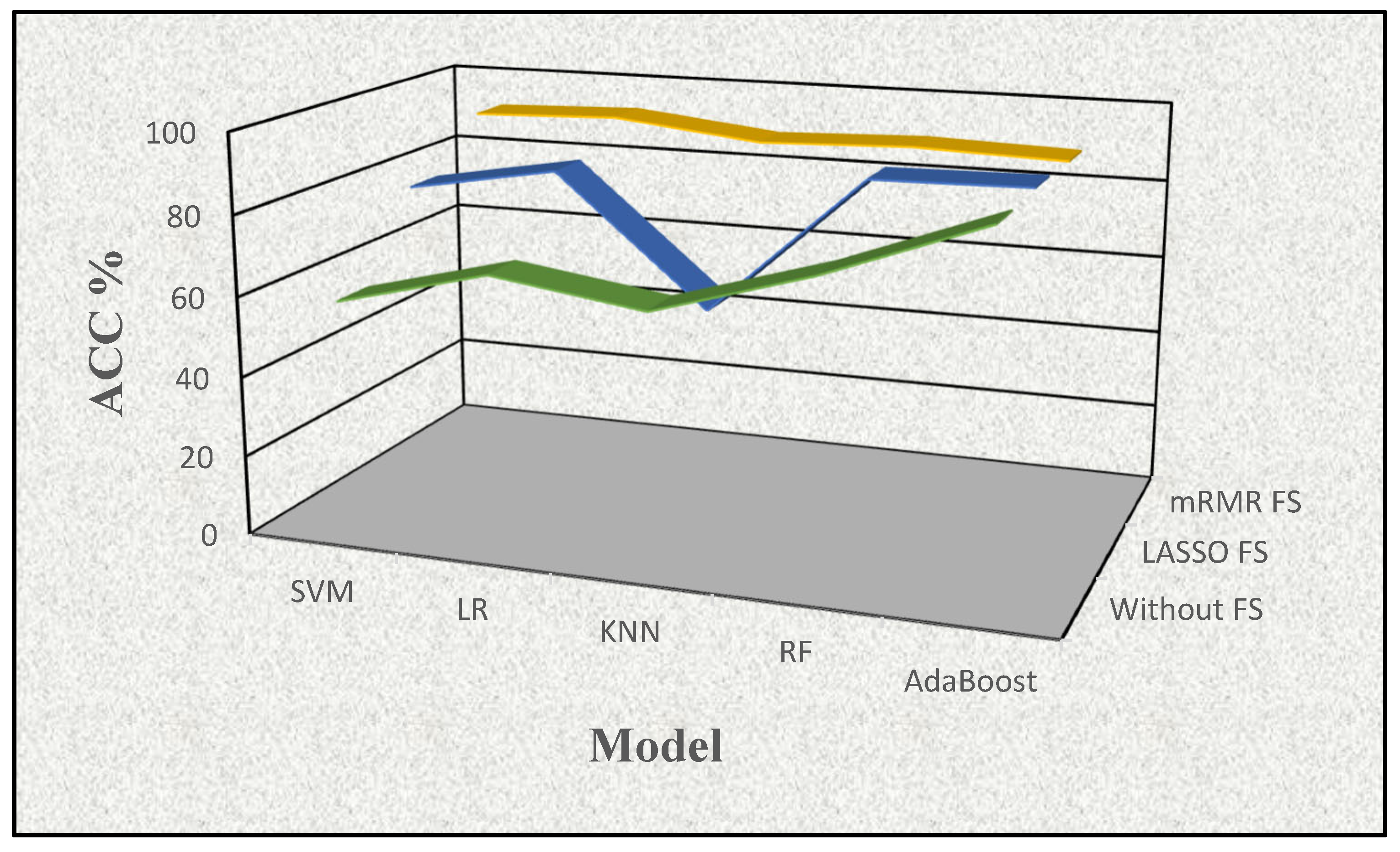
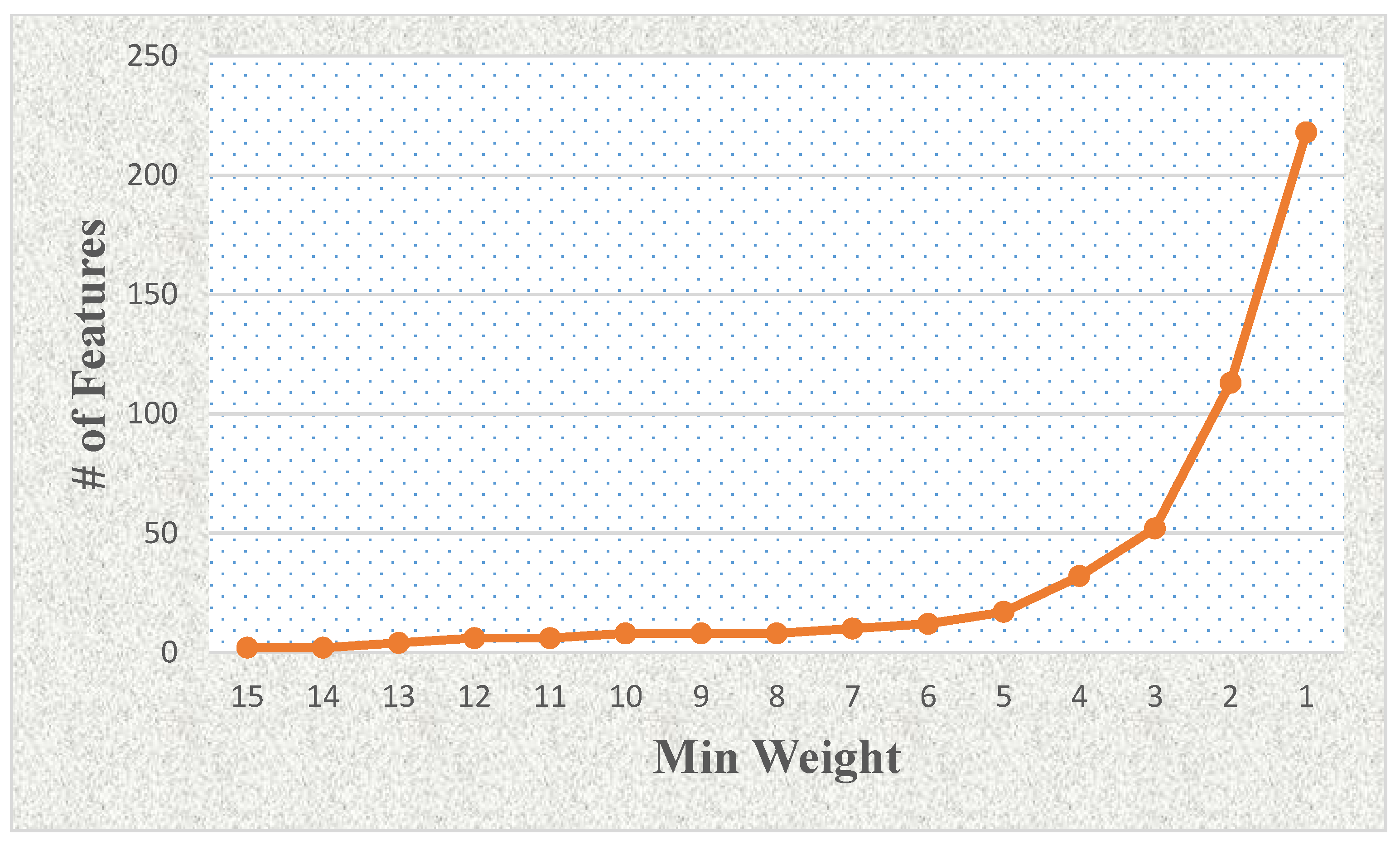
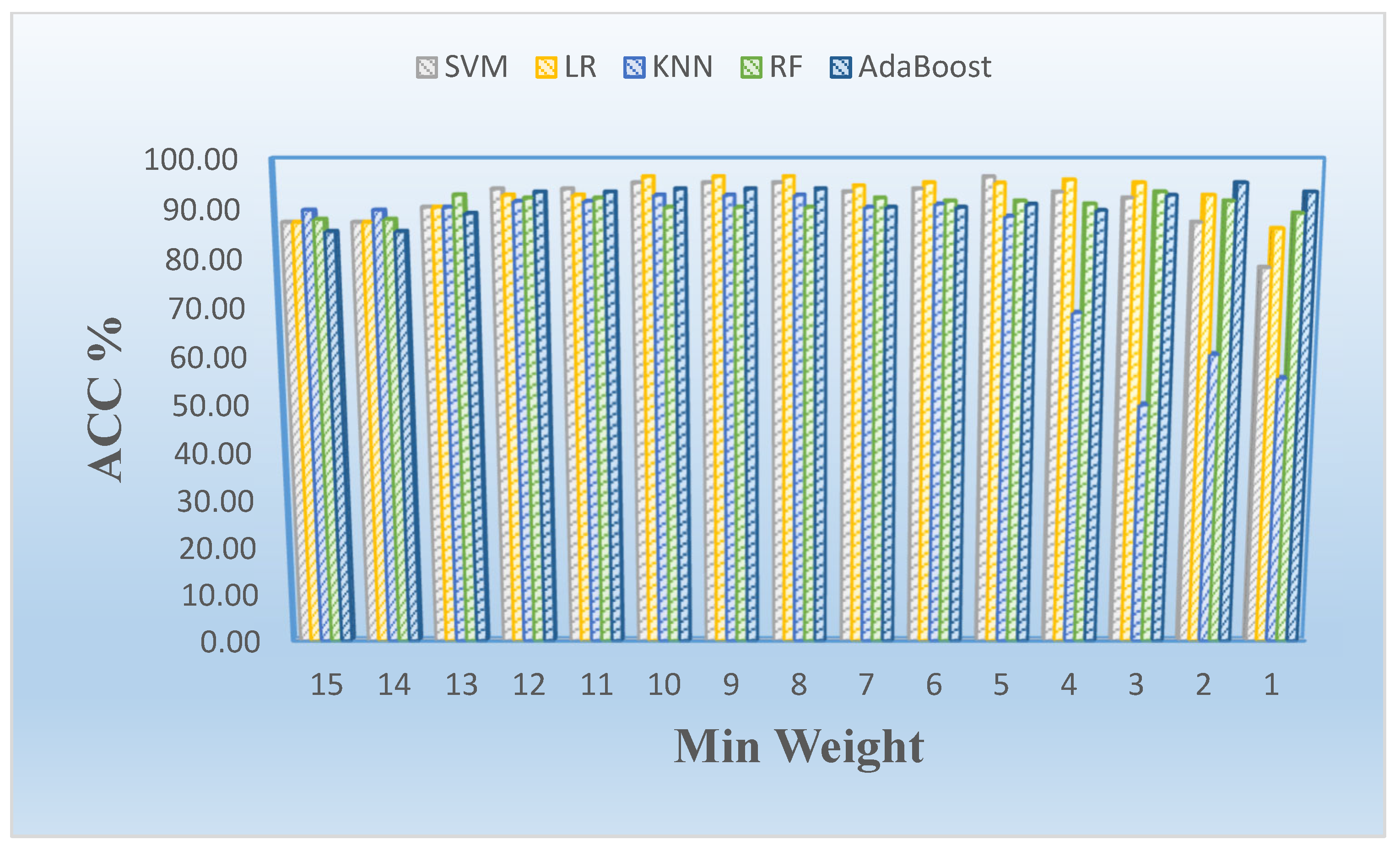

| ML-ACC | Without FS | LASSO FS | mRMR FS |
|---|---|---|---|
| SVM | 57.860 | 78.674 | 91.515 |
| LR | 67.633 | 85.341 | 92.708 |
| KNN | 62.197 | 53.068 | 88.466 |
| RF | 73.826 | 88.466 | 89.659 |
| AdaBoost | 88.409 | 89.072 | 88.447 |
| k | # of Features | SVM | LR | KNN | RF | AdaBoost |
|---|---|---|---|---|---|---|
| 5 | 11 | 93.314 | 92.083 | 82.386 | 91.496 | 91.477 |
| 4 | 26 | 89.640 | 93.314 | 62.197 | 93.939 | 90.890 |
| 3 | 44 | 89.053 | 96.363 | 46.345 | 92.121 | 93.939 |
| 2 | 90 | 85.985 | 92.064 | 60.379 | 91.496 | 93.901 |
| 1 | 197 | 76.269 | 85.966 | 54.868 | 87.841 | 87.822 |
| k | # of Features | SVM | LR | KNN | RF | AdaBoost |
|---|---|---|---|---|---|---|
| 5 | 5 | 86.004 | 88.428 | 87.235 | 87.841 | 87.197 |
| 4 | 7 | 90.890 | 92.708 | 90.265 | 91.496 | 91.496 |
| 3 | 8 | 95.152 | 96.364 | 92.708 | 90.871 | 93.920 |
| 2 | 11 | 92.708 | 96.364 | 92.689 | 90.284 | 94.508 |
| 1 | 34 | 92.102 | 92.102 | 87.254 | 92.121 | 90.871 |
| k | # of Features | SVM | LR | KNN | RF | AdaBoost |
|---|---|---|---|---|---|---|
| 15 | 2 | 87.216 | 87.216 | 89.659 | 87.841 | 85.379 |
| 14 | 2 | 87.216 | 87.216 | 89.659 | 87.841 | 85.379 |
| 13 | 4 | 90.265 | 90.265 | 90.284 | 92.727 | 89.034 |
| 12 | 6 | 93.920 | 92.708 | 91.477 | 92.102 | 93.314 |
| 11 | 6 | 93.920 | 92.708 | 91.477 | 92.102 | 93.314 |
| 10 | 8 | 95.152 | 96.364 | 92.708 | 90.265 | 93.920 |
| 9 | 8 | 95.152 | 96.364 | 92.708 | 90.265 | 93.920 |
| 8 | 8 | 95.152 | 96.364 | 92.708 | 90.265 | 93.920 |
| 7 | 10 | 93.333 | 94.546 | 90.284 | 92.102 | 90.284 |
| 6 | 12 | 93.939 | 95.152 | 90.909 | 91.496 | 90.246 |
| 5 | 17 | 96.345 | 95.114 | 88.447 | 91.496 | 90.871 |
| 4 | 32 | 93.295 | 95.739 | 68.921 | 90.890 | 89.640 |
| 3 | 52 | 92.121 | 95.151 | 49.962 | 93.333 | 92.670 |
| 2 | 113 | 87.216 | 92.670 | 60.379 | 91.496 | 95.152 |
| 1 | 218 | 78.087 | 85.966 | 55.492 | 89.053 | 93.314 |
| k | # of Features | SVM | LR | KNN | RF | AdaBoost |
|---|---|---|---|---|---|---|
| 15 | 2 | 5.175 | 4.408 | 4.072 | 4.232 | 2.191 |
| 14 | 2 | 5.175 | 4.408 | 4.072 | 4.232 | 2.191 |
| 13 | 4 | 4.423 | 4.821 | 4.425 | 4.924 | 2.384 |
| 12 | 6 | 5.060 | 5.269 | 3.509 | 4.088 | 3.515 |
| 11 | 6 | 5.060 | 5.269 | 3.509 | 4.088 | 3.515 |
| 10 | 8 | 3.636 | 4.454 | 5.269 | 3.496 | 4.272 |
| 9 | 8 | 3.636 | 4.454 | 5.269 | 3.496 | 4.272 |
| 8 | 8 | 3.636 | 4.454 | 5.269 | 3.496 | 4.272 |
| 7 | 10 | 4.848 | 5.555 | 4.425 | 4.088 | 4.425 |
| 6 | 12 | 4.285 | 3.636 | 6.357 | 3.995 | 3.526 |
| 5 | 17 | 2.966 | 2.444 | 3.476 | 4.827 | 5.400 |
| 4 | 32 | 6.177 | 4.105 | 6.384 | 4.259 | 1.442 |
| 3 | 52 | 4.535 | 2.424 | 7.329 | 4.020 | 2.469 |
| 2 | 113 | 4.807 | 1.557 | 4.954 | 4.430 | 4.924 |
| 1 | 218 | 4.720 | 3.133 | 5.559 | 3.031 | 3.515 |
| ML | ACC% | AUC | F1 | PRE | REC | SPEC |
|---|---|---|---|---|---|---|
| SVM | 57.860 | 0.415 | 0.518 | 0.698 | 0.515 | 0.690 |
| LR | 67.633 | 0.755 | 0.676 | 0.681 | 0.681 | 0.673 |
| KNN | 62.197 | 0.647 | 0.581 | 0.662 | 0.527 | 0.722 |
| RF | 73.826 | 0.808 | 0.744 | 0.768 | 0.746 | 0.737 |
| AdaBoost | 88.409 | 0.951 | 0.886 | 0.882 | 0.893 | 0.873 |
| ML | ACC% | AUC | F1 | PRE | REC | SPEC |
|---|---|---|---|---|---|---|
| SVM | 95.152 | 0.989 | 0.949 | 0.975 | 0.928 | 0.976 |
| LR | 96.364 | 0.987 | 0.964 | 0.963 | 0.965 | 0.965 |
| KNN | 92.708 | 0.965 | 0.930 | 0.929 | 0.932 | 0.923 |
| RF | 90.265 | 0.978 | 0.902 | 0.885 | 0.928 | 0.876 |
| AdaBoost | 93.920 | 0.979 | 0.941 | 0.941 | 0.942 | 0.935 |
| Entrez Gene Symbol | Target Full Name | Biological Relevance to Glioma |
|---|---|---|
| K2C5 | Keratin, type II cytoskeletal 5 | Yes, evolving biomarker/target [61] |
| Keratin-1 | Keratin, type II cytoskeletal 1 | Yes, evolving biomarker/target [61] |
| STRATIFIN (SFN) | 14-3-3 protein sigma | Yes, tumor suppressor gene expression pattern correlates with glioma grade and prognosis [62] |
| MIC-1 (GDF15) | Growth/differentiation factor 15 | Yes, biomarker, novel immune checkpoint [63] |
| GFAP | Glial fibrillary acidic protein | Yes, evolving biomarker/target [64] |
| CSPG3 (NCAN) | Neurocan core protein | Yes, glycoproteomic profiles of GBM subtypes, differential expression versus control tissue [65] |
| Cystatin M (CST6) | Cystatin M | Yes, cell type-specific expression in normal brain and epigenetic silencing in glioma [66] |
| Proteinase-3 (PRTN3) | Proteinase-3 | Yes, evolving role, may relate to pyroptosis, oxidative stress and immune response [59] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tasci, E.; Jagasia, S.; Zhuge, Y.; Sproull, M.; Cooley Zgela, T.; Mackey, M.; Camphausen, K.; Krauze, A.V. Correction: Tasci et al. RadWise: A Rank-Based Hybrid Feature Weighting and Selection Method for Proteomic Categorization of Chemoirradiation in Patients with Glioblastoma. Cancers 2023, 15, 2672. Cancers 2024, 16, 2744. https://doi.org/10.3390/cancers16152744
Tasci E, Jagasia S, Zhuge Y, Sproull M, Cooley Zgela T, Mackey M, Camphausen K, Krauze AV. Correction: Tasci et al. RadWise: A Rank-Based Hybrid Feature Weighting and Selection Method for Proteomic Categorization of Chemoirradiation in Patients with Glioblastoma. Cancers 2023, 15, 2672. Cancers. 2024; 16(15):2744. https://doi.org/10.3390/cancers16152744
Chicago/Turabian StyleTasci, Erdal, Sarisha Jagasia, Ying Zhuge, Mary Sproull, Theresa Cooley Zgela, Megan Mackey, Kevin Camphausen, and Andra Valentina Krauze. 2024. "Correction: Tasci et al. RadWise: A Rank-Based Hybrid Feature Weighting and Selection Method for Proteomic Categorization of Chemoirradiation in Patients with Glioblastoma. Cancers 2023, 15, 2672" Cancers 16, no. 15: 2744. https://doi.org/10.3390/cancers16152744
APA StyleTasci, E., Jagasia, S., Zhuge, Y., Sproull, M., Cooley Zgela, T., Mackey, M., Camphausen, K., & Krauze, A. V. (2024). Correction: Tasci et al. RadWise: A Rank-Based Hybrid Feature Weighting and Selection Method for Proteomic Categorization of Chemoirradiation in Patients with Glioblastoma. Cancers 2023, 15, 2672. Cancers, 16(15), 2744. https://doi.org/10.3390/cancers16152744






