The Roles of RAC1 and RAC1B in Colorectal Cancer and Their Potential Contribution to Cetuximab Resistance
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Colorectal Cancer Treatment—Surgery, Chemotherapy, and Targeted Therapies
1.2. Cancer Treatment Resistance
1.3. Epidermal Growth Factor Receptor and Cetuximab
1.4. CRC Resistance to Cetuximab
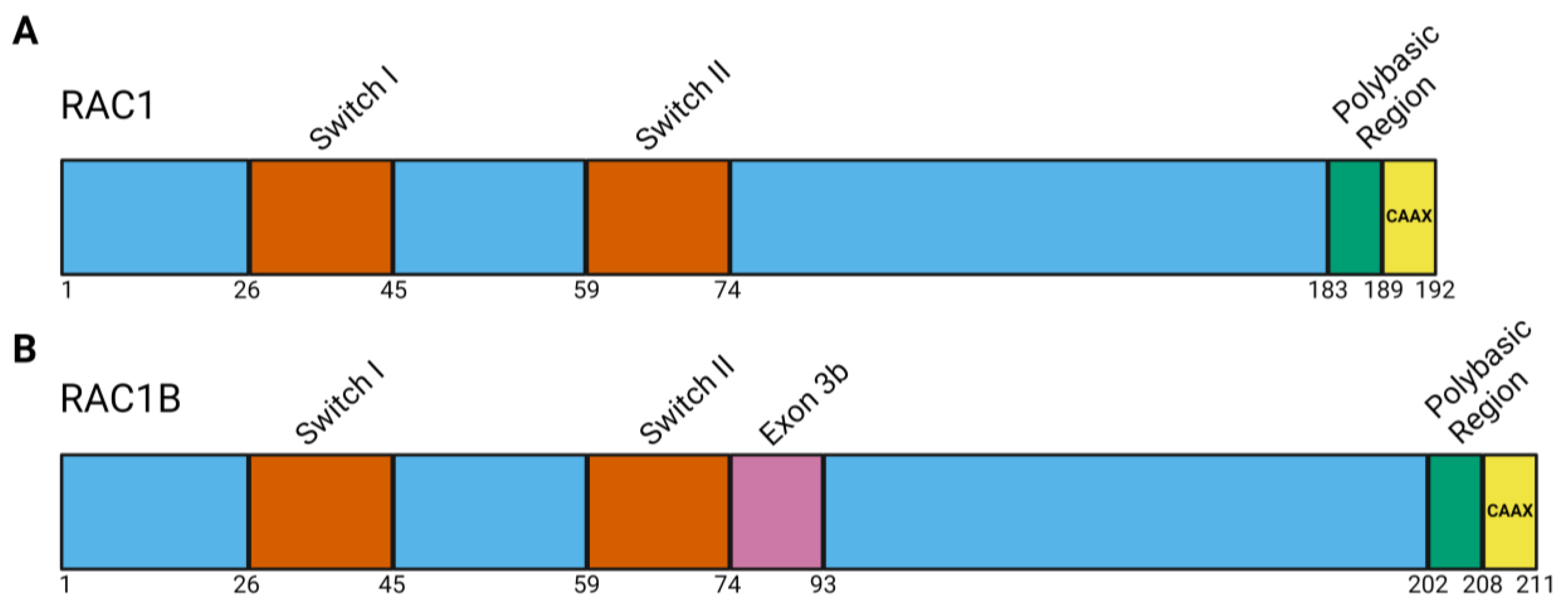
2. RAC1 and RAC1B Structure and Expression
2.1. RAC1 Structure and Function
2.2. RAC1B Protein Structure
2.3. Regulation of RAC1 and RAC1B Isoform Expression
3. RAC1 and RAC1B in Colorectal Cancer and Drug Resistance
3.1. RAC1 in Colorectal Cacner
3.2. RAC1B in Colorectal Cancer
3.3. Comparison of RAC1 and RAC1B in CRC
| Feature | RAC1 | RAC1B | Ref. |
|---|---|---|---|
| Roles in Normal Intestinal Epithelium | Maintain epithelial cell polarity, tight junction integrity, and stem cell adhesion, regulate migration and wound healing, regulate cell extrusion, | N/A RAC1B is predominantly expressed in tumor tissue, thus a role in normal tissue is unknown | [46,71] |
| Roles in CRC | Overexpressed in CRC LGR5+ stem cell expansion following APC loss; ROS production; NFκB signaling; accelerate tumorigenesis and tumor progression; promote cell proliferation, migration, and invasion; alter cellular metabolism; promote EMT | Promote cell survival and decrease apoptosis, increase cell cycle progression and cell proliferation, NFκB signaling, ROS production, mediate BRAF(V600E)-oncogene-induced senescence, promote EMT | [46,71] |
| Correlation with Patient Outcomes | Increased expression correlated to worse overall survival, advanced disease stage, metastasis Expression in liver metastases higher than primary tumor | Expression related to worse overall survival, advanced disease stage, and worse disease-free survival | [77,78,79,93,94] |
| Overexpression sufficient to initiate tumorigenesis? | Yes | No | [76,91] |
| Roles in CRC Therapy Resistance | GAP ARHGAP17 expression increases CRC sensitivity to 5-FU | Expression promotes 5-FU and oxaliplatin resistance through NFκB signaling | [90,105] |
| Mechanisms for RAC1/RAC1B targeting for combination therapies | GEF-interaction inhibition, GTP-binding inhibition, downstream protein interaction inhibition, post-translational modification inhibition | GEF-interaction inhibition, GTP-binding inhibition, downstream protein interaction inhibition, post-translational modification inhibition | [51] |
3.4. RAC1 and RAC1B Contribution to CRC Chemotherapy Resistance
4. Cetuximab Resistance Mechanisms and Potential for RAC1/RAC1B to Contribute to Cetuximab Resistance
4.1. EGFR Signaling
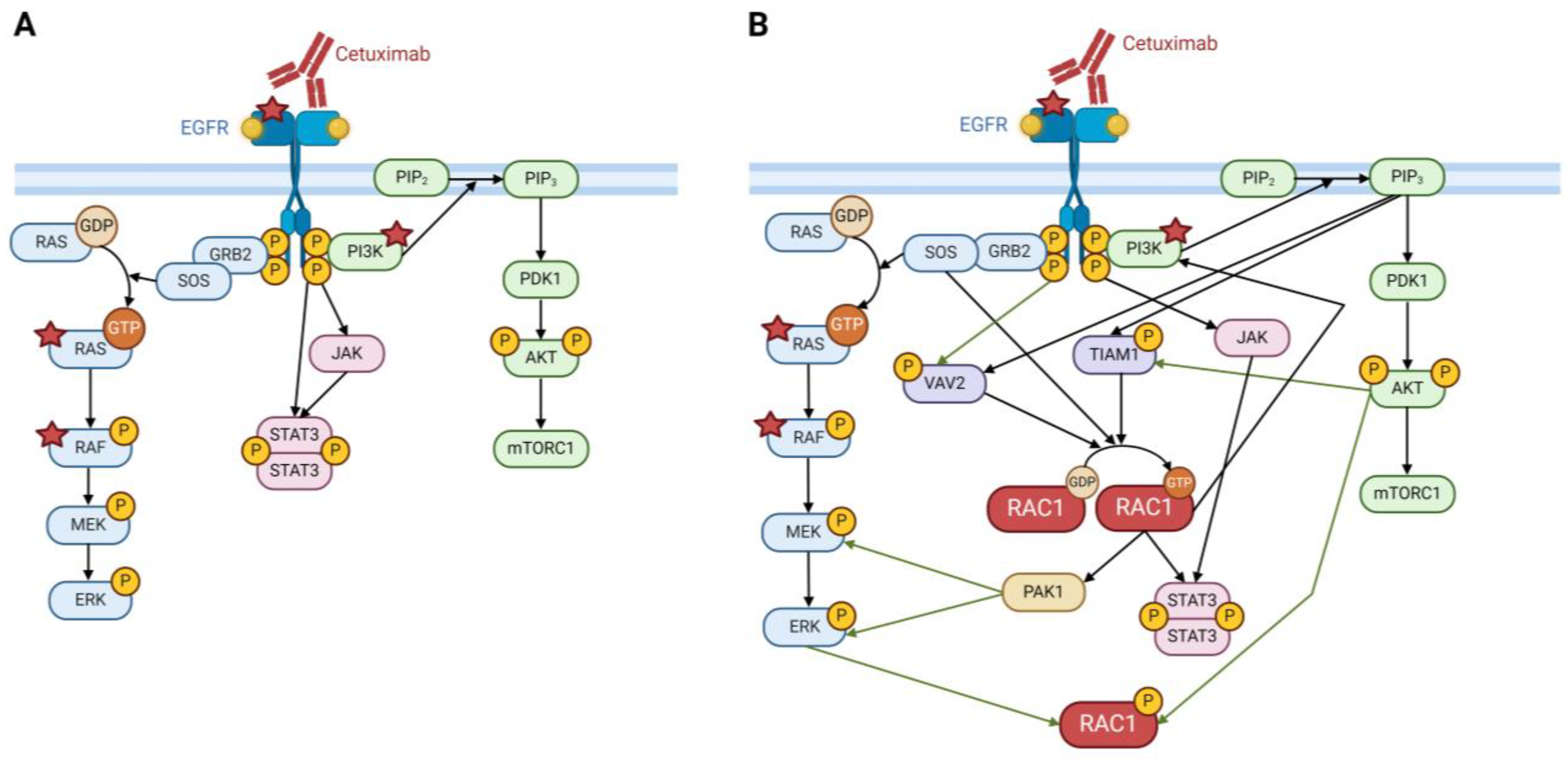
4.2. Activation by Other Receptor Tyrosine Kinases
4.3. EGFR Expression and Activation
4.4. Epithelial-to-Mesenchymal Transition
4.5. CRC Clonal Molecular Subtype and CMS Switching
5. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| GTPase | guanosine triphosphatase |
| 5-FU | 5-fluorouracil |
| EGFR | epidermal growth factor receptor |
| VEGF | vascular endothelial growth factor |
| RTK | receptor tyrosine kinase |
| FDA | Food and Drug Administration |
| KRAS | Kirsten rat sarcoma viral oncogene homolog |
| BRAF | v-raf murine sarcoma viral oncogene homolog B1 |
| PI3K | phosphatidylinositol-4,5-bisphosphate 3-kinase |
| PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| EMT | epithelial-to-mesenchymal transition |
| RAC1 | Rac family small GTPase 1, Ras-related C3 botulinum toxin substrate 1 |
| GDP | guanosine diphosphate |
| GTP | guanosine triphosphate |
| GEF | guanine nucleotide exchange factor |
| GAP | GTPase activating proteins |
| GDI | guanine nucleotide-dissociation inhibitors |
| SRSF1 | serine/arginine rich splicing factor 1, SF/AF2 |
| SRp20 | serine/arginine rich protein 20, SRSF3 |
| EGF | epidermal growth factor |
| hnRNP A1 | heterogeneous nuclear ribonucleoprotein A1 |
| ESRP1 | epithelial splicing regulatory protein 1 |
| LGR5 | leucine-rich repeat-containing G protein-coupled receptor 5 |
| APC | adenomatous polyposis coli |
| ROS | reactive oxygen species |
| SOX9 | SRY-box transcription factor 9 |
| IQGAP1 | IQ motif-containing GTPase-activating protein 1 |
| NOX1 | NADPH oxidase 1 |
| FOLFOX | leucovorin calcium (folinic acid), fluorouracil, and oxaliplatin |
| XELOX | capecitabine and oxaliplatin |
| TCGA | The Cancer Genome Atlas |
| PAK1 | p21 activated kinase |
| RACK1 | receptor for activated kinase 1 |
| STAT | signal transducers and activators of transcription |
| SOS | son of sevenless |
| PIP3 | phosphatidylinositol (3,4,5)-triphosphate |
| MET | Mesenchymal–epithelial transition factor receptor |
| IGF-1R | insulin-like growth factor receptor |
| HGF | hepatocyte growth factor |
| 3D | three dimensional |
| LIMK | LIM kinase 1 |
| MMP-3 | matrix metalloproteinase 3 |
| CMS | consensus molecular subtypes |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer Statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Colorectal Cancer Facts & Figures 2023–2025; American Cancer Society, Inc.: Atlanta, GA, USA, 2023. [Google Scholar]
- National Cancer Institute SEER Cancer Stat Facts: Colorectal Cancer. Available online: https://seer.cancer.gov/statfacts/html/colorect.html (accessed on 2 March 2024).
- National Cancer Institute, Surveillance Research Program. SEER*Explorer: An Interactive Website for SEER Cancer Statistics. Available online: https://seer.cancer.gov/statistics-network/explorer/application.html?site=20&data_type=1&graph_type=1&compareBy=sex&chk_sex_1=1&chk_sex_3=3&chk_sex_2=2&rate_type=2&race=1&age_range=1&advopt_precision=1&advopt_show_ci=on&hdn_view=0&advopt_show_apc=on&advopt_display=1#resultsRegion0 (accessed on 2 March 2024).
- National Cancer Institute, Surveillance Research Program. SEER*Explorer: An Interactive Website for SEER Cancer Statistics. Available online: https://seer.cancer.gov/statistics-network/explorer/application.html?site=20&data_type=4&graph_type=2&compareBy=sex&chk_sex_1=1&chk_sex_3=3&chk_sex_2=2&relative_survival_interval=5&race=1&age_range=1&stage=101&advopt_precision=1&advopt_show_ci=on&hdn_view=0&advopt_show_apc=on&advopt_display=1#resultsRegion0 (accessed on 2 March 2024).
- Kumar, A.; Gautam, V.; Sandhu, A.; Rawat, K.; Sharma, A.; Saha, L. Current and Emerging Therapeutic Approaches for Colorectal Cancer: A Comprehensive Review. World J. Gastrointest. Surg. 2023, 15, 495–519. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Baselga, J.; Hyman, D.M. A View on Drug Resistance in Cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Cabanos, H.F.; Hata, A.N. Emerging Insights into Targeted Therapy-Tolerant Persister Cells in Cancer. Cancers 2021, 13, 2666. [Google Scholar] [CrossRef] [PubMed]
- Martincorena, I.; Campbell, P.J. Somatic Mutation in Cancer and Normal Cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Holohan, C.; Van Schaeybroeck, S.; Longley, D.B.; Johnston, P.G. Cancer Drug Resistance: An Evolving Paradigm. Nat. Rev. Cancer 2013, 13, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Johnston, P.G. Molecular Mechanisms of Drug Resistance. J. Pathol. 2005, 205, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Pisco, A.O.; Huang, S. Non-Genetic Cancer Cell Plasticity and Therapy-Induced Stemness in Tumour Relapse: ‘What Does Not Kill Me Strengthens Me’. Br. J. Cancer 2015, 112, 1725–1732. [Google Scholar] [CrossRef] [PubMed]
- Pogrebniak, K.L.; Curtis, C. Harnessing Tumor Evolution to Circumvent Resistance. Trends Genet. 2018, 34, 639–651. [Google Scholar] [CrossRef]
- Spano, J.-P.; Lagorce, C.; Atlan, D.; Milano, G.; Domont, J.; Benamouzig, R.; Attar, A.; Benichou, J.; Martin, A.; Morere, J.-F.; et al. Impact of EGFR Expression on Colorectal Cancer Patient Prognosis and Survival. Ann. Oncol. 2005, 16, 102–108. [Google Scholar] [CrossRef]
- Citri, A.; Yarden, Y. EGF–ERBB Signalling: Towards the Systems Level. Nat. Rev. Mol. Cell Biol. 2006, 7, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Seshacharyulu, P.; Ponnusamy, M.P.; Haridas, D.; Jain, M.; Ganti, A.K.; Batra, S.K. Targeting the EGFR Signaling Pathway in Cancer Therapy. Expert. Opin. Ther. Targets 2012, 16, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Sato, J.D.; Kawamoto, T.; Le, A.D.; Mendelsohn, J.; Polikoff, J.; Sato, G.H. Biological Effects in Vitro of Monoclonal Antibodies to Human Epidermal Growth Factor Receptors. Mol. Biol. Med. 1983, 1, 511–529. [Google Scholar] [PubMed]
- Masui, H.; Kawamoto, T.; Sato, J.D.; Wolf, B.; Sato, G.; Mendelsohn, J. Growth Inhibition of Human Tumor Cells in Athymic Mice by Anti-Epidermal Growth Factor Receptor Monoclonal Antibodies. Cancer Res. 1984, 44, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Gill, G.N.; Kawamoto, T.; Cochet, C.; Le, A.; Sato, J.D.; Masui, H.; McLeod, C.; Mendelsohn, J. Monoclonal Anti-Epidermal Growth Factor Receptor Antibodies Which Are Inhibitors of Epidermal Growth Factor Binding and Antagonists of Epidermal Growth Factor-Stimulated Tyrosine Protein Kinase Activity. J. Biol. Chem. 1984, 259, 7755–7760. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, N.I.; Prewett, M.; Zuklys, K.; Rockwell, P.; Mendelsohn, J. Biological Efficacy of a Chimeric Antibody to the Epidermal Growth Factor Receptor in a Human Tumor Xenograft Model. Clin. Cancer Res. 1995, 1, 1311–1318. [Google Scholar] [PubMed]
- Mendelsohn, J.; Prewett, M.; Rockwell, P.; Goldstein, N.I. CCR 20th Anniversary Commentary: A Chimeric Antibody, C225, Inhibits EGFR Activation and Tumor Growth. Clin. Cancer Res. 2015, 21, 227–229. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mendelsohn, J.; Baselga, J. The EGF Receptor Family as Targets for Cancer Therapy. Oncogene 2000, 19, 6550–6565. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, D.; Humblet, Y.; Siena, S.; Khayat, D.; Bleiberg, H.; Santoro, A.; Bets, D.; Mueser, M.; Harstrick, A.; Verslype, C.; et al. Cetuximab Monotherapy and Cetuximab plus Irinotecan in Irinotecan-Refractory Metastatic Colorectal Cancer. N. Engl. J. Med. 2004, 351, 337–345. [Google Scholar] [CrossRef]
- Lièvre, A.; Bachet, J.B.; Le Corre, D.; Boige, V.; Landi, B.; Emile, J.-F.; Côté, J.-F.; Tomasic, G.; Penna, C.; Ducreux, M.; et al. KRAS Mutation Status Is Predictive of Response to Cetuximab Therapy in Colorectal Cancer. Cancer Res. 2006, 66, 3992–3995. [Google Scholar] [CrossRef]
- Benvenuti, S.; Sartore-Bianchi, A.; Di Nicolantonio, F.; Zanon, C.; Moroni, M.; Veronese, S.; Siena, S.; Bardelli, A. Oncogenic Activation of the RAS/RAF Signaling Pathway Impairs the Response of Metastatic Colorectal Cancers to Anti–Epidermal Growth Factor Receptor Antibody Therapies. Cancer Res. 2007, 67, 2643–2648. [Google Scholar] [CrossRef] [PubMed]
- Di Nicolantonio, F.; Martini, M.; Molinari, F.; Sartore-Bianchi, A.; Arena, S.; Saletti, P.; De Dosso, S.; Mazzucchelli, L.; Frattini, M.; Siena, S.; et al. Wild-Type BRAF Is Required for Response to Panitumumab or Cetuximab in Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 5705–5712. [Google Scholar] [CrossRef] [PubMed]
- Eng, C.; Yoshino, T.; Ruíz-García, E.; Mostafa, N.; Cann, C.G.; O’Brian, B.; Benny, A.; Perez, R.O.; Cremolini, C. Colorectal Cancer. Lancet, 2024; online first. [Google Scholar] [CrossRef]
- Bardelli, A.; Siena, S. Molecular Mechanisms of Resistance to Cetuximab and Panitumumab in Colorectal Cancer. J. Clin. Oncol. 2010, 28, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.D.; Jia, X.C.; Corvalan, J.R.F.; Wang, P.; Davis, C.G.; Jakobovits, A. Eradication of Established Tumors by a Fully Human Monoclonal Antibody to the Epidermal Growth Factor Receptor without Concomitant Chemotherapy. Cancer Res. 1999, 59, 1236–1243. [Google Scholar] [PubMed]
- Stremitzer, S.; Sebio, A.; Stintzing, S.; Lenz, H.-J. Panitumumab Safety for Treating Colorectal Cancer. Expert Opin. Drug Saf. 2014, 13, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Voigt, M.; Braig, F.; Göthel, M.; Schulte, A.; Lamszus, K.; Bokemeyer, C.; Binder, M. Functional Dissection of the Epidermal Growth Factor Receptor Epitopes Targeted by Panitumumab and Cetuximab. Neoplasia 2012, 14, 1023–1031. [Google Scholar] [CrossRef]
- Montagut, C.; Dalmases, A.; Bellosillo, B.; Crespo, M.; Pairet, S.; Iglesias, M.; Salido, M.; Gallen, M.; Marsters, S.; Tsai, S.P.; et al. Identification of a Mutation in the Extracellular Domain of the Epidermal Growth Factor Receptor Conferring Cetuximab Resistance in Colorectal Cancer. Nat. Med. 2012, 18, 221–223. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.H.; Mirakhur, B.; Chan, E.; Le, Q.-T.; Berlin, J.; Morse, M.; Murphy, B.A.; Satinover, S.M.; Hosen, J.; Mauro, D.; et al. Cetuximab-Induced Anaphylaxis and IgE Specific for Galactose-α-1,3-Galactose. N. Engl. J. Med. 2008, 358, 1109–1117. [Google Scholar] [CrossRef]
- Yaeger, R.; Weiss, J.; Pelster, M.S.; Spira, A.I.; Barve, M.; Ou, S.-H.I.; Leal, T.A.; Bekaii-Saab, T.S.; Paweletz, C.P.; Heavey, G.A.; et al. Adagrasib with or without Cetuximab in Colorectal Cancer with Mutated KRAS G12C. N. Engl. J. Med. 2023, 388, 44–54. [Google Scholar] [CrossRef]
- Fakih, M.G.; Salvatore, L.; Esaki, T.; Modest, D.P.; Lopez-Bravo, D.P.; Taieb, J.; Karamouzis, M.V.; Ruiz-Garcia, E.; Kim, T.-W.; Kuboki, Y.; et al. Sotorasib plus Panitumumab in Refractory Colorectal Cancer with Mutated KRAS G12C. N. Engl. J. Med. 2023, 389, 2125–2139. [Google Scholar] [CrossRef]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E–Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef] [PubMed]
- Amodio, V.; Yaeger, R.; Arcella, P.; Cancelliere, C.; Lamba, S.; Lorenzato, A.; Arena, S.; Montone, M.; Mussolin, B.; Bian, Y.; et al. EGFR Blockade Reverts Resistance to KRASG12C Inhibition in Colorectal Cancer. Cancer Discov. 2020, 10, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Saoudi González, N.; Ros, J.; Baraibar, I.; Salvà, F.; Rodríguez-Castells, M.; Alcaraz, A.; García, A.; Tabernero, J.; Élez, E. Cetuximab as a Key Partner in Personalized Targeted Therapy for Metastatic Colorectal Cancer. Cancers 2024, 16, 412. [Google Scholar] [CrossRef] [PubMed]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of KRAS Mutations and Acquired Resistance to Anti-EGFR Therapy in Colorectal Cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef] [PubMed]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA Mutations on the Efficacy of Cetuximab plus Chemotherapy in Chemotherapy-Refractory Metastatic Colorectal Cancer: A Retrospective Consortium Analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Misale, S.; Arena, S.; Lamba, S.; Siravegna, G.; Lallo, A.; Hobor, S.; Russo, M.; Buscarino, M.; Lazzari, L.; Sartore-Bianchi, A.; et al. Blockade of EGFR and MEK Intercepts Heterogeneous Mechanisms of Acquired Resistance to Anti-EGFR Therapies in Colorectal Cancer. Sci. Transl. Med. 2014, 6, 224ra26. [Google Scholar] [CrossRef] [PubMed]
- Parseghian, C.M.; Sun, R.; Woods, M.; Napolitano, S.; Lee, H.M.; Alshenaifi, J.; Willis, J.; Nunez, S.; Raghav, K.P.; Morris, V.K.; et al. Resistance Mechanisms to Anti–Epidermal Growth Factor Receptor Therapy in RAS/RAF Wild-Type Colorectal Cancer Vary by Regimen and Line of Therapy. J. Clin. Oncol. 2023, 41, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Brand, T.M.; Iida, M.; Wheeler, D.L. Molecular Mechanisms of Resistance to the EGFR Monoclonal Antibody Cetuximab. Cancer Biol. Ther. 2011, 11, 777–792. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ji, Q.; Li, Q. Resistance to Anti-EGFR Therapies in Metastatic Colorectal Cancer: Underlying Mechanisms and Reversal Strategies. J. Exp. Clin. Cancer Res. 2021, 40, 328. [Google Scholar] [CrossRef]
- Liang, J.; Oyang, L.; Rao, S.; Han, Y.; Luo, X.; Yi, P.; Lin, J.; Xia, L.; Hu, J.; Tan, S.; et al. Rac1, A Potential Target for Tumor Therapy. Front. Oncol. 2021, 11, 674426. [Google Scholar] [CrossRef]
- Melzer, C.; Hass, R.; Lehnert, H.; Ungefroren, H. RAC1B: A Rho GTPase with Versatile Functions in Malignant Transformation and Tumor Progression. Cells 2019, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Bid, H.K.; Roberts, R.D.; Manchanda, P.K.; Houghton, P.J. RAC1: An Emerging Therapeutic Option for Targeting Cancer Angiogenesis and Metastasis. Mol. Cancer Ther. 2013, 12, 1925–1934. [Google Scholar] [CrossRef] [PubMed]
- Heasman, S.J.; Ridley, A.J. Mammalian Rho GTPases: New Insights into Their Functions from in Vivo Studies. Nat. Rev. Mol. Cell Biol. 2008, 9, 690–701. [Google Scholar] [CrossRef]
- Porter, A.P.; Papaioannou, A.; Malliri, A. Deregulation of Rho GTPases in Cancer. Small GTPases 2016, 7, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Xu, E.; Luo, Q.; Song, G. Rac1: A Regulator of Cell Migration and a Potential Target for Cancer Therapy. Molecules 2023, 28, 2976. [Google Scholar] [CrossRef] [PubMed]
- Marei, H.; Malliri, A. Rac1 in Human Diseases: The Therapeutic Potential of Targeting Rac1 Signaling Regulatory Mechanisms. Small GTPases 2017, 8, 139–163. [Google Scholar] [CrossRef] [PubMed]
- Matos, P.; Skaug, J.; Marques, B.; Beck, S.; Veríssimo, F.; Gespach, C.; Boavida, M.G.; Scherer, S.W.; Jordan, P. Small GTPase Rac1: Structure, Localization, and Expression of the Human Gene. Biochem. Biophys. Res. Commun. 2000, 277, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Michaelson, D.; Silletti, J.; Murphy, G.; D’Eustachio, P.; Rush, M.; Philips, M.R. Differential Localization of Rho GTPases in Live Cells: Regulation by Hypervariable Regions and RhoGDI Binding. J. Cell Biol. 2001, 152, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Li, L.; Ballermann, B.; Wang, Z. Phosphorylation of Rac1 T108 by Extracellular Signal-Regulated Kinase in Response to Epidermal Growth Factor: A Novel Mechanism To Regulate Rac1 Function. Mol. Cell. Biol. 2013, 33, 4538–4551. [Google Scholar] [CrossRef]
- Roberts, P.J.; Mitin, N.; Keller, P.J.; Chenette, E.J.; Madigan, J.P.; Currin, R.O.; Cox, A.D.; Wilson, O.; Kirschmeier, P.; Der, C.J. Rho Family GTPase Modification and Dependence on CAAX Motif-Signaled Posttranslational Modification. J. Biol. Chem. 2008, 283, 25150–25163. [Google Scholar] [CrossRef]
- Esufali, S.; Charames, G.S.; Pethe, V.V.; Buongiorno, P.; Bapat, B. Activation of Tumor-Specific Splice Variant Rac1b by Dishevelled Promotes Canonical Wnt Signaling and Decreased Adhesion of Colorectal Cancer Cells. Cancer Res. 2007, 67, 2469–2479. [Google Scholar] [CrossRef] [PubMed]
- Jordan, P.; Brazã£o, R.; Boavida, M.G.; Gespach, C.; Chastre, E. Cloning of a Novel Human Rac1b Splice Variant with Increased Expression in Colorectal Tumors. Oncogene 1999, 18, 6835–6839. [Google Scholar] [CrossRef]
- Schnelzer, A.; Prechtel, D.; Knaus, U.; Dehne, K.; Gerhard, M.; Graeff, H.; Harbeck, N.; Schmitt, M.; Lengyel, E. Rac1 in Human Breast Cancer: Overexpression, Mutation Analysis, and Characterization of a New Isoform, Rac1b. Oncogene 2000, 19, 3013–3020. [Google Scholar] [CrossRef]
- Abdrabou, A.; Wang, Z. Post-Translational Modification and Subcellular Distribution of Rac1: An Update. Cells 2018, 7, 263. [Google Scholar] [CrossRef]
- Olson, M.F. Rho GTPases, Their Post-Translational Modifications, Disease-Associated Mutations and Pharmacological Inhibitors. Small GTPases 2018, 9, 203–215. [Google Scholar] [CrossRef]
- Schiller, M.R. Coupling Receptor Tyrosine Kinases to Rho GTPases—GEFs What’s the Link. Cell. Signal. 2006, 18, 1834–1843. [Google Scholar] [CrossRef] [PubMed]
- Fiegen, D.; Haeusler, L.C.; Blumenstein, L.; Herbrand, U.; Dvorsky, R.; Vetter, I.R.; Ahmadian, M.R. Alternative Splicing of Rac1 Generates Rac1b, a Self-Activating GTPase. J. Biol. Chem. 2004, 279, 4743–4749. [Google Scholar] [CrossRef]
- Haeusler, L.C.; Hemsath, L.; Fiegen, D.; Blumenstein, L.; Herbrand, U.; Stege, P.; Dvorsky, R.; Ahmadian, M.R. Purification and Biochemical Properties of Rac1, 2, 3 and the Splice Variant Rac1b. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2006; Volume 406, pp. 1–11. [Google Scholar]
- Matos, P.; Collard, J.G.; Jordan, P. Tumor-Related Alternatively Spliced Rac1b Is Not Regulated by Rho-GDP Dissociation Inhibitors and Exhibits Selective Downstream Signaling. J. Biol. Chem. 2003, 278, 50442–50448. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, V.; Matos, P.; Jordan, P. Antagonistic SR Proteins Regulate Alternative Splicing of Tumor-Related Rac1b Downstream of the PI3-Kinase and Wnt Pathways. Hum. Mol. Genet. 2009, 18, 3696–3707. [Google Scholar] [CrossRef]
- Bordonaro, M. Crosstalk between Wnt Signaling and RNA Processing in Colorectal Cancer. J. Cancer 2013, 4, 96–103. [Google Scholar] [CrossRef]
- Gonçalves, V.; Henriques, A.; Pereira, J.; Costa, A.N.; Moyer, M.P.; Moita, L.F.; Gama-Carvalho, M.; Matos, P.; Jordan, P. Phosphorylation of SRSF1 by SRPK1 Regulates Alternative Splicing of Tumor-Related Rac1b in Colorectal Cells. RNA 2014, 20, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Pelisch, F.; Khauv, D.; Risso, G.; Stallings-Mann, M.; Blaustein, M.; Quadrana, L.; Radisky, D.C.; Srebrow, A. Involvement of HnRNP A1 in the Matrix Metalloprotease-3-Dependent Regulation of Rac1 Pre-MRNA Splicing. J. Cell Biochem. 2012, 113, 2319–2329. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Fu, X.; Chen, P.; Wu, P.; Fan, X.; Li, N.; Zhu, H.; Jia, T.-T.; Ji, H.; Wang, Z.; et al. SPSB1-Mediated HnRNP A1 Ubiquitylation Regulates Alternative Splicing and Cell Migration in EGF Signaling. Cell Res. 2017, 27, 540–588. [Google Scholar] [CrossRef] [PubMed]
- Manco, M.; Ala, U.; Cantarella, D.; Tolosano, E.; Medico, E.; Altruda, F.; Fagoonee, S. The RNA-Binding Protein ESRP1 Modulates the Expression of RAC1b in Colorectal Cancer Cells. Cancers 2021, 13, 4092. [Google Scholar] [CrossRef] [PubMed]
- Kotelevets, L.; Chastre, E. Rac1 Signaling: From Intestinal Homeostasis to Colorectal Cancer Metastasis. Cancers 2020, 12, 665. [Google Scholar] [CrossRef] [PubMed]
- Carmon, K.S.; Gong, X.; Yi, J.; Wu, L.; Thomas, A.; Moore, C.M.; Masuho, I.; Timson, D.J.; Martemyanov, K.A.; Liu, Q.J. LGR5 Receptor Promotes Cell–Cell Adhesion in Stem Cells and Colon Cancer Cells via the IQGAP1–Rac1 Pathway. J. Biol. Chem. 2017, 292, 14989–15001. [Google Scholar] [CrossRef] [PubMed]
- Dise, R.S.; Frey, M.R.; Whitehead, R.H.; Polk, D.B. Epidermal Growth Factor Stimulates Rac Activation through Src and Phosphatidylinositol 3-Kinase to Promote Colonic Epithelial Cell Migration. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 294, G276–G285. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Sánchez, L.D.C.; Ngo, P.A.; Pradhan, R.; Becker, L.S.; Boehringer, D.; Soteriou, D.; Kubankova, M.; Schweitzer, C.; Koch, T.; Thonn, V.; et al. Epithelial RAC1-Dependent Cytoskeleton Dynamics Controls Cell Mechanics, Cell Shedding and Barrier Integrity in Intestinal Inflammation. Gut 2023, 72, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.M.; Zilz, N.; Beazer-Barclay, Y.; Bryan, T.M.; Hamilton, S.R.; Thibodeau, S.N.; Vogelstein, B.; Kinzler, K.W. APC Mutations Occur Early during Colorectal Tumorigenesis. Nature 1992, 359, 235–237. [Google Scholar] [CrossRef]
- Myant, K.B.; Cammareri, P.; McGhee, E.J.; Ridgway, R.A.; Huels, D.J.; Cordero, J.B.; Schwitalla, S.; Kalna, G.; Ogg, E.L.; Athineos, D.; et al. ROS Production and NF-ΚB Activation Triggered by RAC1 Facilitate WNT-Driven Intestinal Stem Cell Proliferation and Colorectal Cancer Initiation. Cell Stem Cell 2013, 12, 761–773. [Google Scholar] [CrossRef]
- Lou, S.; Wang, P.; Yang, J.; Ma, J.; Liu, C.; Zhou, M. Prognostic and Clinicopathological Value of Rac1 in Cancer Survival: Evidence from a Meta-Analysis. J. Cancer 2018, 9, 2571–2579. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Xu, W.; Lu, T.; Zhou, J.; Ge, X.; Hua, D. MicroRNA-142-3p Promotes Cellular Invasion of Colorectal Cancer Cells by Activation of RAC1. Technol. Cancer Res. Treat. 2018, 17, 1533033818790508. [Google Scholar] [CrossRef]
- Xia, L.; Lin, J.; Su, J.; Oyang, L.; Wang, H.; Tan, S.; Tang, Y.; Chen, X.; Liu, W.; Luo, X.; et al. Diallyl Disulfide Inhibits Colon Cancer Metastasis by Suppressing Rac1-Mediated Epithelial-Mesenchymal Transition. Onco Targets Ther. 2019, 12, 5713–5728. [Google Scholar] [CrossRef] [PubMed]
- Krauthammer, M.; Kong, Y.; Ha, B.H.; Evans, P.; Bacchiocchi, A.; McCusker, J.P.; Cheng, E.; Davis, M.J.; Goh, G.; Choi, M.; et al. Exome Sequencing Identifies Recurrent Somatic RAC1 Mutations in Melanoma. Nat. Genet. 2012, 44, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Espina, C.; Céspedes, M.V.; García-Cabezas, M.A.; Del Pulgar, M.T.G.; Boluda, A.; Oroz, L.G.; Cejas, P.; Nistal, M.; Mangues, R.; Lacal, J.C. A Critical Role for Rac1 in Tumor Progression of Human Colorectal Adenocarcinoma Cells. Am. J. Pathol. 2008, 172, 156–166. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xie, X.; Lu, Y.; Wang, B.; Yin, X.; Chen, J. DOCK4 Is a Novel Prognostic Biomarker and Correlated with Immune Infiltrates in Colon Adenocarcinoma. Comb. Chem. High. Throughput Screen. 2023, 27, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhou, M.; Guan, B.; Xie, B.; Liu, Y.; He, J.; Zhao, J.; Zhao, Q.; Yan, D. Tumour-associated Macrophage-derived DOCK7-enriched Extracellular Vesicles Drive Tumour Metastasis in Colorectal Cancer via the RAC1/ABCA1 Axis. Clin. Transl. Med. 2024, 14, e1591. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Hu, F.; Huang, C.; Lan, J.; She, X.; Zhao, C.; Wu, H.; Liu, A.; Wu, Q.; Chen, Y.; et al. Tiam1 Methylation by NSD2 Promotes Rac1 Signaling Activation and Colon Cancer Metastasis. Proc. Natl. Acad. Sci. USA 2023, 120, e2305684120. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Liu, Q.; Xia, L.; Lin, J.; Oyang, L.; Tan, S.; Peng, Q.; Jiang, X.; Xu, X.; Wu, N.; et al. Rac1 Promotes the Reprogramming of Glucose Metabolism and the Growth of Colon Cancer Cells through Upregulating SOX9. Cancer Sci. 2023, 114, 822–836. [Google Scholar] [CrossRef]
- Singh, A.; Karnoub, A.E.; Palmby, T.R.; Lengyel, E.; Sondek, J.; Der, C.J. Rac1b, a Tumor Associated, Constitutively Active Rac1 Splice Variant, Promotes Cellular Transformation. Oncogene 2004, 23, 9369–9380. [Google Scholar] [CrossRef]
- Li, G.; Ying, L.; Wang, H.; Wei, S.-S.; Chen, J.; Chen, Y.-H.; Xu, W.-P.; Jie, Q.-Q.; Zhou, Q.; Li, Y.-G.; et al. Rac1b Enhances Cell Survival through Activation of the JNK2/c-JUN/Cyclin-D1 and AKT2/MCL1 Pathways. Oncotarget 2016, 7, 17970–17985. [Google Scholar] [CrossRef] [PubMed]
- Matos, P.; Jordan, P. Expression of Rac1b Stimulates NF-ΚB-Mediated Cell Survival and G1/S Progression. Exp. Cell Res. 2005, 305, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Matos, P.; Jordan, P. Increased Rac1b Expression Sustains Colorectal Tumor Cell Survival. Mol. Cancer Res. 2008, 6, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Goka, E.T.; Chaturvedi, P.; Lopez, D.T.M.; De La Garza, A.; Lippman, M.E. RAC1b Overexpression Confers Resistance to Chemotherapy Treatment in Colorectal Cancer. Mol. Cancer Ther. 2019, 18, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Kotelevets, L.; Walker, F.; Mamadou, G.; Lehy, T.; Jordan, P.; Chastre, E. The Rac1 Splice Form Rac1b Favors Mouse Colonic Mucosa Regeneration and Contributes to Intestinal Cancer Progression. Oncogene 2018, 37, 6054–6068. [Google Scholar] [CrossRef] [PubMed]
- Matos, P.; Oliveira, C.; Velho, S.; Gonçalves, V.; da Costa, L.T.; Moyer, M.P.; Seruca, R.; Jordan, P. B-Raf V600E Cooperates With Alternative Spliced Rac1b to Sustain Colorectal Cancer Cell Survival. Gastroenterology 2008, 135, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Espinaco, V.; Cuatrecasas, M.; Alonso, V.; Escudero, P.; Marmol, M.; Horndler, C.; Ortego, J.; Gallego, R.; Codony-Servat, J.; Garcia-Albeniz, X.; et al. RAC1b Overexpression Correlates with Poor Prognosis in KRAS/BRAF WT Metastatic Colorectal Cancer Patients Treated with First-Line FOLFOX/XELOX Chemotherapy. Eur. J. Cancer 2014, 50, 1973–1981. [Google Scholar] [CrossRef] [PubMed]
- Gudiño, V.; Pohl, S.Ö.G.; Billard, C.V.; Cammareri, P.; Bolado, A.; Aitken, S.; Stevenson, D.; Hall, A.E.; Agostino, M.; Cassidy, J.; et al. RAC1B Modulates Intestinal Tumourigenesis via Modulation of WNT and EGFR Signalling Pathways. Nat. Commun. 2021, 12, 2335. [Google Scholar] [CrossRef] [PubMed]
- Matos, P.; Kotelevets, L.; Gonçalves, V.; Henriques, A.; Zerbib, P.; Moyer, M.P.; Chastre, E.; Jordan, P. Ibuprofen Inhibits Colitis-Induced Overexpression of Tumor-Related Rac1b. Neoplasia 2013, 15, 102–111. [Google Scholar] [CrossRef]
- Gonçalves, V.; Henriques, A.F.A.; Matos, P.; Jordan, P. Ibuprofen Disrupts a WNK1/GSK3β/SRPK1 Protein Complex Required for Expression of Tumor-Related Splicing Variant RAC1B in Colorectal Cells. Oncotarget 2020, 11, 4421–4437. [Google Scholar] [CrossRef]
- Pereira, J.F.S.; Bessa, C.; Matos, P.; Jordan, P. Pro-Inflammatory Cytokines Trigger the Overexpression of Tumour-Related Splice Variant RAC1B in Polarized Colorectal Cells. Cancers 2022, 14, 1393. [Google Scholar] [CrossRef] [PubMed]
- Orlichenko, L.; Geyer, R.; Yanagisawa, M.; Khauv, D.; Radisky, E.S.; Anastasiadis, P.Z.; Radisky, D.C. The 19-Amino Acid Insertion in the Tumor-Associated Splice Isoform Rac1b Confers Specific Binding to P120 Catenin. J. Biol. Chem. 2010, 285, 19153–19161. [Google Scholar] [CrossRef]
- Radisky, D.C.; Levy, D.D.; Littlepage, L.E.; Liu, H.; Nelson, C.M.; Fata, J.E.; Leake, D.; Godden, E.L.; Albertson, D.G.; Nieto, M.A.; et al. Rac1b and Reactive Oxygen Species Mediate MMP-3-Induced EMT and Genomic Instability. Nature 2005, 436, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.A.; Chen, Q.K.; Lui, C.; Cichon, M.A.; Radisky, D.C.; Nelson, C.M. Matrix Compliance Regulates Rac1b Localization, NADPH Oxidase Assembly, and Epithelial–Mesenchymal Transition. Mol. Biol. Cell 2012, 23, 4097–4108. [Google Scholar] [CrossRef] [PubMed]
- Pethe, V.V.; Charames, G.S.; Bapat, B. Rac1b Recruits Dishevelled and β-Catenin to Wnt Target Gene Promoters Independent of Wnt3A Stimulation. Int. J. Oncol. 2011, 39, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Cizmecioglu, O.; Ni, J.; Xie, S.; Zhao, J.J.; Roberts, T.M. Rac1-Mediated Membrane Raft Localization of PI3K/P110β Is Required for Its Activation by GPCRs or PTEN Loss. Elife 2016, 5, e17635. [Google Scholar] [CrossRef]
- Niba, E.T.E.; Nagaya, H.; Kanno, T.; Tsuchiya, A.; Gotoh, A.; Tabata, C.; Kuribayashi, K.; Nakano, T.; Nishizaki, T. Crosstalk between PI3 Kinase/PDK1/Akt/Rac1 and Ras/Raf/MEK/ERK Pathways Downstream PDGF Receptor. Cell. Physiol. Biochem. 2013, 31, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Gao, N.; Chen, J.; Fan, L.; Zeng, Z.; Gao, G.; Li, L.; Fang, G.; Hu, K.; Pang, X.; et al. Erk and MAPK Signaling Is Essential for Intestinal Development through Wnt Pathway Modulation. Development 2020, 147, dev185678. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.-L.; Deng, Y.-Y.; Fu, J.; Zhang, Y.-H.; Zhang, Z.-J.; Qin, X.-J. ARHGAP17 Enhances 5-Fluorouracil-Induced Apoptosis in Colon Cancer Cells by Suppressing Rac1. Neoplasma 2022, 69, 640–647. [Google Scholar] [CrossRef]
- Li, Q.; Qin, T.; Bi, Z.; Hong, H.; Ding, L.; Chen, J.; Wu, W.; Lin, X.; Fu, W.; Zheng, F.; et al. Rac1 Activates Non-Oxidative Pentose Phosphate Pathway to Induce Chemoresistance of Breast Cancer. Nat. Commun. 2020, 11, 1456. [Google Scholar] [CrossRef]
- De, P.; Rozeboom, B.J.; Aske, J.C.; Dey, N. Active RAC1 Promotes Tumorigenic Phenotypes and Therapy Resistance in Solid Tumors. Cancers 2020, 12, 1541. [Google Scholar] [CrossRef] [PubMed]
- Bagheri-Yarmand, R.; Busaidy, N.L.; McBeath, E.; Danysh, B.P.; Evans, K.W.; Moss, T.J.; Akcakanat, A.; Ng, P.K.S.; Knippler, C.M.; Golden, J.A.; et al. RAC1 Alterations Induce Acquired Dabrafenib Resistance in Association with Anaplastic Transformation in a Papillary Thyroid Cancer Patient. Cancers 2021, 13, 4950. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Takagi, K.; Sato, A.; Miki, Y.; Miyashita, M.; Sasano, H.; Suzuki, T. Rac1 Activation in Human Breast Carcinoma as a Prognostic Factor Associated with Therapeutic Resistance. Breast Cancer 2020, 27, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.-J.; Zheng, C.-W.; Gu, J.-E.; Zhang, H.-X.; Xie, L.; Xu, L.-Y.; Li, E.-M. RAC1 Inhibition Reverses Cisplatin Resistance in Esophageal Squamous Cell Carcinoma and Induces Downregulation of Glycolytic Enzymes. Mol. Oncol. 2019, 13, 2010–2030. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Hu, H.; Ma, Y.; Xiong, S.; Zhu, D. Vav1-Dependent Rac1 Activation Mediates Hypoxia-Induced Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma Cells through Upregulation of HIF-1α Expression. Cell Biol. Int. 2023, 47, 1835–1842. [Google Scholar] [CrossRef] [PubMed]
- Zhu, E.Y.; Riordan, J.D.; Vanneste, M.; Henry, M.D.; Stipp, C.S.; Dupuy, A.J. SRC-RAC1 Signaling Drives Drug Resistance to BRAF Inhibition in de-Differentiated Cutaneous Melanomas. NPJ Precis. Oncol. 2022, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yin, L.; Qiao, G.; Li, Y.; Li, B.; Bai, Y.; Feng, F. Inhibition of Rac1 Reverses Enzalutamide Resistance in Castration-Resistant Prostate Cancer. Oncol. Lett. 2020, 20, 2997–3005. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Ren, W.; Cui, Q.; Liang, H.; Yang, C.; Liu, W.; Wang, X.; Liu, X.; Shi, Y.; Feng, J.; et al. Integrin Β4 Promotes DNA Damage-Related Drug Resistance in Triple-Negative Breast Cancer via TNFAIP2/IQGAP1/RAC1. Elife 2023, 12, RP88483. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Gurler, S.B.; Novo, D.; Selli, C.; Alferez, D.G.; Eroglu, S.; Pavlou, K.; Zhang, J.; Sims, A.H.; Humphreys, N.E.; et al. RAC1B Function Is Essential for Breast Cancer Stem Cell Maintenance and Chemoresistance of Breast Tumor Cells. Oncogene 2023, 42, 679–692. [Google Scholar] [CrossRef]
- Wieduwilt, M.J.; Moasser, M.M. The Epidermal Growth Factor Receptor Family: Biology Driving Targeted Therapeutics. Cell. Mol. Life Sci. 2008, 65, 1566–1584. [Google Scholar] [CrossRef]
- Nimnual, A.S.; Yatsula, B.A.; Bar-Sagi, D. Coupling of Ras and Rac Guanosine Triphosphatases through the Ras Exchanger Sos. Science 1998, 279, 560–563. [Google Scholar] [CrossRef] [PubMed]
- Eblen, S.T.; Slack, J.K.; Weber, M.J.; Catling, A.D. Rac-PAK Signaling Stimulates Extracellular Signal-Regulated Kinase (ERK) Activation by Regulating Formation of MEK1-ERK Complexes. Mol. Cell. Biol. 2002, 22, 6023–6033. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.-H.; Wang, A.-X.; Chen, Y. Radixin Enhances Colon Cancer Cell Invasion by Increasing MMP-7 Production via Rac1-ERK Pathway. Sci. World J. 2014, 2014, 340271. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Rao, J.; Zhou, Z.-H.; Yao, X.-H.; Wu, F.; Yang, J.; Yang, L.; Zhang, X.; Cui, Y.-H.; Bian, X.-W.; et al. RAC1-GTP Promotes Epithelial-Mesenchymal Transition and Invasion of Colorectal Cancer by Activation of STAT3. Lab. Investig. 2018, 98, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Podtelejnikov, A.V.; Blagoev, B.; Bustelo, X.R.; Mann, M.; Lodish, H.F. Analysis of Receptor Signaling Pathways by Mass Spectrometry: Identification of Vav-2 as a Substrate of the Epidermal and Platelet-Derived Growth Factor Receptors. Proc. Natl. Acad. Sci. USA 2000, 97, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Tamás, P.; Solti, Z.; Bauer, P.; Illés, A.; Sipeki, S.; Bauer, A.; Faragó, A.; Downward, J.; Buday, L. Mechanism of Epidermal Growth Factor Regulation of Vav2, a Guanine Nucleotide Exchange Factor for Rac. J. Biol. Chem. 2003, 278, 5163–5171. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Luby-Phelps, K.; Das, B.; Shu, X.; Xia, Y.; Mosteller, R.D.; Krishna, U.M.; Falck, J.R.; White, M.A.; Broek, D. Role of Substrates and Products of PI 3-Kinase in Regulating Activation of Rac-Related Guanosine Triphosphatases by Vav. Science 1998, 279, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Kwon, D.Y.; Chun, J.; Kim, J.H.; Kang, S.S. Akt Protein Kinase Inhibits Rac1-GTP Binding through Phosphorylation at Serine 71 of Rac1. J. Biol. Chem. 2000, 275, 423–428. [Google Scholar] [CrossRef]
- Saci, A.; Cantley, L.C.; Carpenter, C.L. Rac1 Regulates the Activity of MTORC1 and MTORC2 and Controls Cellular Size. Mol. Cell 2011, 42, 50–61. [Google Scholar] [CrossRef]
- Gulhati, P.; Bowen, K.A.; Liu, J.; Stevens, P.D.; Rychahou, P.G.; Chen, M.; Lee, E.Y.; Weiss, H.L.; O’Connor, K.L.; Gao, T.; et al. MTORC1 and MTORC2 Regulate EMT, Motility, and Metastasis of Colorectal Cancer via RhoA and Rac1 Signaling Pathways. Cancer Res. 2011, 71, 3246–3256. [Google Scholar] [CrossRef]
- Marcoux, N.; Vuori, K. EGF Receptor Mediates Adhesion-Dependent Activation of the Rac GTPase: A Role for Phosphatidylinositol 3-Kinase and Vav2. Oncogene 2003, 22, 6100–6106. [Google Scholar] [CrossRef] [PubMed]
- Campa, C.C.; Ciraolo, E.; Ghigo, A.; Germena, G.; Hirsch, E. Crossroads of PI3K and Rac Pathways. Small GTPases 2015, 6, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Fan, Z.; Ding, M.; Zhang, H.; Mu, L.; Ding, Y.; Zhang, Y.; Jia, B.; Chen, L.; Chang, Z.; et al. An EGFR/PI3K/AKT Axis Promotes Accumulation of the Rac1-GEF Tiam1 That Is Critical in EGFR-Driven Tumorigenesis. Oncogene 2015, 34, 5971–5982. [Google Scholar] [CrossRef] [PubMed]
- Fleming, I.N.; Gray, A.; Downes, C.P. Regulation of the Rac1-Specific Exchange Factor Tiam1 Involves Both Phosphoinositide 3-Kinase-Dependent and -Independent Components. Biochem. J. 2000, 351, 173–182. [Google Scholar] [CrossRef]
- Malliri, A.; Rygiel, T.P.; Van Der Kammen, R.A.; Song, J.-Y.; Engers, R.; Hurlstone, A.F.L.; Clevers, H.; Collard, J.G. The Rac Activator Tiam1 Is a Wnt-Responsive Gene That Modifies Intestinal Tumor Development. J. Biol. Chem. 2006, 281, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Buongiorno, P.; Pethe, V.V.; Charames, G.S.; Esufali, S.; Bapat, B. Rac1 GTPase and the Rac1 Exchange Factor Tiam1 Associate with Wnt-Responsive Promoters to Enhance Beta-Catenin/TCF-Dependent Transcription in Colorectal Cancer Cells. Mol. Cancer 2008, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Henriques, A.F.A.; Barros, P.; Moyer, M.P.; Matos, P.; Jordan, P. Expression of Tumor-Related Rac1b Antagonizes B-Raf-Induced Senescence in Colorectal Cells. Cancer Lett. 2015, 369, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Yonesaka, K.; Zejnullahu, K.; Okamoto, I.; Satoh, T.; Cappuzzo, F.; Souglakos, J.; Ercan, D.; Rogers, A.; Roncalli, M.; Takeda, M.; et al. Activation of ERBB2 Signaling Causes Resistance to the EGFR-Directed Therapeutic Antibody Cetuximab. Sci. Transl. Med. 2011, 3, 99ra86. [Google Scholar] [CrossRef] [PubMed]
- Bertotti, A.; Migliardi, G.; Galimi, F.; Sassi, F.; Torti, D.; Isella, C.; Corà, D.; di Nicolantonio, F.; Buscarino, M.; Petti, C.; et al. A Molecularly Annotated Platform of Patient- Derived Xenografts (“xenopatients”) Identifies HER2 as an Effective Therapeutic Target in Cetuximab-Resistant Colorectal Cancer. Cancer Discov. 2011, 1, 508–523. [Google Scholar] [CrossRef]
- Bardelli, A.; Corso, S.; Bertotti, A.; Hobor, S.; Valtorta, E.; Siravegna, G.; Sartore-Bianchi, A.; Scala, E.; Cassingena, A.; Zecchin, D.; et al. Amplification of the MET Receptor Drives Resistance to Anti-EGFR Therapies in Colorectal Cancer. Cancer Discov. 2013, 3, 658–673. [Google Scholar] [CrossRef]
- Cappuzzo, F.; Varella-Garcia, M.; Finocchiaro, G.; Skokan, M.; Gajapathy, S.; Carnaghi, C.; Rimassa, L.; Rossi, E.; Ligorio, C.; Di Tommaso, L.; et al. Primary Resistance to Cetuximab Therapy in EGFR FISH-Positive Colorectal Cancer Patients. Br. J. Cancer 2008, 99, 83–89. [Google Scholar] [CrossRef]
- Luraghi, P.; Reato, G.; Cipriano, E.; Sassi, F.; Orzan, F.; Bigatto, V.; De Bacco, F.; Menietti, E.; Han, M.; Rideout, W.M.; et al. MET Signaling in Colon Cancer Stem-like Cells Blunts the Therapeutic Response to EGFR Inhibitors. Cancer Res. 2014, 74, 1857–1869. [Google Scholar] [CrossRef]
- Scartozzi, M.; Mandolesi, A.; Giampieri, R.; Pierantoni, C.; Loupakis, F.; Zaniboni, A.; Galizia, E.; Giustini, L.; Silva, R.R.; Bisonni, R.; et al. Insulin-like Growth Factor 1 Expression Correlates with Clinical Outcome in K-RAS Wild Type Colorectal Cancer Patients Treated with Cetuximab and Irinotecan. Int. J. Cancer 2010, 127, 1941–1947. [Google Scholar] [CrossRef] [PubMed]
- Winder, T.; Zhang, W.; Yang, D.; Ning, Y.; Bohanes, P.; Gerger, A.; Wilson, P.M.; Pohl, A.; Mauro, D.J.; Langer, C.; et al. Germline Polymorphisms in Genes Involved in the IGF1 Pathway Predict Efficacy of Cetuximab in Wild-Type KRAS MCRC Patients. Clin. Cancer Res. 2010, 16, 5591–5602. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wang, L.; Qiu, H.; Zhang, M.; Sun, L.; Peng, P.; Yu, Q.; Yuan, X. Mechanisms of Resistance to Anti-EGFR Therapy in Colorectal Cancer. Oncotarget 2017, 8, 3980–4000. [Google Scholar] [CrossRef] [PubMed]
- Jones, V.T.; Graves-Deal, R.; Cao, Z.; Bogatcheva, G.; Ramirez, M.A.; Harmych, S.J.; Higginbotham, J.N.; Sharma, V.; Damalanka, V.C.; Wahoski, C.C.; et al. Inhibition of Autocrine HGF Maturation Overcomes Cetuximab Resistance in Colorectal Cancer. Cell. Mol. Life Sci. 2024, 81, 28. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J.; Comoglio, P.M.; Hall, A. Regulation of Scatter Factor/Hepatocyte Growth Factor Responses by Ras, Rac, and Rho in MDCK Cells. Mol. Cell Biol. 1995, 15, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Royal, I.; Lamarche-Vane, N.; Lamorte, L.; Kaibuchi, K.; Park, M. Activation of Cdc42, Rac, PAK, and Rho-Kinase in Response to Hepatocyte Growth Factor Differentially Regulates Epithelial Cell Colony Spreading and Dissociation. Mol. Biol. Cell 2000, 11, 1709–1725. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.-H.; Chen, Y.-X.; Fang, J.-Y. Comprehensive Review of Targeted Therapy for Colorectal Cancer. Signal Transduct. Target. Ther. 2020, 5, 22. [Google Scholar] [CrossRef]
- Meric-Bernstam, F.; Hurwitz, H.; Raghav, K.P.S.; McWilliams, R.R.; Fakih, M.; VanderWalde, A.; Swanton, C.; Kurzrock, R.; Burris, H.; Sweeney, C.; et al. Pertuzumab Plus Trastuzumab for HER2-Amplified Metastatic Colorectal Cancer (MyPathway): An Updated Report from a Multicentre, Open-Label, Phase 2a Multiple Basket Study. Lancet Oncol. 2019, 20, 518–530. [Google Scholar] [CrossRef]
- Strickler, J.H.; Cercek, A.; Siena, S.; André, T.; Ng, K.; Van Cutsem, E.; Wu, C.; Paulson, A.S.; Hubbard, J.M.; Coveler, A.L.; et al. Tucatinib plus Trastuzumab for Chemotherapy-Refractory, HER2-Positive, RAS Wild-Type Unresectable or Metastatic Colorectal Cancer (MOUNTAINEER): A Multicentre, Open-Label, Phase 2 Study. Lancet Oncol. 2023, 24, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Dokmanovic, M.; Hirsch, D.S.; Shen, Y.; Wu, W.J. Rac1 Contributes to Trastuzumab Resistance of Breast Cancer Cells: Rac1 as a Potential Therapeutic Target for the Treatment of Trastuzumab-Resistant Breast Cancer. Mol. Cancer Ther. 2009, 8, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Kimura, T.; Nakagawa, T.; Okamoto, K.; Fukuya, A.; Goji, T.; Fujimoto, S.; Sogabe, M.; Miyamoto, H.; Muguruma, N.; et al. EGFR Downregulation after Anti-EGFR Therapy Predicts the Antitumor Effect in Colorectal Cancer. Mol. Cancer Res. 2017, 15, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.Y.; Shia, J.; Kemeny, N.E.; Shah, M.; Schwartz, G.K.; Tse, A.; Hamilton, A.; Pan, D.; Schrag, D.; Schwartz, L.; et al. Cetuximab Shows Activity in Colorectal Cancer Patients with Tumors That Do Not Express the Epidermal Growth Factor Receptor by Immunohistochemistry. J. Clin. Oncol. 2005, 23, 1803–1810. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, X.; Liang, K.; Luwor, R.; Siddik, Z.H.; Mills, G.B.; Mendelsohn, J.; Fan, Z. Epidermal Growth Factor Receptor (EGFR) Ubiquitination as a Mechanism of Acquired Resistance Escaping Treatment by the Anti-EGFR Monoclonal Antibody Cetuximab. Cancer Res. 2007, 67, 8240–8247. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Teeuwssen, M.; Fodde, R. Cell Heterogeneity and Phenotypic Plasticity in Metastasis Formation: The Case of Colon Cancer. Cancers 2019, 11, 1368. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.-W.; Hsu, S.-C.; Xia, W.; Cao, X.; Shih, J.-Y.; Wei, Y.; Abbruzzese, J.L.; Hortobagyi, G.N.; Hung, M.-C. Epidermal Growth Factor Receptor Cooperates with Signal Transducer and Activator of Transcription 3 to Induce Epithelial-Mesenchymal Transition in Cancer Cells via Up-Regulation of TWIST Gene Expression. Cancer Res. 2007, 67, 9066–9076. [Google Scholar] [CrossRef] [PubMed]
- Buck, E.; Eyzaguirre, A.; Barr, S.; Thompson, S.; Sennello, R.; Young, D.; Iwata, K.K.; Gibson, N.W.; Cagnoni, P.; Haley, J.D. Loss of Homotypic Cell Adhesion by Epithelial-Mesenchymal Transition or Mutation Limits Sensitivity to Epidermal Growth Factor Receptor Inhibition. Mol. Cancer Ther. 2007, 6, 532–541. [Google Scholar] [CrossRef]
- Schmitz, S.; Bindea, G.; Albu, R.I.; Mlecnik, B.; Machiels, J.-P. Cetuximab Promotes Epithelial to Mesenchymal Transition and Cancer Associated Fibroblasts in Patients with Head and Neck Cancer. Oncotarget 2015, 6, 34288–34299. [Google Scholar] [CrossRef]
- Weng, C.-H.; Chen, L.-Y.; Lin, Y.-C.; Shih, J.-Y.; Lin, Y.-C.; Tseng, R.-Y.; Chiu, A.-C.; Yeh, Y.-H.; Liu, C.; Lin, Y.-T.; et al. Epithelial-Mesenchymal Transition (EMT) beyond EGFR Mutations per Se Is a Common Mechanism for Acquired Resistance to EGFR TKI. Oncogene 2018, 38, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ma, H.; Wang, Y.; Cao, Z.; Graves-Deal, R.; Powell, A.E.; Starchenko, A.; Ayers, G.D.; Washington, M.K.; Kamath, V.; et al. Excess PLAC8 Promotes an Unconventional ERK2-Dependent EMT in Colon Cancer. J. Clin. Investig. 2014, 124, 2172–2187. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Singh, B.; Graves-Deal, R.; Ma, H.; Starchenko, A.; Fry, W.H.; Lu, Y.; Wang, Y.; Bogatcheva, G.; Khan, M.P.; et al. Three-Dimensional Culture System Identifies a New Mode of Cetuximab Resistance and Disease-Relevant Genes in Colorectal Cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E2852–E2861. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhou, Y.; Pan, Z.; Shi, L.; Yang, J.; Liao, A.; Liao, Q.; Su, Q. Downregulation of LIMK1–ADF/Cofilin by DADS Inhibits the Migration and Invasion of Colon Cancer. Sci. Rep. 2017, 7, 45624. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Su, J.; Shi, L.; Liao, Q.; Su, Q. DADS Downregulates the Rac1-ROCK1/PAK1-LIMK1-ADF/Cofilin Signaling Pathway, Inhibiting Cell Migration and Invasion. Oncol. Rep. 2013, 29, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.-Y.; Sun, Y.; Lai, Z.-S.; Nan, Q.-Z.; Li, K.; Zhang, Z.-S. Inhibition of Migration and Invasion of Colorectal Cancer Cells via Deletion of Rac1 with RNA Interference. Mol. Cell. Biochem. 2009, 322, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Li, C.; Rajoka, M.S.R.; He, Z.; Huang, J.; Wang, J.; Zhang, J. P21-Activated Kinase 1: Emerging Biological Functions and Potential Therapeutic Targets in Cancer. Theranostics 2020, 10, 9741–9766. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Guo, H.; Lu, Y.; Feng, W.; Sun, X.; Tang, C.; Wang, X.; Shen, M. Blocking Hepatic Metastases of Colon Cancer Cells Using an ShRNA against Rac1 Delivered by Activatable Cell-Penetrating Peptide. Oncotarget 2016, 7, 77183–77195. [Google Scholar] [CrossRef][Green Version]
- Misra, A.; Pandey, C.; Sze, S.K.; Thanabalu, T. Hypoxia Activated EGFR Signaling Induces Epithelial to Mesenchymal Transition (EMT). PLoS ONE 2012, 7, e49766. [Google Scholar] [CrossRef]
- Tátrai, E.; Ranđelović, I.; Surguta, S.E.; Tóvári, J. Role of Hypoxia and Rac1 Inhibition in the Metastatic Cascade. Cancers 2024, 16, 1872. [Google Scholar] [CrossRef]
- Eiden, C.; Ungefroren, H. The Ratio of RAC1B to RAC1 Expression in Breast Cancer Cell Lines as a Determinant of Epithelial/Mesenchymal Differentiation and Migratory Potential. Cells 2021, 10, 351. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The Consensus Molecular Subtypes of Colorectal Cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Gupta, R.; Millstein, J.; Lin, K.; Haridas, V.; Zeineddine, M.A.; Parseghian, C.; Lenz, H.-J.; Kopetz, S.; Shen, J.P. Transcriptional Profiling and Consensus Molecular Subtype Assignment to Understand Response and Resistance to Anti–Epidermal Growth Factor Receptor Therapy in Colorectal Cancer. JCO Precis. Oncol. 2023, 7, e2200422. [Google Scholar] [CrossRef] [PubMed]
- Sadanandam, A.; Lyssiotis, C.A.; Homicsko, K.; Collisson, E.A.; Gibb, W.J.; Wullschleger, S.; Ostos, L.C.G.; Lannon, W.A.; Grotzinger, C.; Del Rio, M.; et al. A Colorectal Cancer Classification System That Associates Cellular Phenotype and Responses to Therapy. Nat. Med. 2013, 19, 619–625. [Google Scholar] [CrossRef]
- Linnekamp, J.F.; Van Hooff, S.R.; Prasetyanti, P.R.; Kandimalla, R.; Buikhuisen, J.Y.; Fessler, E.; Ramesh, P.; Lee, K.A.S.T.; Bochove, G.G.W.; De Jong, J.H.; et al. Consensus Molecular Subtypes of Colorectal Cancer Are Recapitulated in in Vitro and in Vivo Models. Cell Death Differ. 2018, 25, 616–633. [Google Scholar] [CrossRef] [PubMed]
- Woolston, A.; Khan, K.; Spain, G.; Barber, L.J.; Griffiths, B.; Gonzalez-Exposito, R.; Hornsteiner, L.; Punta, M.; Patil, Y.; Newey, A.; et al. Genomic and Transcriptomic Determinants of Therapy Resistance and Immune Landscape Evolution during Anti-EGFR Treatment in Colorectal Cancer. Cancer Cell 2019, 36, 35–50.e9. [Google Scholar] [CrossRef]
- Del Mar Maldonado, M.; Dharmawardhane, S. Targeting Rac and Cdc42 GTPases in Cancer. Cancer Res. 2018, 78, 3101–3111. [Google Scholar] [CrossRef]
- Gao, Y.; Dickerson, J.B.; Guo, F.; Zheng, J.; Zheng, Y. Rational Design and Characterization of a Rac GTPase-Specific Small Molecule Inhibitor. Proc. Natl. Acad. Sci. USA 2004, 101, 7618–7623. [Google Scholar] [CrossRef]
- Medina, J.I.; Cruz-Collazo, A.; Maldonado, M.D.M.; Gascot, T.M.; Borrero-Garcia, L.D.; Cooke, M.; Kazanietz, M.G.; O’Farril, E.H.; Vlaar, C.P.; Dharmawardhane, S. Characterization of Novel Derivatives of MBQ-167, an Inhibitor of the GTP-Binding Proteins Rac/Cdc42. Cancer Res. Commun. 2022, 2, 1711–1726. [Google Scholar] [CrossRef]
- Montalvo-Ortiz, B.L.; Castillo-Pichardo, L.; Hernández, E.; Humphries-Bickley, T.; De La Mota-Peynado, A.; Cubano, L.A.; Vlaar, C.P.; Dharmawardhane, S. Characterization of EHop-016, Novel Small Molecule Inhibitor of Rac GTPase. J. Biol. Chem. 2012, 287, 13238. [Google Scholar] [CrossRef]
- Humphries-Bickley, T.; Castillo-Pichardo, L.; Hernandez-O’Farrill, E.; Borrero-Garcia, L.D.; Forestier-Roman, I.; Gerena, Y.; Blanco, M.; Rivera-Robles, M.J.; Rodriguez-Medina, J.R.; Cubano, L.A.; et al. Characterization of a Dual Rac/Cdc42 Inhibitor MBQ-167 in Metastatic Cancer. Mol. Cancer Ther. 2017, 16, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Shutes, A.; Onesto, C.; Picard, V.; Leblond, B.; Schweighoffer, F.; Der, C.J. Specificity and Mechanism of Action of EHT 1864, a Novel Small Molecule Inhibitor of Rac Family Small GTPases. J. Biol. Chem. 2007, 282, 35666–35678. [Google Scholar] [CrossRef] [PubMed]
- Dütting, S.; Heidenreich, J.; Cherpokova, D.; Amin, E.; Zhang, S.-C.; Ahmadian, M.R.; Brakebusch, C.; Nieswandt, B. Critical Off-target Effects of the Widely Used Rac1 Inhibitors NSC23766 and EHT1864 in Mouse Platelets. J. Thromb. Haemost. 2015, 13, 827–838. [Google Scholar] [CrossRef] [PubMed]


| Feature | RAC1 | RAC1B | Refs. |
|---|---|---|---|
| Protein Length (amino acids) | 192 | 211 | [52,57,58] |
| Post-translational modifications | Ubiquitination: K147 and K166 Phosphorylation: S71, T108, and Y64 Geranylgeranylation: CAAX motif (AA 189–192) | Minimal ubiquitination Phosphorylation: unknown Geranylgeranylation: CAAX motif (AA 208–211) | [54,55,59,60] |
| GTPase Dynamics | GEF-dependent activation Regulated by RHO-GDIs Regulated by GAPs | GEF-independent activation Not regulated by RHO-GDIs Regulated by GAPs Impaired GTP hydrolysis Decreased GDP affinity Increased flexibility between switch I and switch II domains | [53,61,62,63,64] |
| Predominant State at Baseline | GDP-bound | GTP-bound | [62] |
| Protein Expression | Ubiquitous | Almost exclusively expressed in tumor tissue (breast, colon, lung) | [57,58] |
| Cellular Localization | Plasma Membrane Cytoplasm Nucleus | Plasma Membrane (predominantly) Nucleus | [53,56] |
| Splicing Factors that Favor Isoform Expression | SRSF3 hnRNP A1 | SRSF1 ESRP1 | [65,66,67,68,69,70] |
| Signaling Pathways Regulating Splicing Factor Expression and Activity | WNT/β-catenin/TCF4 | EGFR/PI3K/AKT MMP3 IL-6/STAT3 | [65,67,68,69] |
| Downstream Effector Proteins and Pathways | PAK1, WAVE, JNK, IQGAP1, RelB-NF-κB2/p100, β-catenin/TCF4, NOX1, EGFR and EGFR downstream signaling proteins | NOX1, SmgGDS, RACK1, p120(ctn), disheveled/ β-catenin, NF-κB, EGFR and EGFR downstream signaling proteins (MEK, ERK) | [45,46,47,50] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wahoski, C.C.; Singh, B. The Roles of RAC1 and RAC1B in Colorectal Cancer and Their Potential Contribution to Cetuximab Resistance. Cancers 2024, 16, 2472. https://doi.org/10.3390/cancers16132472
Wahoski CC, Singh B. The Roles of RAC1 and RAC1B in Colorectal Cancer and Their Potential Contribution to Cetuximab Resistance. Cancers. 2024; 16(13):2472. https://doi.org/10.3390/cancers16132472
Chicago/Turabian StyleWahoski, Claudia C., and Bhuminder Singh. 2024. "The Roles of RAC1 and RAC1B in Colorectal Cancer and Their Potential Contribution to Cetuximab Resistance" Cancers 16, no. 13: 2472. https://doi.org/10.3390/cancers16132472
APA StyleWahoski, C. C., & Singh, B. (2024). The Roles of RAC1 and RAC1B in Colorectal Cancer and Their Potential Contribution to Cetuximab Resistance. Cancers, 16(13), 2472. https://doi.org/10.3390/cancers16132472







