High M2-TAM Infiltration and STAT3/NF-κB Signaling Pathway as a Predictive Factor for Tumor Progression and Death in Cervical Cancer
Abstract
Simple Summary
Abstract
1. Introduction
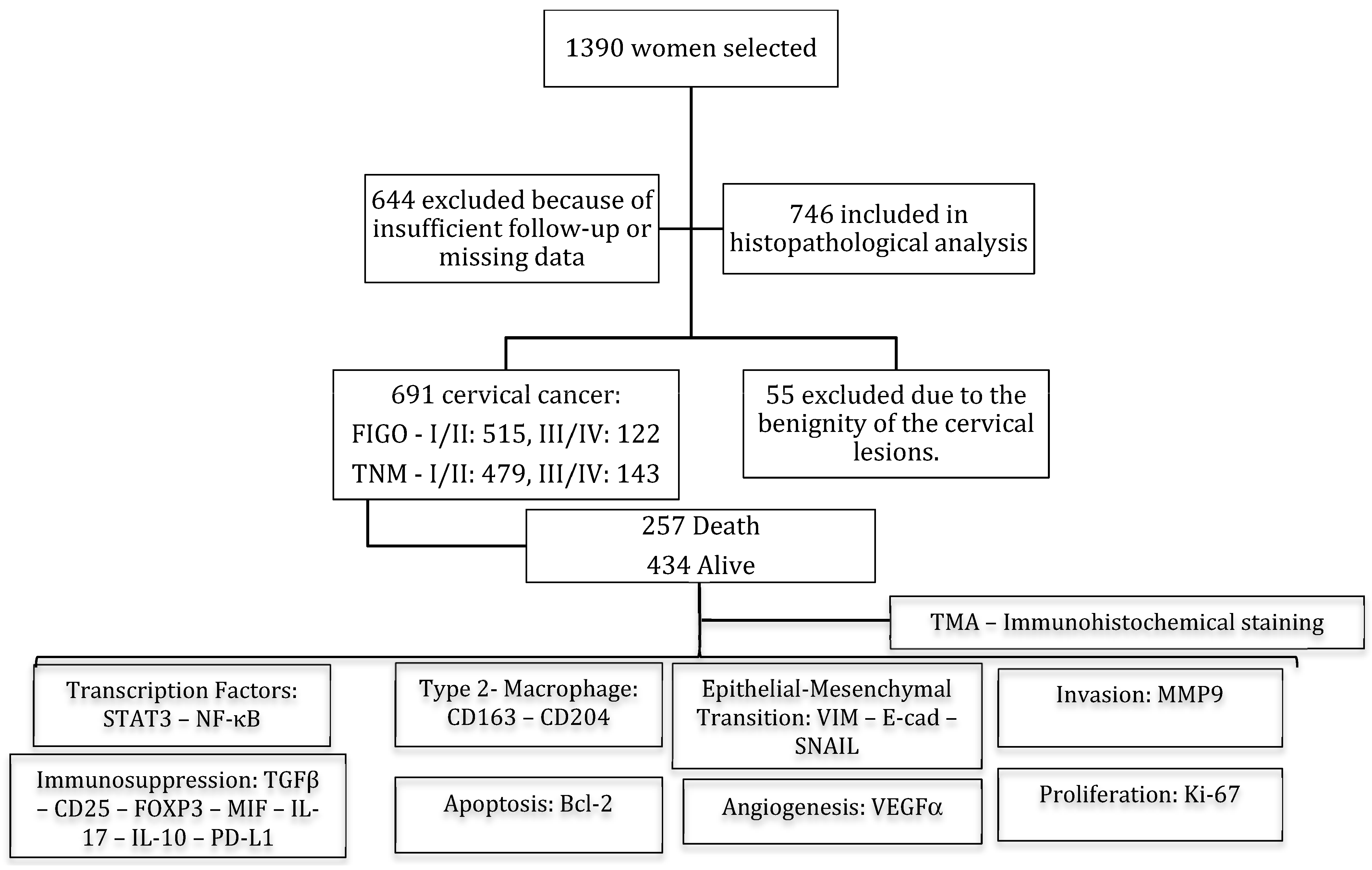
2. Material and Methods
2.1. Tissue Samples and Data Collection
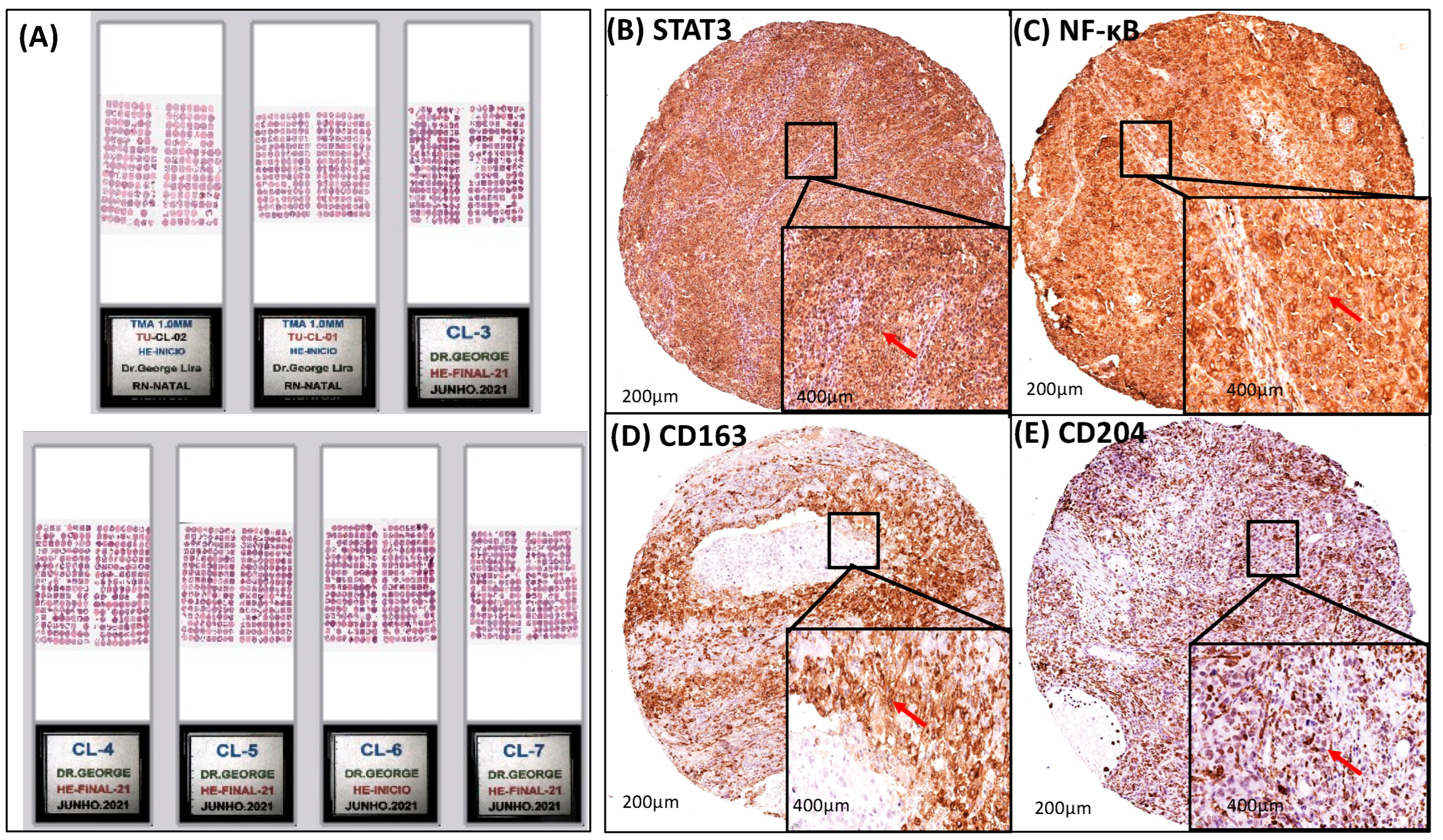
2.2. Tissue Microarray (TMA) Block Construction and Staining by Hematoxylin and Eosin (HE) and Immunohistochemistry (IHC)
Immunohistochemistry (IHC)
2.3. Quantitative Evaluation of Immunostaining
2.4. Follow-Up
2.5. Statistical Analysis
3. Results
3.1. Clinicopathologic Features of Patients
3.2. Analysis of Survival Rate, Clinicopathological Features, and Age
3.3. A Strong Association Interaction between CD204+ and CD163+ M2-TAM with STAT3 and NF-κB Signaling Pathways
3.4. Transcription Factors (STAT3 and NF-κB) and M2-TAM (CD204 and CD163) Modulated EMT (Vimentin, E-Cadherin, and SNAIL) and Invasion (MMP9)
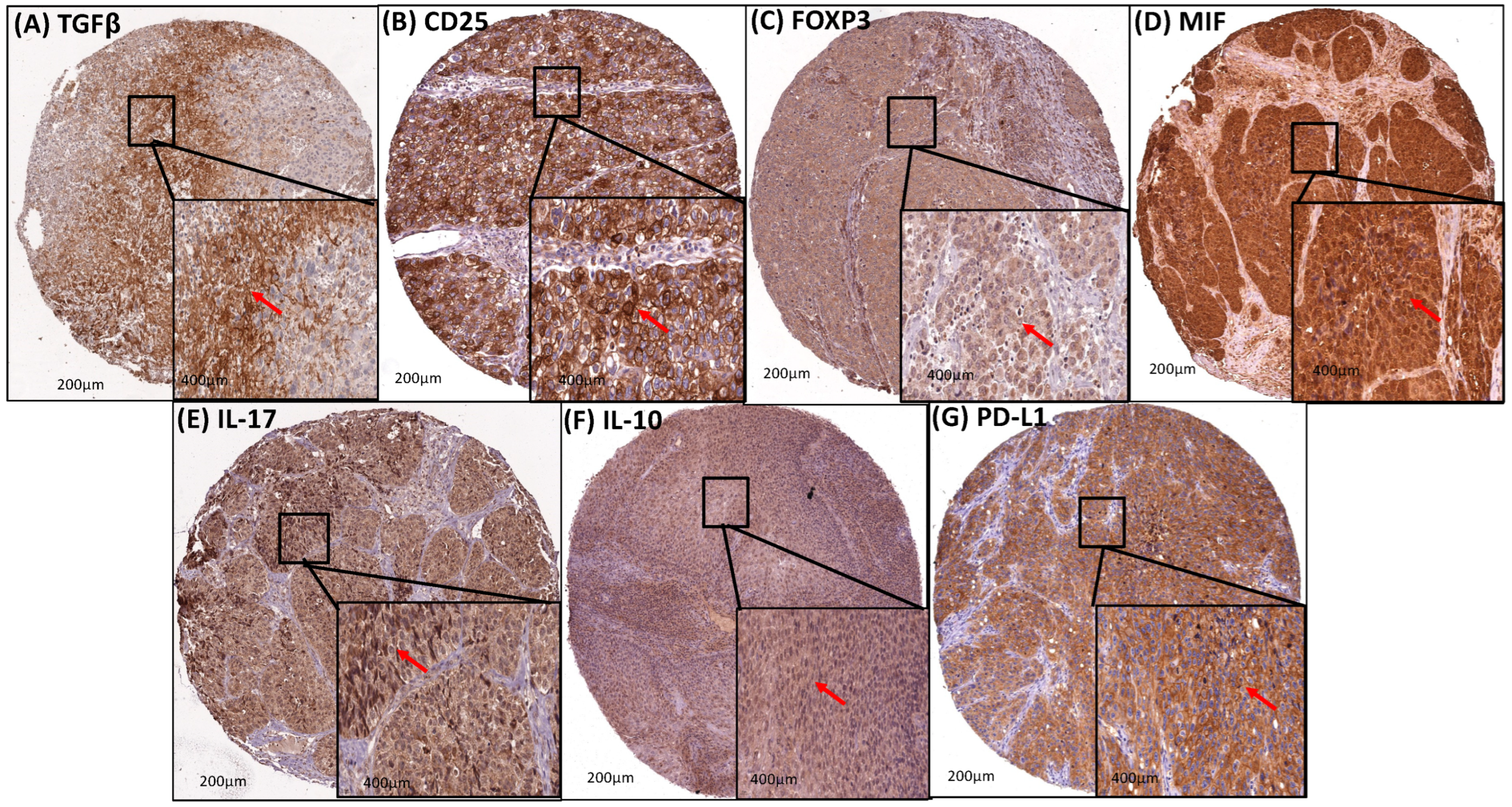
3.5. Transcription Factors (STAT3 and NF-κB) and M2-TAM (CD204 and CD163) Upregulated the Immunosuppression in the Cervical Cancer TME
3.6. Transcription Factors (STAT3 and NF-κB) and M2-TAM (CD204 and CD163) Had a Strong Association Correlation with Apoptosis, Angiogenesis, and Proliferation
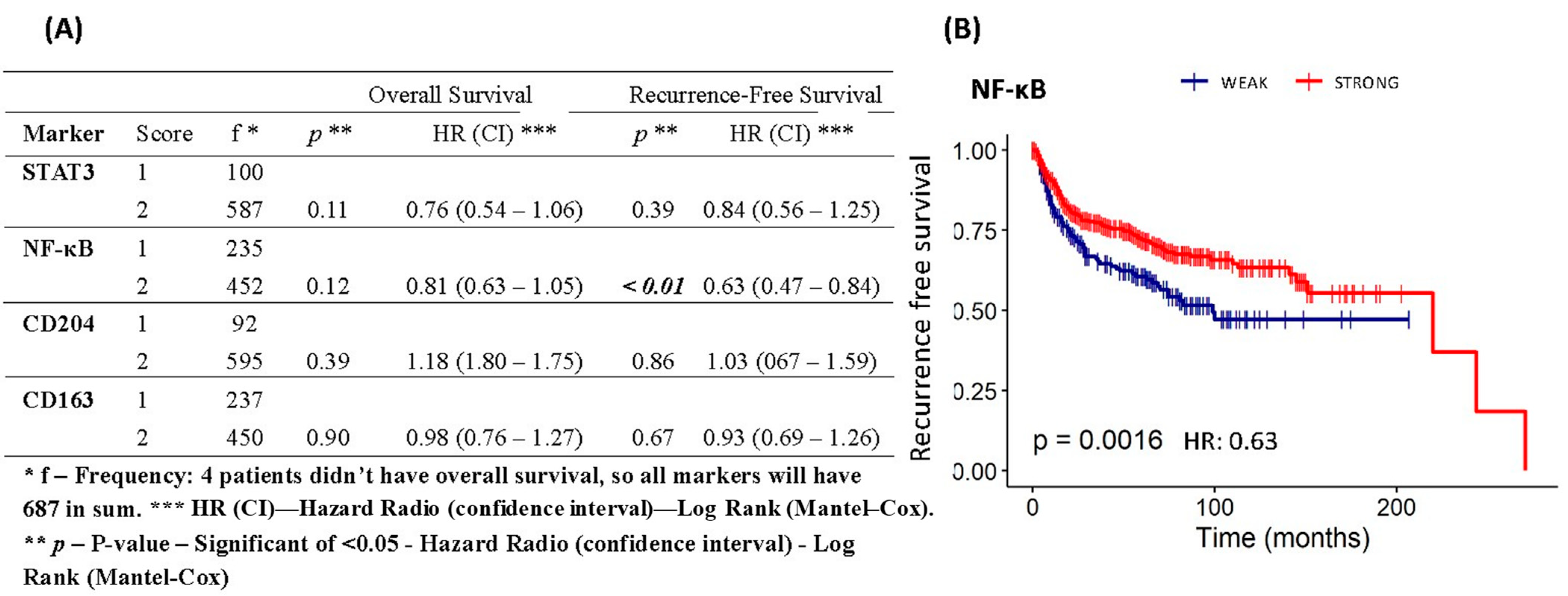
3.7. Assessment of Overall Survival and Recurrence-Free Survival of Patients with Cervical Cancer Based on Protein Expression of STAT3, NF-κB, CD163, and CD204
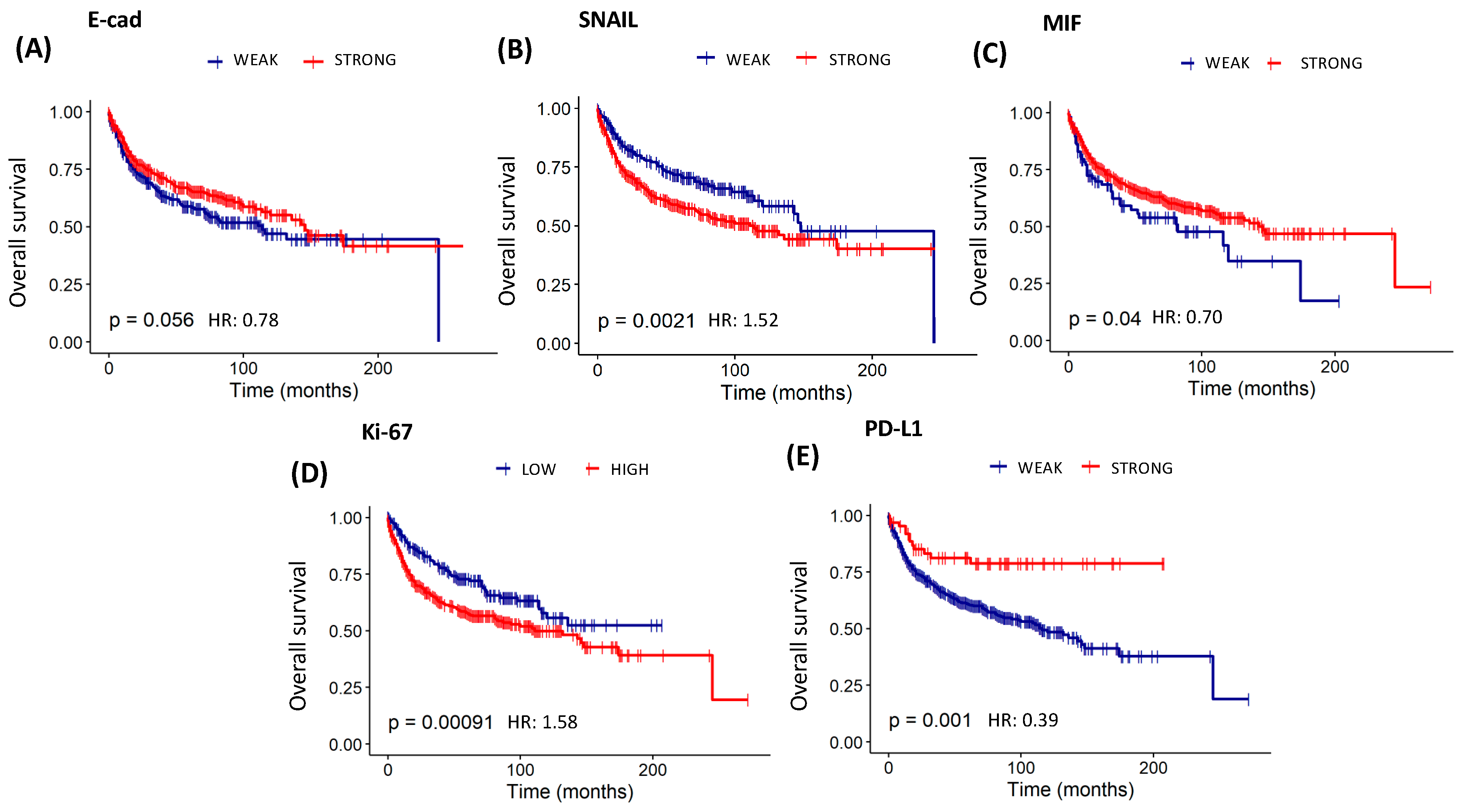
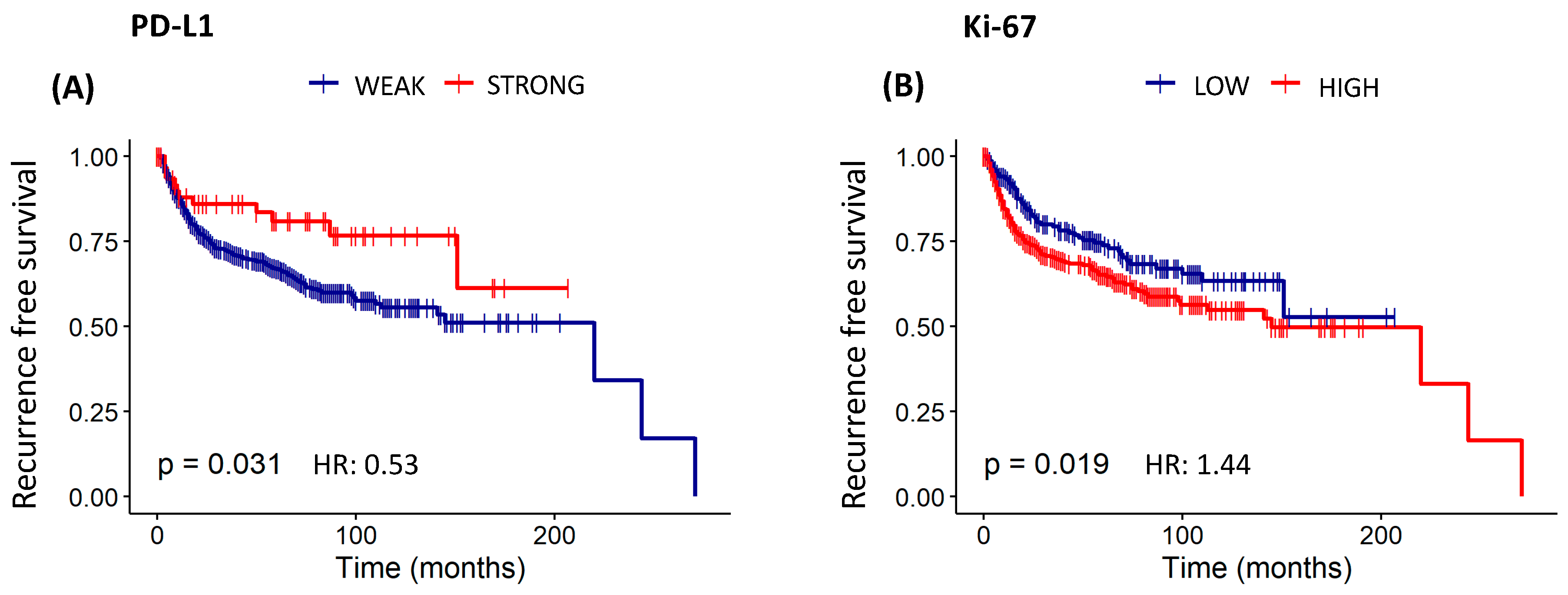
3.8. Assessment of Overall Survival and Recurrence-Free Survival Evaluations of Patients with Cervical Cancer Based on Protein Expression of EMT, Invasion, Immunosuppression, Resistance to Apoptosis, Angiogenesis, and Proliferation
3.9. Association Correlation between Transcription Factors, EMT, Invasion, Immunosuppression, Apoptosis Resistance, Angiogenesis, and Proliferation with Clinical TNM Staging
3.10. Prognostic Significance of Patients by TNM Clinic Staging
3.11. Overall Survival of Patients with Cervical Cancer Based on Lifestyle Database, Laboratory Analysis, Treatment, and Clinicopathological Characteristics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F.; Bsc, M.F.B.; Me, J.F.; Soerjomataram, M.I.; et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Santos, M.d.O.; Lima, F.C.d.S.d.; Martins, L.F.L.; Oliveira, J.F.P.; Almeida, L.M.d.; Cancela, M.d.C. Estimativa de Incidência de Câncer no Brasil, 2023–2025. Rev. Bras. Cancerol. 2023, 69, e213700. [Google Scholar] [CrossRef]
- Lei, J.; Ploner, A.; Elfström, K.M.; Wang, J.; Roth, A.; Fang, F.; Sundström, K.; Dillner, J.; Sparén, P. HPV Vaccination and the Risk of Invasive Cervical Cancer. N. Engl. J. Med. 2020, 383, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Wang, L.; Lin, J.-C.; Jan, J.-S. The prognostic factors for locally advanced cervical cancer patients treated by intensity-modulated radiation therapy with concurrent chemotherapy. J. Formos. Med. Assoc. 2015, 114, 231–237. [Google Scholar] [CrossRef][Green Version]
- Gadducci, A.; Cosio, S. Pharmacological Treatment of Patients with Metastatic, Recurrent or Persistent Cervical Cancer Not Amenable by Surgery or Radiotherapy: State of Art and Perspectives of Clinical Research. Cancers 2020, 12, 2678. [Google Scholar] [CrossRef] [PubMed]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-Associated Macrophages as Major Players in the Tumor Microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Wang, J.; Lu, D.; Xu, X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduct. Target. Ther. 2021, 6, 75. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, S.; Zhang, M.; Zhen, L.; Pang, D.; Zhang, Q.; Li, Z. High-Infiltration of Tumor-Associated Macrophages Predicts Unfavorable Clinical Outcome for Node-Negative Breast Cancer. PLoS ONE 2013, 8, e76147. [Google Scholar] [CrossRef] [PubMed]
- Poh, A.R.; Ernst, M. Targeting Macrophages in Cancer: From Bench to Bedside. Front. Oncol. 2018, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Tong, L.; Liu, F.L.; Liu, A.; Zeng, S.; Xiong, Q.; Yang, Z.; He, X.; Sun, Y.; Xu, C. Tumor-infiltrating M2 macrophages driven by specific genomic alterations are associated with prognosis in bladder cancer. Oncol. Rep. 2019, 42, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.T.; Knops, A.; Swendseid, B.; Martinez-Outschoom, U.; Harshyne, L.; Philp, N.; Rodeck, U.; Luginbuhl, A.; Cognetti, D.; Johnson, J.; et al. Prognostic Significance of Tumor-Associated Macrophage Content in Head and Neck Squamous Cell Carcinoma: A Meta-Analysis. Front. Oncol. 2019, 9, 656. [Google Scholar] [CrossRef]
- Guo, F.; Kong, W.; Zhao, G.; Cheng, Z.Z.; Ai, L.; Lv, J.; Feng, Y.C.; Ma, X.M. The correlation between tumor-associated macrophage infiltration and progression in cervical carcinoma. Biosci. Rep. 2021, 41. [Google Scholar] [CrossRef] [PubMed]
- Horta, B.; Pereira, T.; Medeiros, R.; Cerqueira, F. Cervical Cancer Outcome and Tumor-Associated Macrophages: Research Evidence. Immuno 2022, 2, 460–468. [Google Scholar] [CrossRef]
- Kanda, N.; Seno, H.; Konda, Y.; Marusawa, H.; Kanai, M.; Nakajima, T.; Kawashima, T.; Nanakin, A.; Sawabu, T.; Uenoyama, Y.; et al. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene 2004, 23, 4921–4929. [Google Scholar] [CrossRef]
- Scholz, A.; Heinze, S.; Detjen, K.M.; Peters, M.; Welzel, M.; Hauff, P.; Schirner, M.; Wiedenmann, B.; Rosewicz, S. Activated signal transducer and activator of transcription 3 (STAT3) supports the malignant phenotype of human pancreatic cancer. Gastroenterology 2003, 125, 891–905. [Google Scholar] [CrossRef]
- Júnior, R.F.d.A.; A Lira, G.; Schomann, T.; Cavalcante, R.S.; Vilar, N.F.; de Paula, R.C.M.; Gomes, R.F.; Chung, C.K.; Jorquera-Cordero, C.; Vepris, O.; et al. Retinoic acid-loaded PLGA nanocarriers targeting cell cholesterol potentialize the antitumour effect of PD-L1 antibody by preventing epithelial-mesenchymal transition mediated by M2-TAM in colorectal cancer. Transl. Oncol. 2023, 31, 101647. [Google Scholar] [CrossRef]
- Junior, R.F.d.A.; Eich, C.; Jorquera, C.; Schomann, T.; Baldazzi, F.; Chan, A.B.; Cruz, L.J. Ceramide and palmitic acid inhibit macrophage-mediated epithelial–mesenchymal transition in colorectal cancer. Mol. Cell. Biochem. 2020, 468, 153–168. [Google Scholar] [CrossRef]
- Cavalcante, R.S.; Ishikawa, U.; Silva, E.S.; Silva-Júnior, A.A.; Araújo, A.A.; Cruz, L.J.; Chan, A.B.; de Araújo Júnior, R.F. STAT3/NF-κB signalling disruption in M2 tumour-associated macrophages is a major target of PLGA nanocarriers/PD-L1 antibody immunomodulatory therapy in breast cancer. Br. J. Pharmacol. 2021, 178, 2284–2304. [Google Scholar] [CrossRef] [PubMed]
- Hideki, T.; Muneaki, S.; Mitsuya, I.; Nobuo, Y. TNM classification of gynaecological malignant tumours, eighth edition: Changes between the seventh and eighth editions. Jpn. J. Clin. Oncol. 2019, 49, 311–320. [Google Scholar]
- Araújo RFJr Lira, G.A.; Vilaça, J.A.; Guedes, H.G.; Leitão, M.C.; Lucena, H.F.; Ramos, C.C. Prognostic and diagnostic implications of MMP-2, MMP-9, and VEGF-α expressions in colorectal cancer. Pathol. Res. Pract. 2015, 211, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Araújo Júnior, R.F.; Garcia, V.B.; Leitão, R.F.; Brito, G.A.; Miguel Ede, C.; Guedes, P.M.; de Araújo, A.A. Carvedilol Improves Inflammatory Response, Oxidative Stress and Fibrosis in the Alcohol-Induced Liver Injury in Rats by Regulating Kuppfer Cells and Hepatic Stellate Cells. PLoS ONE 2016, 11, e0148868. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Dong, X.; Estrella, J.S.; Correa, A.M.; Xu, Y.; Hofstetter, W.L.; Sudo, K.; Onodera, H.; Suzuki, K.; Suzuki, A.; et al. Tumor-associated macrophage infiltration is highly associated with PD-L1 expression in gastric adenocarcinoma. Gastric Cancer Off. J. Int. Gastric Cancer Assoc. Jpn. Gastric Cancer Assoc. 2018, 21, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Staniszewski, W. Virtual microscopy, data management and image analysis in Aperio ScanScope system. Folia Histochem. Cytobiol. 2010, 47, 699–701. [Google Scholar] [CrossRef][Green Version]
- Fasanella, S.; Leonardi, E.; Cantaloni, C.; Eccher, C.; Bazzanella, I.; Aldovini, D.; Bragantini, E.; Morelli, L.; Cuorvo, L.; Ferro, A.; et al. Proliferative activity in human breast cancer: Ki-67 automated evaluation and the influence of different Ki-67 equivalent antibodies. Diagn. Pathol. 2011, 6, S7. [Google Scholar] [CrossRef] [PubMed]
- García-Rojo, M.; Sánchez, J.R.; de la Santa, E.; Durán, E.; Ruiz, J.L.; Silva, A.; Rubio, F.J.; Rodríguez, A.M.; Meléndez, B.; González, L.; et al. Automated image analysis in the study of lymphocyte subpopulation in eosinophilic oesophagitis. Diagn. Pathol. 2014, 9, S7. [Google Scholar] [CrossRef] [PubMed]
- Barricelli, B.R.; Casiraghi, E.; Gliozzo, J.; Huber, V.; Leone, B.E.; Rizzi, A.; Vergani, B. ki67 nuclei detection and ki67-index estimation: A novel automatic approach based on human vision modeling. BMC Bioinform. 2019, 20, 733. [Google Scholar] [CrossRef]
- Lashen, A.; Toss, M.S.; Green, A.R.; Mongan, N.P.; Rakha, E. Ki67 assessment in invasive luminal breast cancer: A comparative study between different scoring methods. Histopathology 2022, 81, 786–798. [Google Scholar] [CrossRef]
- Sakakibara, A.; Matsui, K.; Katayama, T.; Higuchi, T.; Terakawa, K.; Konishi, I. Age-related survival disparity in stage IB and IIB cervical cancer patients: Survival disparity in cervical cancer. J. Obstet. Gynaecol. Res. 2019, 45, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.; Park, S.-Y.; Seo, S.-S.; Lim, M.C.; Kim, J.-Y.; Kang, S. Predicting the risk of the distant recurrence of cervical cancer after concurrent chemoradiation: A validation study of the Korean Gynecologic Oncologic Group (KGOG)-1024 model. Gynecol. Oncol. 2021, 164, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- McAllister, S.S.; Weinberg, R.A. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nature 2014, 16, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Wang, S.; Wang, N.; Zheng, Y.; Zhou, J.; Yang, B.; Wang, X.; Zhang, J.; Guo, L.; Wang, S.; et al. CCL5 derived from tumor-associated macrophages promotes prostate cancer stem cells and metastasis via activating β-catenin/STAT3 signaling. Cell Death Dis. 2020, 11, 234. [Google Scholar] [CrossRef] [PubMed]
- Kusaba, T.; Nakayama, T.; Yamazumi, K.; Yakata, Y.; Yoshizaki, A.; Inoue, K.; Nagayasu, T.; Sekine, I. Activation of STAT3 is a marker of poor prognosis in human colorectal cancer. Oncol. Rep. 2006, 15, 1445–1451. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Lowe, G.; Roberts, C.M.; Finlay, J.; Han, E.S.; Glackin, C.A.; Dellinger, T.H. Pterostilbene Suppresses Ovarian Cancer Growth via Induction of Apoptosis and Blockade of Cell Cycle Progression Involving Inhibition of the STAT3 Pathway. Int. J. Mol. Sci. 2018, 19, 1983. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Ma, H.-Y.; Zhang, Z.-G. Clinicopathological and prognostic value of STAT3/p-STAT3 in cervical cancer: A meta and bioinformatics analysis. Pathol. Res. Pract. 2021, 227, 153624. [Google Scholar] [CrossRef]
- Wu, P.; Wu, D.; Zhao, L.; Huang, L.; Shen, G.; Huang, J.; Chai, Y. Prognostic role of STAT3 in solid tumors: A systematic review and meta-analysis. Oncotarget 2016, 7, 19863–19883. [Google Scholar] [CrossRef]
- Naugler, W.E.; Karin, M. NF-κB and cancer—Identifying targets and mechanisms. Curr. Opin. Genet. Dev. 2008, 18, 19–26. [Google Scholar] [CrossRef]
- El-Arabey, A.A.; Abdalla, M.; Abd-Allah, A.R. Ovarian cancer immunotherapy of NF-κB may have a dark side. Hum. Cell Off. J. Hum. Cell Res. Soc. 2021, 34, 1019–1020. [Google Scholar] [CrossRef] [PubMed]
- Van Antwerp, D.J.; Martin, S.J.; Kafri, T.; Green, D.R.; Verma, I.M. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science 1996, 274, 787–789. [Google Scholar] [CrossRef] [PubMed]
- Valeta-Magara, A.; Gadi, A.; Volta, V.; Walters, B.; Arju, R.; Giashuddin, S.; Zhong, H.; Schneider, R.J. Inflammatory Breast Cancer Promotes Development of M2 Tumor-Associated Macrophages and Cancer Mesenchymal Cells through a Complex Chemokine Network. Cancer Res. 2019, 79, 3360–3371. [Google Scholar] [CrossRef] [PubMed]
- Mancino, A.; Lawrence, T. Nuclear Factor-κB and Tumor-Associated Macrophages. Clin. Cancer Res. 2010, 16, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Debnath, P.; Huirem, R.S.; Dutta, P.; Palchaudhuri, S. Epithelial–mesenchymal transition and its transcription factors. Biosci. Rep. 2021, 42. [Google Scholar] [CrossRef]
- Qureshi, R.; Arora, H.; Rizvi, M. EMT in cervical cancer: Its role in tumour progression and response to therapy. Cancer Lett. 2015, 356, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sastre, M.A.; González-Maya, L.; Delgado, R.; Lizano, M.; Tsubaki, G.; Mohar, A.; García-Carrancá, A. Abnormal distribution of E-cadherin and β-catenin in different histologic types of cancer of the uterine cervix. Gynecol. Oncol. 2005, 97, 330–336. [Google Scholar] [CrossRef] [PubMed]
- E Koay, M.H.; Crook, M.; Stewart, C.J.R. Cyclin D1, E-cadherin and beta-catenin expression in FIGO Stage IA cervical squamous carcinoma: Diagnostic value and evidence for epithelial-mesenchymal transition. Histopathology 2012, 61, 1125–1133. [Google Scholar] [CrossRef] [PubMed]
- Mendonsa, A.M.; Na, T.-Y.; Gumbiner, B.M. E-cadherin in contact inhibition and cancer. Oncogene 2018, 37, 4769–4780. [Google Scholar] [CrossRef]
- Zhao, W.; Zhou, Y.; Xu, H.; Cheng, Y.; Kong, B. Snail family proteins in cervical squamous carcinoma: Expression and significance. Clin. Investig. Med. 2013, 36, E223–E233. [Google Scholar] [CrossRef]
- Kaufhold, S.; Bonavida, B. Central role of Snail1 in the regulation of EMT and resistance in cancer: A target for therapeutic intervention. J. Exp. Clin. Cancer Res. 2014, 33, 62. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Qi, P.; Niu, Q.; Hu, X. Combined Snail and E-cadherin Predicts Overall Survival of Cervical Carcinoma Patients: Comparison Among Various Epithelial-Mesenchymal Transition Proteins. Front. Mol. Biosci. 2020, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, T.; Zhang, B.; Yao, Y.; Yin, G. Matrix metalloproteinase-9 is a prognostic marker for patients with cervical cancer. Med Oncol. 2012, 29, 3394–3399. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.M.A.; Rabelo-Santos, S.H.; Westin, M.C.D.A.; Zeferino, L.C. Tumoral and stromal expression of MMP-2, MMP-9, MMP-14, TIMP-1, TIMP-2, and VEGF-A in cervical cancer patient survival: A competing risk analysis. BMC Cancer 2020, 20, 660. [Google Scholar]
- Ruffell, B.; Affara, N.I.; Coussens, L.M. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012, 33, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Neophytou, C.M.; Trougakos, I.P.; Erin, N.; Papageorgis, P. Apoptosis Deregulation and the Development of Cancer Multi-Drug Resistance. Cancers 2021, 13, 4363. [Google Scholar] [CrossRef] [PubMed]
- Li, L.T.; Jiang, G.; Chen, Q.; Zheng, J.N. Ki67 is a promising molecular target in the diagnosis of cancer (Review). Mol. Med. Rep. 2015, 11, 1566–1572. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Zhang, S.; Gong, Z.; Li, X.; Cao, K.; Deng, H.; He, Y.; et al. The role of microenvironment in tumor angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 204. [Google Scholar] [CrossRef]
- Fathi, N.; Rashidi, G.; Khodadadi, A.; Shahi, S.; Sharifi, S. STAT3 and apoptosis challenges in cancer. Int. J. Biol. Macromol. 2018, 117, 993–1001. [Google Scholar] [CrossRef]
- Wang, Y.; Chang, T.; Wu, T.; Xu, W.; Dou, G.; Wang, Y.; Guo, C. M2 macrophages promote vasculogenesis during retinal neovascularization by regulating bone marrow-derived cells via SDF-1/VEGF. Cell Tissue Res. 2020, 380, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, J.; Ding, M.; Xu, K.; Li, L.; Mao, L.; Zheng, J. Ki67 targeted strategies for cancer therapy. Clin. Transl. Oncol. 2017, 20, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, T.; Farolfi, A.; Scarpi, E.; Mercatali, L.; Medri, L.; Ricci, M.; Nanni, O.; Serra, L.; Amadori, D. Hormonal Receptor, Human Epidermal Growth Factor Receptor-2, and Ki67 Discordance between Primary Breast Cancer and Paired Metastases: Clinical Impact. Oncology 2012, 84, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zheng, M.; Zheng, X.; Fang, X.; Dong, J.; Wang, C.; Wang, T. Predictive Ki-67 Proliferation Index of Cervical Squamous Cell Carcinoma Based on IVIM-DWI Combined with Texture Features. Contrast Media Mol. Imaging 2021, 2021, 8873065. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Xu, L.; Yang, R.; Meng, Y.; Qiu, L. Ki-67 and P16 proteins in cervical cancer and precancerous lesions of young women and the diagnostic value for cervical cancer and precancerous lesions. Oncol. Lett. 2019, 18, 1351–1355. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Jiang, P.; Zhang, J.; Jiang, S.; Yi, Q.; Yuan, R. The positive threshold of the immunohistochemical parameter Ki67 for predicting the recurrence of cervical cancer. Int. J. Gynecol. Obstet. 2021, 158, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Hensler, M.; Kasikova, L.; Fiser, K.; Rakova, J.; Skapa, P.; Laco, J.; Lanickova, T.; Pecen, L.; Truxova, I.; Vosahlikova, S.; et al. M2-like macrophages dictate clinically relevant immunosuppression in metastatic ovarian cancer. J. Immunother. Cancer 2020, 8, e000979. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, Y.; Du, W.; Gong, L.; Chang, H.; Zou, Z. Tumor-associated macrophages: An accomplice in solid tumor progression. J. Biomed. Sci. 2019, 26, 78. [Google Scholar] [CrossRef]
- Zhang, H.; Li, R.; Cao, Y.; Gu, Y.; Lin, C.; Liu, X.; Lv, K.; He, X.; Fang, H.; Jin, K.; et al. Poor Clinical Outcomes and Immunoevasive Contexture in Intratumoral IL-10-Producing Macrophages Enriched Gastric Cancer Patients. Ann. Surg. 2020, 275, e626–e635. [Google Scholar] [CrossRef]
- Fu, X.-L.; Duan, W.; Su, C.-Y.; Mao, F.-Y.; Lv, Y.-P.; Teng, Y.-S.; Yu, P.-W.; Zhuang, Y.; Zhao, Y.-L. Interleukin 6 induces M2 macrophage differentiation by STAT3 activation that correlates with gastric cancer progression. Cancer Immunol. Immunother. 2017, 66, 1597–1608. [Google Scholar] [CrossRef]
- Hagemann, T.; Lawrence, T.; McNeish, I.; Charles, K.A.; Kulbe, H.; Thompson, R.G.; Robinson, S.C.; Balkwill, F.R. “Re-educating” tumor-associated macrophages by targeting NF-kappa B. J. Exp. Med. 2008, 205, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shen, Y.; Wang, S.; Shen, Q.; Zhou, X. The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 2018, 415, 117–128. [Google Scholar] [CrossRef]
- Munir, M.T.; Kay, M.K.; Kang, M.H.; Rahman, M.; Al-Harrasi, A.; Choudhury, M.; Moustaid-Moussa, N.; Hussain, F.; Rahman, S.M. Tumor-Associated Macrophages as Multifaceted Regulators of Breast Tumor Growth. Int. J. Mol. Sci. 2021, 22, 6526. [Google Scholar] [CrossRef] [PubMed]
- Nizar, S.; Meyer, B.; Galustian, C.; Kumar, D.; Dalgleish, A. T regulatory cells, the evolution of targeted immunotherapy. Biochim. Biophys. Acta Rev. Cancer 2010, 1806, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Murai, M.; Turovskaya, O.; Kim, G.; Madan, R.; Karp, C.L.; Cheroutre, H.; Kronenberg, M. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat. Immunol. 2009, 10, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Yamagiwa, S.; Gray, J.D.; Hashimoto, S.; Horwitz, D.A. A role for TGF-beta in the generation and expansion of CD4(+)CD25(+) regulatory T cells from human peripheral blood. J. Immunol. 2001, 166, 7282–7289. [Google Scholar] [CrossRef] [PubMed]
- Que, Y.; Xiao, W.; Guan, Y.-X.; Liang, Y.; Yan, S.-M.; Chen, H.-Y.; Li, Q.-Q.; Xu, B.-S.; Zhou, Z.-W.; Zhang, X. PD-L1 Expression Is Associated with FOXP3+ Regulatory T-Cell Infiltration of Soft Tissue Sarcoma and Poor Patient Prognosis. J. Cancer 2017, 8, 2018–2025. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A. From phagocyte diversity and activation to probiotics: Back to Metchnikoff. Eur. J. Immunol. 2008, 38, 3269–3273. [Google Scholar] [CrossRef]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef]
- Venkatesulu, B.P.; Mallick, S.; Rath, G.K. Patterns of care of cervical cancer in the elderly: A qualitative literature review. J. Geriatr. Oncol. 2017, 8, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.N.; Java, J.J.; Slaughter, K.N.; Rose, P.G.; Lanciano, R.; DiSilvestro, P.A.; Thigpen, J.T.; Lee, Y.-C.; Tewari, K.S.; Chino, J.; et al. Is age a prognostic biomarker for survival among women with locally advanced cervical cancer treated with chemoradiation? An NRG Oncology/Gynecologic Oncology Group ancillary data analysis. Gynecol. Oncol. 2016, 143, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Quinn, B.A.; Deng, X.; Colton, A.; Bandyopadhyay, D.; Carter, J.S.; Fields, E.C. Increasing age predicts poor cervical cancer prognosis with subsequent effect on treatment and overall survival. Brachytherapy 2018, 18, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Miyahara, S.; Tsuji, K.; Shimada, M.; Shibuya, Y.; Shigeta, S.; Nagai, T.; Umezawa, R.; Tokunaga, H.; Jingu, K.; Yaegashi, N. The Impact of Histological Subtype on Survival Outcome of Patients with Stage IIB-IVA Cervical Cancer Who Received Definitive Radiotherapy. Tohoku J. Exp. Med. 2021, 255, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Gien, L.; Covens, A. Lymph node assessment in cervical cancer: Prognostic and therapeutic implications. J. Surg. Oncol. 2008, 99, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Eifel, P.J.; Jhingran, A.; Levenback, C.F.; Tucker, S. Predictive Value of a Proposed Subclassification of Stages I and II Cervical Cancer Based on Clinical Tumor Diameter. Int. J. Gynecol. Cancer 2009, 19, 2–7. [Google Scholar] [CrossRef]
- Liu, Y.-M.; Ni, L.-Q.; Wang, S.-S.; Lv, Q.-L.; Chen, W.-J.; Ying, S.-P. Outcome and prognostic factors in cervical cancer patients treated with surgery and concurrent chemoradiotherapy: A retrospective study. World J. Surg. Oncol. 2018, 16, 18. [Google Scholar] [CrossRef]
- Madeddu, C.; Sanna, E.; Nemolato, S.; Mulas, O.; Oppi, S.; Scartozzi, M.; La Nasa, G.; Maccio, A. Pathogenic and Prognostic Roles of Paraneoplastic Leukocytosis in Cervical Cancer: Can Genomic-Based Targeted Therapies Have a Role? A Literature Review and an Emblematic Case Report. Diagnostics 2022, 12, 1910. [Google Scholar] [CrossRef]


| Clinicopathological Features | Category | Frequency | Percentage |
|---|---|---|---|
| Age (Years) | ≤40 | 184 | 26.6% |
| >40 and ≤60 | 278 | 40.2% | |
| >60 and ≤80 | 201 | 29.1% | |
| >80 | 27 | 3.9% | |
| Not available | 1 | 0.1% | |
| Total Number | 691 | 100.0% | |
| FIGO Staging | I | 393 | 56.9% |
| II | 122 | 17.7% | |
| III | 101 | 14.6% | |
| IV | 21 | 3.0% | |
| Not available | 54 | 7.8% | |
| Total Number | 691 | 100.0% | |
| TNM Staging | I | 370 | 53.5% |
| II | 109 | 15.8% | |
| III | 106 | 15.3% | |
| IV | 37 | 5.4% | |
| Not available | 69 | 10.0% | |
| Total Number | 691 | 100.0% | |
| Recurrence | No | 460 | 66.6% |
| Yes | 194 | 28.1% | |
| Not available | 37 | 5.4% | |
| Total Number | 691 | 100.0% | |
| Death | No | 417 | 60.3% |
| Yes | 257 | 37.2% | |
| Not available | 17 | 2.5% | |
| Total Number | 691 | 100.0% |
| Clinicopathological Features | Category | Freq * | Significance ** | Hazard Ratio of Exp(B) *** |
|---|---|---|---|---|
| Age (Years) | <=40 | 182 | <0.0001 | |
| >40 and <=60 | 277 | <0.05 | 1.50 (1.06–2.12) | |
| >60 and <=80 | 200 | <0.0001 | 2.33 (1.63–3.31) | |
| >80 | 27 | <0.0001 | 3.67 (1.98–6.80) | |
| FIGO Staging | I | 391 | <0.0001 | |
| System | II | 122 | <0.0001 | 2.44 (1.74–3.41) |
| III | 100 | <0.0001 | 5.68 (4.15–7.79) | |
| IV | 20 | <0.0001 | 8.89 (5.31–14.88) | |
| TNM Staging System | I | 369 | <0.0001 | |
| II | 109 | <0.0001 | 2.29 (1.60–3.28) | |
| III | 105 | <0.0001 | 4.47 (3.23–6.17) | |
| IV | 36 | <0.0001 | 10.95 (7.22–16.62) |
| Marker | STAT3 | NF-κB | ||||||
|---|---|---|---|---|---|---|---|---|
| Immunoexpression | 1 | 2 | 1 | 2 | ||||
| Weak n [%] = 101 [14.6] | Strong n [%] = 590 [85.4] | p * | Weak n [%] = 236 [34.2] | Strong n [%] = 455 [65.8] | p * | |||
| NF-κB | 1 | Weak n [%] = 236 [34.2] | 62 [9.0] | 174 [25.2] | <0.0001 | - | - | |
| 2 | Strong n [%] = 455 [65.8] | 39 [5.6] | 416 [60.2] | |||||
| CD163 | 1 | Weak n [%] = 238 [34.4] | 68 [9.8] | 170 [24.6] | <0.0001 | 118 [17.1] | 120 [17.4] | <0.0001 |
| 2 | Strong n [%] = 453 [65.6] | 33 [4.8] | 420 [60.8] | 118 [17.1] | 335 [48.5] | |||
| CD204 | 1 | Weak n [%] = 93 [13.5] | 51 [7.4] | 42 [6.1] | <0.0001 | 48 [6.9] | 45 [6.5] | <0.0001 |
| 2 | Strong n [%] = 598 [86.5] | 50 [7.2] | 548 [79.3] | 188 [27.2] | 410 [59.3] | |||
| Marker | STAT3 | NF-κB | ||||||
|---|---|---|---|---|---|---|---|---|
| Immunoexpression | 1 | 2 | 1 | 2 | ||||
| Weak n [%] = 101 [14.6] | Strong n [%] = 590 [85.4] | p * | Weak n [%] = 236 [34.2] | Strong n [%] = 455 [65.8] | p * | |||
| VIM | 1 | Weak n [%] = 110 [15.9] | 58 [8.4] | 52 [7.5] | <0.0001 | 68 [9.8] | 42 [6.1] | <0.0001 |
| 2 | Strong n [%] = 581 [84.1] | 43 [6.2] | 538 [77.9] | 168 [24.3] | 413 [59.8] | |||
| E-cad | 1 | Weak n [%] = 311 [45.0] | 79 [11.4] | 232 [33.6] | <0.0001 | 122 [17.7] | 189 [27.4] | <0.05 |
| 2 | Strong n [%] = 380 [55.0] | 22 [3.2] | 358 [51.8] | 114 [16.5] | 266 [38.5] | |||
| MMP9 | 1 | Weak n [%] = 73 [10.6] | 47 [6.8] | 26 [3.8] | <0.0001 | 44 [6.4] | 29 [4.2] | <0.0001 |
| 2 | Strong n [%] = 618 [89.4] | 54 [7.8] | 564 [81.6] | 192 [27.8] | 426 [61.6] | |||
| SNAIL | 1 | Weak n [%] = 238 [34.4] | 47 [6.8] | 191 [27.6] | <0.01 | 78 [11.3] | 160 [23.2] | 0.61 |
| 2 | Strong n [%] = 453 [65.6] | 54 [7.8] | 399 [57.7] | 158 [22.9] | 295 [42.7] | |||
| Marker | CD163 | CD204 | ||||||
|---|---|---|---|---|---|---|---|---|
| Immunoexpression | 1 | 2 | 1 | 2 | ||||
| Weak n [%] = 238 [34.4] | Strong n [%] = 453 [65.6] | p * | Weak n [%] = 93 [13.5] | Strong n [%] = 598 [86.5] | p * | |||
| VIM | 1 | Weak n [%] = 110 [15.9] | 77 [11.1] | 33 [4.8] | <0.0001 | 56 [8.1] | 54 [7.8] | <0.0001 |
| 2 | Strong n [%] = 581 [84.1] | 161 [23.3] | 420 [60.8] | 37 [5.4] | 544 [78.7] | |||
| E-cad | 1 | Weak n [%] = 311 [45.0] | 131 [19.0] | 180 [26.0] | <0.0001 | 68 [9.8] | 243 [35.2] | <0.0001 |
| 2 | Strong n [%] = 380 [55.0] | 107 [15.5] | 273 [39.5] | 25 [3.6] | 355 [51.4] | |||
| MMP9 | 1 | Weak n [%] = 73 [10.6] | 53 [7.7] | 20 [2.9] | <0.0001 | 44 [6.4] | 29 [4.2] | <0.0001 |
| 2 | Strong n [%] = 618 [89.4] | 185 [26.8] | 433 [62.7] | 49 [7.1] | 569 [82.3] | |||
| SNAIL | 1 | Weak n [%] = 238 [34.4] | 92 [13.3] | 146 [21.1] | 0.09 | 41 [5.9] | 197 [28.5] | <0.05 |
| 2 | Strong n [%] = 453 [65.6] | 146 [21.1] | 307 [44.4] | 52 [7.5] | 401 [8.0] | |||
| Marker | STAT3 | NF-κB | ||||||
|---|---|---|---|---|---|---|---|---|
| Immunoexpression | 1 | 2 | 1 | 2 | ||||
| Weak n [%] = 101 [14.6] | Strong n [%] = 590 [85.4] | p * | Weak n [%] = 236 [34.2] | Strong n [%] = 455 [65.8] | p * | |||
| TGFβ | 1 | Weak n [%] = 518 [75.0] | 80 [11.6] | 438 [63.4] | 0.25 | 180 [26.0] | 338 [48.9] | 0.58 |
| 2 | Strong n [%] = 173 [25.0] | 21 [3.0] | 152 [22.0] | 56 [8.1] | 117 [16.9] | |||
| CD25 | 1 | Weak n [%] = 303 [43.8] | 74 [10.7] | 229 [33.1] | <0.0001 | 137 [19.8] | 166 [24.0] | <0.0001 |
| 2 | Strong n [%] = 388 [56.2] | 27 [3.9] | 361 [52.2] | 99 [14.3] | 289 [41.8] | |||
| FOXP3 | 1 | Weak n [%] = 164 [23.7] | 66 [9.6] | 98 [14.2] | <0.0001 | 72 [10.4] | 92 [13.3] | <0.0001 |
| 2 | Strong n [%] = 527 [76.3] | 35 [5.1] | 492 [71.2] | 164 [23.7] | 363 [52.5] | |||
| MIF | 1 | Weak n [%] = 102 [14.8] | 60 [8.7] | 42 [6.1] | <0.0001 | 51 [7.4] | 51 [7.4] | <0.0001 |
| 2 | Strong n [%] = 589 [85.2] | 41 [5.9] | 548 [79.3] | 185 [26.8] | 404 [58.5] | |||
| IL-17 | 1 | Weak n [%] = 96 [13.9] | 56 [8.1] | 40 [5.8] | <0.0001 | 57 [8.2] | 39 [5.6] | <0.0001 |
| 2 | Strong n [%] = 595 [86.1] | 45 [6.5] | 550 [79.6] | 179 [25.9] | 416 [60.2] | |||
| IL-10 | 1 | Weak n [%] = 641 [92.8] | 101 [14.6] | 540 [78.1] | <0.001 | 220 [31.8] | 421 [60.9] | 0.87 |
| 2 | Strong n [%] = 50 [7.2] | 0 [0.0] | 50 [7.2] | 16 [2.3] | 34 [4.9] | |||
| PD-L1 | 1 | Weak n [%] = 626 [90.6] | 100 [14.5] | 526 [76.1] | <0.001 | 210 [30.4] | 416 [60.2] | 0.33 |
| 2 | Strong n [%] = 65 [9.4] | 1 [0.1] | 64 [9.3] | 26 [3.8] | 39 [5.6] | |||
| Marker | CD163 | CD204 | ||||||
|---|---|---|---|---|---|---|---|---|
| Immunoexpression | 1 | 2 | 1 | 2 | ||||
| Weak n [%] = 238 [34.4] | Strong n [%] = 453 [65.6] | p * | Weak n [%] = 93 [13.5] | Strong n [%] = 598 [86.5] | p * | |||
| TGFβ | 1 | Weak n [%] = 518 [75.0] | 191 [27.6] | 327 [47.3] | <0.05 | 80 [11.6] | 438 [63.4] | <0.01 |
| 2 | Strong n [%] = 173 [25.0] | 47 [6.8] | 126 [18.2] | 13 [1.9] | 160 [23.2] | |||
| CD25 | 1 | Weak n [%] = 303 [43.8] | 137 [19.8] | 166 [24.0] | <0.0001 | 69 [10.0] | 234 [33.9] | <0.0001 |
| 2 | Strong n [%] = 388 [56.2] | 101 [14.6] | 287 [41.5] | 24 [3.5] | 364 [52.7] | |||
| FOXP3 | 1 | Weak n [%] = 164 [23.7] | 85 [12.3] | 79 [11.4] | <0.0001 | 64 [9.3] | 100 [14.5] | <0.0001 |
| 2 | Strong n [%] = 527 [76.3] | 153 [22.1] | 374 [54.1] | 29 [4.2] | 498 [72.1] | |||
| MIF | 1 | Weak n [%] = 102 [14.8] | 59 [8.5] | 43 [6.2] | <0.0001 | 44 [6.4] | 58 [8.4] | <0.0001 |
| 2 | Strong n [%] = 589 [85.2] | 179 [25.9] | 410 [59.3] | 49 [7.1] | 540 [78.1] | |||
| IL-17 | 1 | Weak n [%] = 96 [13.9] | 69 [10.0] | 27 [3.9] | <0.0001 | 55 [8.0] | 41 [5.9] | <0.0001 |
| 2 | Strong n [%] = 595 [86.1] | 169 [24.5] | 426 [61.6] | 38 [5.5] | 557 [80.6] | |||
| IL-10 | 1 | Weak n [%] = 641 [92.8] | 228 [33.0] | 413 [59.8] | <0.05 | 91 [13.2] | 550 [79.6] | <0.05 |
| 2 | Strong n [%] = 50 [7.2] | 10 [1.4] | 40 [5.8] | 2 [0.3] | 48 [6.9] | |||
| PD-L1 | 1 | Weak n [%] = 626 [90.6] | 223 [32.3] | 403 [58.3] | 0.054 | 91 [13.2] | 535 [77.4] | <0.01 |
| 2 | Strong n [%] = 65 [9.4] | 15 [2.2] | 50 [7.2] | 2 [0.3] | 63 [9.1] | |||
| Marker | STAT3 | NF-κB | ||||||
|---|---|---|---|---|---|---|---|---|
| Immunoexpression | 1 | 2 | 1 | 2 | ||||
| Weak n [%] = 101 [14.6] | Strong n [%] = 590 [85.4] | p * | Weak n [%] = 236 [34.2] | Strong n [%] = 455 [65.8] | p * | |||
| Bcl-2 | 1 | Weak n [%] = 156 [22.6] | 51 [7.4] | 105 [15.2] | <0.0001 | 67 [9.7] | 89 [12.9] | <0.05 |
| 2 | Strong n [%] = 535 [77.4] | 50 [7.2] | 485 [70.2] | 169 [24.7] | 366 [53.0] | |||
| VEGFα | 1 | Weak n [%] = 149 [21.6] | 71 [10.3] | 78 [11.3] | <0.0001 | 80 [11.6] | 69 [10.0] | <0.0001 |
| 2 | Strong n [%] = 542 [78.4] | 30 [4.3] | 512 [74.1] | 156 [22.6] | 386 [55.9] | |||
| Ki-67 | 1 | Low n [%] = 223 [33.7] | 39 [5.6] | 194 [28.1] | 0.25 | 60 [8.7] | 173 [25.0] | <0.01 |
| 2 | High n [%] = 458 [66.3] | 62 [9.0] | 396 [57.3] | 176 [25.5] | 282 [40.8] | |||
| Marker | Immunoexpression | CD163 | CD204 | |||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 1 | 2 | |||||
| Weak n [%] = 238 [34.4] | Strong n [%] = 453 [65.6] | p * | Weak n [%] = 93 [13.5] | Strong n [%] = 598 [86.5] | p * | |||
| Bcl-2 | 1 | Weak n [%] = 156 [22.6] | 77 [11.1] | 79 [11.4] | <0.0001 | 46 [6.7] | 110 [15.9] | <0.0001 |
| 2 | Strong n [%] = 535 [77.4] | 161 [23.3] | 374 [54.1] | 47 [6.8] | 488 [70.6] | |||
| VEGFα | 1 | Weak n [%] = 149 [21.6] | 88 [12.7] | 61 [8.8] | <0.0001 | 62 [9.0] | 87 [12.6] | <0.0001 |
| 2 | Strong n [%] = 542 [78.4] | 150 [21.7] | 392 [56.7] | 31 [4.5] | 511 [74.0] | |||
| Ki-67 | 1 | Low n [%] = 223 [33.7] | 85 [12.3] | 148 [21.4] | 0.44 | 35 [5.1] | 198 [28.4] | 0.41 |
| 2 | High n [%] = 458 [66.3] | 62 [9.0] | 396 [57.3] | 176 [25.5] | 282 [40.8] | |||
| Marker | Score | f * | Overall Survival | Recurrence-Free Survival | ||
|---|---|---|---|---|---|---|
| p ** | HR (CI) *** | p ** | HR (CI) *** | |||
| VIM | 1 | 109 | ||||
| 2 | 578 | 0.28 | 0.83 (0.60–1.16) | 0.14 | 0.75 (0.52–1.09) | |
| E-cad | 1 | 309 | ||||
| 2 | 378 | <0.05 | 0.78 (0.61–1.00) | 0.72 | 0.95 (0.71–1.26) | |
| MMP9 | 1 | 72 | ||||
| 2 | 615 | 0.45 | 1.19 (0.75–1.88) | 0.35 | 1.29 (0.74–2.22) | |
| SNAIL | 1 | 237 | ||||
| 2 | 450 | <0.01 | 1.52 (1.16–2.00) | 0.52 | 1.10 (1.81–1.48) | |
| TGFß | 1 | 515 | ||||
| 2 | 172 | 0.07 | 1.27 (0.97–1.67) | 0.41 | 1.14 (0.82–1.56) | |
| CD25 | 1 | 302 | ||||
| 2 | 385 | 0.40 | 0.90 (0.70–1.15) | 0.43 | 0.89 (0.66–1.19) | |
| FOXP3 | 1 | 163 | ||||
| 2 | 524 | 0.82 | 0.96 (0.72–1.29) | 0.96 | 0.99 (0.70–1.39) | |
| MIF | 1 | 101 | ||||
| 2 | 586 | <0.05 | 0.70 (0.50–0.98) | 0.25 | 0.79 (0.53–1.18) | |
| IL-17 | 1 | 95 | ||||
| 2 | 592 | 0.69 | 0.92 (0.64–1.34) | 0.46 | 0.85 (0.56–1.30) | |
| IL-10 | 1 | 637 | ||||
| 2 | 50 | 0.39 | 1.21 (0.77–1.89) | 0.21 | 0.65 (0.33–1.28) | |
| PD-L1 | 1 | 623 | ||||
| 2 | 64 | 0.001 | 0.39 (0.22–0.70) | <0.05 | 0.53 (0.29–0.95) | |
| Bcl-2 | 1 | 154 | ||||
| 2 | 533 | 0.06 | 0.76 (0.58–1.01) | 0.36 | 0.85 (0.56–1.30) | |
| VEGFα | 1 | 148 | ||||
| 2 | 539 | 0.07 | 0.76 (0.57–1.02) | 0.06 | 0.72 (0.51–1.10) | |
| Ki-67 | 1 | 232 | ||||
| 2 | 455 | <0.001 | 1.58 (1.20–2.09) | <0.05 | 1.44 (1.05–1.97) | |
| Marker | Score | TNM | |||||
|---|---|---|---|---|---|---|---|
| I | II | III | IV | Total | p * | ||
| STAT3 | 1 | 42 (6.8%) | 14 (2.3%) | 27 (4.3%) | 6 (1.0%) | 89 (14.3%) | |
| 2 | 328 (52.7%) | 95 (15.3%) | 79 (12.7%) | 31 (5.0%) | 533 (85.7%) | <0.01 | |
| NF-κB | 1 | 109 (17.5%) | 44 (7.1%) | 48 (7.7%) | 13 (2.1%) | 214 (34.4%) | |
| 2 | 261 (42.0%) | 65 (10.5%) | 58 (9.3%) | 24 (3.9%) | 408 (65.6%) | <0.01 | |
| CD204 | 1 | 49 (7.9%) | 10 (1.6%) | 21 (3.4%) | 5 (0.8%) | 85 (13.7%) | |
| 2 | 321 (51.6%) | 99 (15.9%) | 85 (13.7%) | 32 (5.1%) | 537 (86.3%) | 0.15 | |
| CD163 | 1 | 128 (20.6%) | 33 (5.3%) | 39 (6.3%) | 12 (1.9%) | 212 (34.1%) | |
| 2 | 242 (38.9%) | 76 (12.2%) | 67 (10.8%) | 25 (4.0%) | 410 (65.9%) | 0.77 | |
| Overall Survival | Recurrence-Free Survival | |||||
|---|---|---|---|---|---|---|
| Stage | f * | p *** | HR **** | f ** | p *** | HR **** |
| I | 369 | 368 | ||||
| II | 109 | <0.0001 | 2.27 (1.59–3.26) | 109 | <0.0001 | 2.14 (1.44–3.17) |
| III | 105 | <0.0001 | 4.43 (3.20–6.14) | 104 | <0.0001 | 3.48 (2.39–5.06) |
| IV | 36 | <0.0001 | 10.90 (7.17–16.57) | 36 | <0.0001 | 10.82 (6.36–18.41) |
| Variables | Characteristics | Freq * | p ** | Hazard Ratio of Exp(B) *** |
|---|---|---|---|---|
| Hemoglobin Count (g/dL) | until 8 | 29 | <0.0001 | 1.0 |
| Leukocytes Count (leukocytes/mm3) | Leukocytosis | 101 | <0.0001 | 2.55 (1.89–3.44) |
| Platelets Count (platelets/mm3) | Thrombocytosis | 50 | <0.0001 | 2.46 (1.69–3.58) |
| Tumor Size **** | >4 | 192 | <0.0001 | 4.32 (1.98–9.41) |
| Invasion **** | Yes | 201 | 0.05 | 1.71 (1.12–2.62) |
| Neural Invasion **** | Yes | 48 | 0.05 | 2.06 (1.14–3.71) |
| Depth of stromal invasion **** | Total | 243 | <0.0001 | 3.43 (1.94–6.06) |
| Compromised margin **** | Yes | 45 | <0.0001 | 2.98 (1.88–4.74) |
| Lymph node metastasis **** | Positive | 125 | <0.0001 | 2.81 (1.92–4.11) |
| TNM Pathological Stage **** | IV | 34 | <0.0001 | 7.58 (4.50–12.75) |
| External Radiation Therapy | Yes | 393 | <0.0001 | 1.68 (1.28–2.21) |
| Chemotherapy | Yes | 294 | <0.01 | 1.362 (1.06–1.74) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lira, G.A.; de Azevedo, F.M.; Lins, I.G.d.S.; Marques, I.d.L.; Lira, G.A.; Eich, C.; de Araujo Junior, R.F. High M2-TAM Infiltration and STAT3/NF-κB Signaling Pathway as a Predictive Factor for Tumor Progression and Death in Cervical Cancer. Cancers 2024, 16, 2496. https://doi.org/10.3390/cancers16142496
Lira GA, de Azevedo FM, Lins IGdS, Marques IdL, Lira GA, Eich C, de Araujo Junior RF. High M2-TAM Infiltration and STAT3/NF-κB Signaling Pathway as a Predictive Factor for Tumor Progression and Death in Cervical Cancer. Cancers. 2024; 16(14):2496. https://doi.org/10.3390/cancers16142496
Chicago/Turabian StyleLira, George Alexandre, Fábio Medeiros de Azevedo, Ingrid Gabrielle dos Santos Lins, Isabelle de Lima Marques, Giovanna Afonso Lira, Christina Eich, and Raimundo Fernandes de Araujo Junior. 2024. "High M2-TAM Infiltration and STAT3/NF-κB Signaling Pathway as a Predictive Factor for Tumor Progression and Death in Cervical Cancer" Cancers 16, no. 14: 2496. https://doi.org/10.3390/cancers16142496
APA StyleLira, G. A., de Azevedo, F. M., Lins, I. G. d. S., Marques, I. d. L., Lira, G. A., Eich, C., & de Araujo Junior, R. F. (2024). High M2-TAM Infiltration and STAT3/NF-κB Signaling Pathway as a Predictive Factor for Tumor Progression and Death in Cervical Cancer. Cancers, 16(14), 2496. https://doi.org/10.3390/cancers16142496






