Decoding the Dynamics of Circulating Tumor DNA in Liquid Biopsies
Abstract
Simple Summary
Abstract
1. Introduction
2. Biology of ctDNA in Blood
2.1. Apoptosis: Orchestrated Cell-Free DNA Release
2.2. Necrosis: Chaotic but Informative DNA Release
2.3. Active Release: Microvesicles and Exosomes Facilitate Communication
2.4. Inflammation and Immune Responses: Triggers and Modulators of cfDNA Release
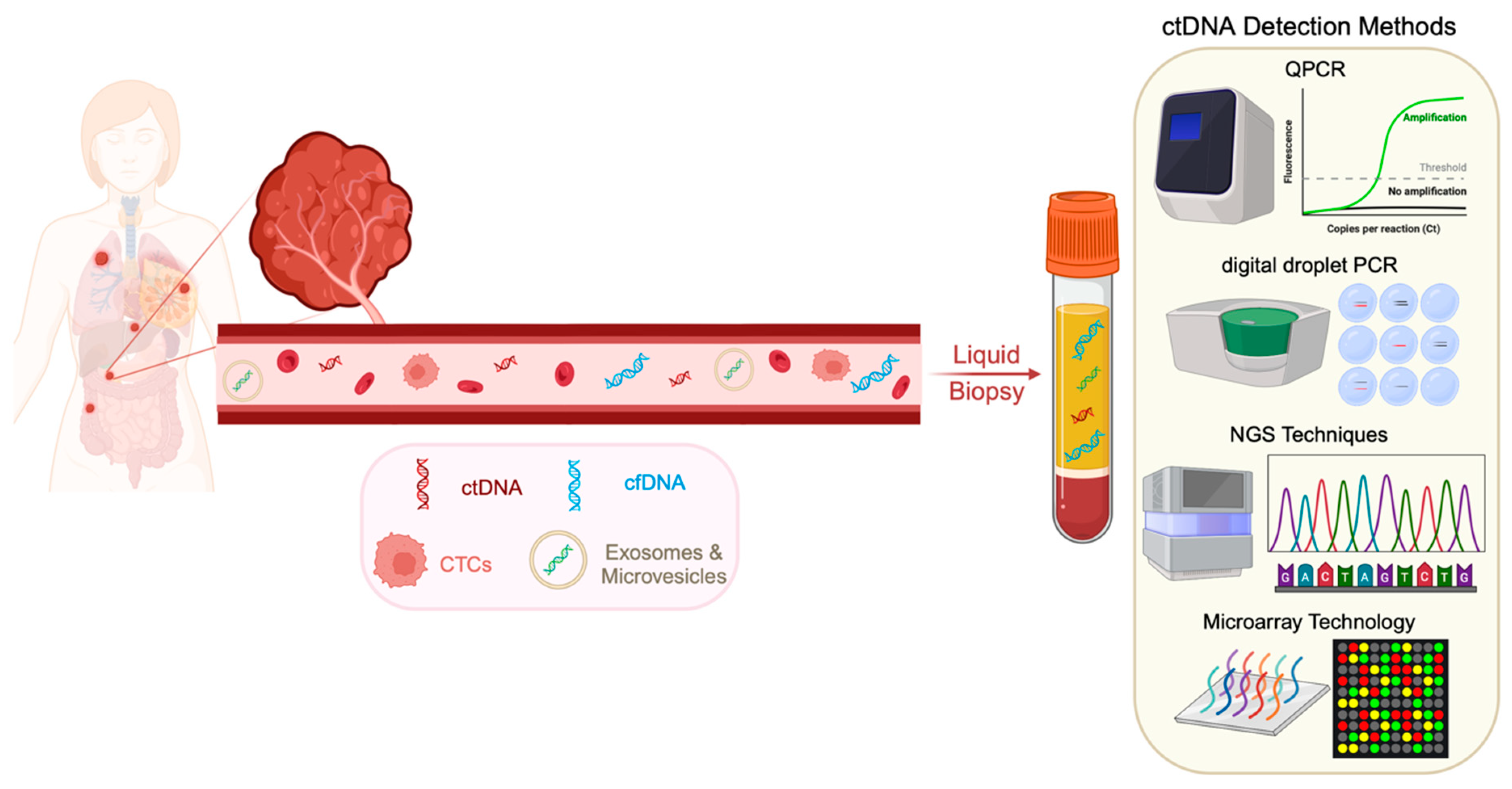
3. Methods of ctDNA Detection
Commercially Available Kits for ctDNA Detection and Analysis
4. Factors Influencing ctDNA Detection
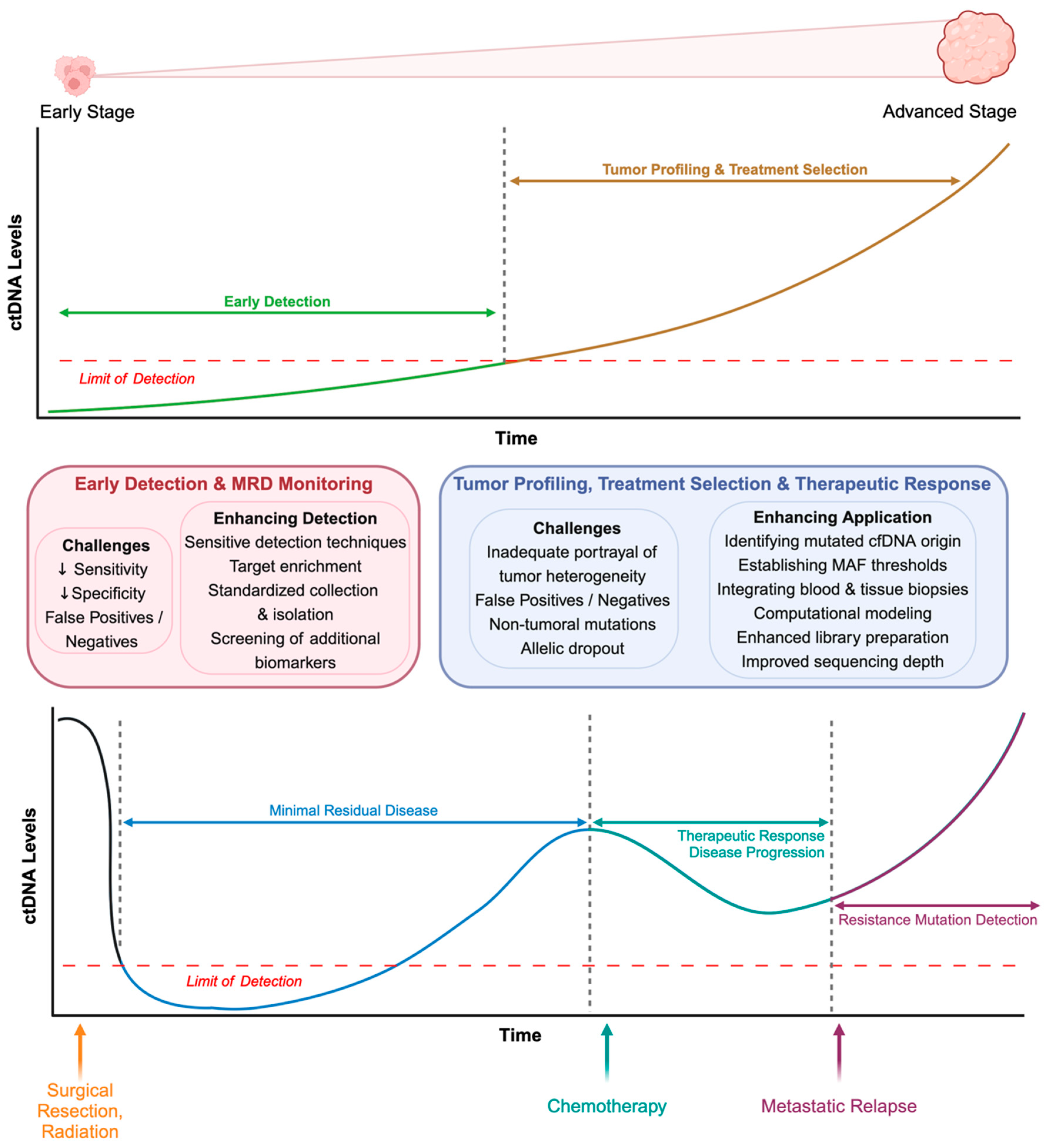
5. Clinical Applications of ctDNA Analysis in Liquid Biopsies
5.1. Early Cancer Detection
5.2. Treatment Selection and Personalized Medicine
5.3. Monitoring Minimal Residual Disease
5.4. Assessment of Therapeutic Response and Disease Progression
6. Overcoming Current Limitations with ctDNA Detection
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AS-PCR | Allele-specific PCR |
| BEAM | Beads, emulsion, amplification, and magnetics |
| CAPP-Seq | Cancer personalized profiling by deep sequencing |
| cfDNA | Cell-free DNA |
| CHIP | Clonal hematopoiesis of indeterminate potential |
| CNVs | Copy number variants |
| COLD-PCR | Co-amplification at lower denaturation temperature-PCR |
| CRC | Colorectal cancer |
| CTC | Circulating tumor cell |
| ctDNA | Circulating tumor DNA |
| ddPCR | Digital droplet PCR |
| dPCR | Digital PCR |
| HR+BC | Hormone receptor-positive breast cancer |
| Lung-CLip | Lung cancer likelihood in plasma |
| MRD | Minimal residual disease |
| NET | Neutrophil extracellular trap |
| NGS | Next generation sequencing |
| NSCLC | Non-small cell lung cancer |
| PDAC | Pancreatic ductal adenocarcinoma |
| Safe-SeqS | Safe-Sequencing System |
| SNVs | Single nucleotide variations |
| TAM-Seq | Tagged-Amplicon deep sequencing |
| TEC-Seq | Targeted error correction sequencing |
| TKIs | Tyrosine Kinase Inhibitors |
| WES | Whole exome sequencing |
| WGS | Whole genome sequencing |
References
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224. [Google Scholar] [CrossRef] [PubMed]
- Dang, D.K.; Park, B.H. Circulating tumor DNA: Current challenges for clinical utility. J. Clin. Investig. 2022, 132, e154941. [Google Scholar] [CrossRef]
- Pascual, J.; Attard, G.; Bidard, F.-C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef] [PubMed]
- Armakolas, A.; Kotsari, M.; Koskinas, J. Liquid Biopsies, Novel Approaches and Future Directions. Cancers 2023, 15, 1579. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, R.; Li, J.; Zhang, R. Size profile of cell-free DNA: A beacon guiding the practice and innovation of clinical testing. Theranostics 2020, 10, 4737–4748. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, H.; Long, Y.; Li, P.; Gu, Y. The main sources of circulating cell-free DNA: Apoptosis, necrosis and active secretion. Crit. Rev. Oncol. 2021, 157, 103166. [Google Scholar] [CrossRef] [PubMed]
- Pessoa, L.S.; Heringer, M.; Ferrer, V.P. ctDNA as a cancer biomarker: A broad overview. Crit. Rev. Oncol. 2020, 155, 103109. [Google Scholar] [CrossRef]
- Davidson, B.A.; Miranda, A.X.; Reed, S.C.; Bergman, R.E.; Kemp, J.D.J.; Reddy, A.P.; Pantone, M.V.; Fox, E.K.; Dorand, R.D.; Hurley, P.J.; et al. An in vitro CRISPR screen of cell-free DNA identifies apoptosis as the primary mediator of cell-free DNA release. Commun. Biol. 2024, 7, 441. [Google Scholar] [CrossRef]
- Gao, Q.; Zeng, Q.; Wang, Z.; Li, C.; Xu, Y.; Cui, P.; Zhu, X.; Lu, H.; Wang, G.; Cai, S.; et al. Circulating cell-free DNA for cancer early detection. Innov. Camb Mass 2022, 3, 100259. [Google Scholar] [CrossRef] [PubMed]
- Stejskal, P.; Goodarzi, H.; Srovnal, J.; Hajdúch, M.; van ’t Veer, L.J.; Magbanua, M.J.M. Circulating tumor nucleic acids: Biology, release mechanisms, and clinical relevance. Mol. Cancer 2023, 22, 15. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Pu, H.; Liu, Q.; Guo, Z.; Luo, D. Circulating Tumor DNA—A Novel Biomarker of Tumor Progression and Its Favorable Detection Techniques. Cancers 2022, 14, 6025. [Google Scholar] [CrossRef] [PubMed]
- Marques, J.F.; Junqueira-Neto, S.; Pinheiro, J.; Machado, J.C.; Costa, J.L. Induction of apoptosis increases sensitivity to detect cancer mutations in plasma. Eur. J. Cancer 2020, 127, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Rostami, A.; Lambie, M.; Yu, C.W.; Stambolic, V.; Waldron, J.N.; Bratman, S.V. Senescence, Necrosis, and Apoptosis Govern Circulating Cell-free DNA Release Kinetics. Cell Rep. 2020, 31, 107830. [Google Scholar] [CrossRef] [PubMed]
- Mair, R.; Mouliere, F.; Smith, C.; Chandrananda, S.; Gale, D.; Marass, F.; Tsui, D.W.; Massie, C.; Wright, A.; Watts, C.; et al. Measurement of Plasma Cell-Free Mitochondrial Tumor DNA Improves Detection of Glioblastoma in Patient-Derived Orthotopic Xenograft Models. Cancer Res. 2019, 79, 220–230. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, Y.; Mouliere, F. Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA. Cancer Cell 2019, 36, 350–368. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef]
- Lee, S.Y.; Ju, M.K.; Jeon, H.M.; Jeong, E.K.; Lee, Y.J.; Kim, C.H.; Park, H.G.; Han, S.I.; Kang, H.S. Regulation of Tumor Progression by Programmed Necrosis. Oxidative Med. Cell. Longev. 2018, 2018, 1–28. [Google Scholar] [CrossRef]
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, F.S.; Barauna, V.G.; dos Santos, L.; Costa, G.; Vassallo, P.F.; Campos, L.C.G. Properties and Application of Cell-Free DNA as a Clinical Biomarker. Int. J. Mol. Sci. 2021, 22, 9110. [Google Scholar] [CrossRef] [PubMed]
- Malkin, E.Z.; Bratman, S.V. Bioactive DNA from extracellular vesicles and particles. Cell Death Dis. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.; Enderle, D.; Noerholm, M.; Breakefield, X.; Skog, J. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. Off J. Eur. Soc. Med. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef]
- Clancy, J.W.; D’Souza-Schorey, C. Tumor-Derived Extracellular Vesicles: Multifunctional Entities in the Tumor Microenvironment. Annu. Rev. Pathol. 2023, 18, 205–229. [Google Scholar] [CrossRef]
- Liu, K.; Gao, X.; Kang, B.; Liu, Y.; Wang, D.; Wang, Y. The Role of Tumor Stem Cell Exosomes in Cancer Invasion and Metastasis. Front. Oncol. 2022, 12, 836548. [Google Scholar] [CrossRef]
- Mondelo-Macía, P.; Castro-Santos, P.; Castillo-García, A.; Muinelo-Romay, L.; Diaz-Peña, R. Circulating Free DNA and Its Emerging Role in Autoimmune Diseases. J. Pers. Med. 2021, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Pastor, B.; Abraham, J.-D.; Pisareva, E.; Sanchez, C.; Kudriavstev, A.; Tanos, R.; Mirandola, A.; Mihalovičová, L.; Pezzella, V.; Adenis, A.; et al. Association of neutrophil extracellular traps with the production of circulating DNA in patients with colorectal cancer. iScience 2022, 25, 103826. [Google Scholar] [CrossRef]
- Duvvuri, B.; Lood, C. Cell-Free DNA as a Biomarker in Autoimmune Rheumatic Diseases. Front. Immunol. 2019, 10, 502. [Google Scholar] [CrossRef] [PubMed]
- Sivapalan, L.; Murray, J.C.; Canzoniero, J.V.; Landon, B.; Jackson, J.; Scott, S.; Lam, V.; Levy, B.P.; Sausen, M.; Anagnostou, V. Liquid biopsy approaches to capture tumor evolution and clinical outcomes during cancer immunotherapy. J. Immunother. Cancer 2023, 11, e005924. [Google Scholar] [CrossRef]
- Saha, S.; Araf, Y.; Promon, S.K. Circulating tumor DNA in cancer diagnosis, monitoring, and prognosis. J. Egypt. Natl. Cancer Inst. 2022, 34, 8. [Google Scholar] [CrossRef] [PubMed]
- Tivey, A.; Church, M.; Rothwell, D.; Dive, C.; Cook, N. Circulating tumour DNA—Looking beyond the blood. Nat. Rev. Clin. Oncol. 2022, 19, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed]
- Dressman, D.; Yan, H.; Traverso, G.; Kinzler, K.W.; Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl. Acad. Sci. USA 2003, 100, 8817–8822. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [PubMed]
- Huerta, M.; Roselló, S.; Sabater, L.; Ferrer, A.; Tarazona, N.; Roda, D.; Gambardella, V.; Alfaro-Cervelló, C.; Garcés-Albir, M.; Cervantes, A.; et al. Circulating Tumor DNA Detection by Digital-Droplet PCR in Pancreatic Ductal Adenocarcinoma: A Systematic Review. Cancers 2021, 13, 994. [Google Scholar] [CrossRef] [PubMed]
- Palacín-Aliana, I.; García-Romero, N.; Asensi-Puig, A.; Carrión-Navarro, J.; González-Rumayor, V.; Ayuso-Sacido, V. Clinical Utility of Liquid Biopsy-Based Actionable Mutations Detected via ddPCR. Biomedicines 2021, 9, 906. [Google Scholar] [CrossRef]
- Kong, S.L.; Liu, X.; Tan, S.J.; Tai, J.A.; Phua, L.Y.; Poh, H.M.; Yeo, T.; Chua, Y.W.; Haw, Y.X.; Ling, W.H.; et al. Complementary Sequential Circulating Tumor Cell (CTC) and Cell-Free Tumor DNA (ctDNA) Profiling Reveals Metastatic Heterogeneity and Genomic Changes in Lung Cancer and Breast Cancer. Front. Oncol. 2021, 11, 698551. [Google Scholar] [CrossRef]
- Freidin, M.B.; Freydina, D.V.; Leung, M.; Fernandez, A.M.; Nicholson, A.G.; Lim, E. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin. Chem. 2015, 61, 1299–1304. [Google Scholar] [CrossRef]
- Luke, J.J.; Oxnard, G.R.; Paweletz, C.P.; Camidge, D.R.; Heymach, J.V.; Solit, D.B.; Johnson, B.E.; for the Cell Free DNA Working Group. Realizing the Potential of Plasma Genotyping in an Age of Genotype-Directed Therapies. JNCI J. Natl. Cancer Inst. 2014, 106, dju214. [Google Scholar] [CrossRef][Green Version]
- Eisen, M.B.; Brown, P.O. (12) DNA arrays for analysis of gene expression. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1999; pp. 179–205. [Google Scholar]
- Galbiati, S.; Damin, F.; Ferraro, L.; Soriani, N.; Burgio, V.; Ronzoni, M.; Gianni, L.; Ferrari, M.; Chiari, M. Microarray Approach Combined with ddPCR: An Useful Pipeline for the Detection and Quantification of Circulating Tumour dna Mutations. Cells 2019, 8, 769. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, K.U. Clinical Circulating Tumor DNA Testing for Precision Oncology. Cancer Res. Treat. 2023, 55, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef] [PubMed]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.Y.; Kaper, F.; Dawson, S.-J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci. Transl. Med. 2012, 4, 136. [Google Scholar] [CrossRef] [PubMed]
- Gale, D.; Heider, K.; Ruiz-Valdepenas, A.; Hackinger, S.; Perry, M.; Marsico, G.; Rundell, V.; Wulff, J.; Sharma, G.; Knock, H.; et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann. Oncol. 2022, 33, 500–510. [Google Scholar] [CrossRef]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. Off J. Eur. Soc. Med. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Merriman, B.; Ion Torrent R&D Team; Rothberg, J.M. Progress in Ion Torrent semiconductor chip based sequencing. Electrophoresis 2012, 33, 3397–3417. [Google Scholar] [CrossRef] [PubMed]
- Köhn, L.; Johansson, M.; Grankvist, K.; Nilsson, J. Liquid biopsies in lung cancer-time to implement research technologies in routine care? Ann. Transl. Med. 2017, 5, 278. [Google Scholar] [CrossRef]
- Guttery, D.S.; Page, K.; Hills, A.; Woodley, L.; Marchese, S.D.; Rghebi, B.; Hastings, R.K.; Luo, J.; Pringle, J.H.; Stebbing, J.; et al. Noninvasive Detection of Activating Estrogen Receptor 1 (ESR1) Mutations in Estrogen Receptor–Positive Metastatic Breast Cancer. Clin. Chem. 2015, 61, 974–982. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef]
- Telekes, A.; Horváth, A. The Role of Cell-Free DNA in Cancer Treatment Decision Making. Cancers 2022, 14, 6115. [Google Scholar] [CrossRef]
- Markou, D.; Londra, D.; Tserpeli, V.; Kollias, E.; Tsaroucha, E.; Vamvakaris, I.; Potaris, K.; Pateras, I.; Kotsakis, E.; Georgoulias, V.; et al. DNA methylation analysis of tumor suppressor genes in liquid biopsy components of early stage NSCLC: A promising tool for early detection. Clin. Epigenetics 2022, 14, 61. [Google Scholar] [CrossRef] [PubMed]
- Locke, W.J.; Guanzon, D.; Ma, C.; Liew, Y.J.; Duesing, K.R.; Fung, K.Y.; Ross, J.P. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 2019, 10, 1150. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Markou, A.; Tzanikou, E.; Lianidou, E. The potential of liquid biopsy in the management of cancer patients. Semin. Cancer Biol. 2022, 84, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Reuther, J.; Scozzaro, K.; Hawley, M.; Metzger, E.; Emery, M.; Chen, I.; Barbosa, M.; Johnson, L.; O’connor, A.; et al. Personalized Cancer Monitoring Assay for the Detection of ctDNA in Patients with Solid Tumors. Mol. Diagn. Ther. 2023, 27, 753–768. [Google Scholar] [CrossRef]
- Sethi, H.; Salari, R.; Navarro, S.; Natarajan, P.; Srinivasan, R.; Dashner, S.; Tin, T.; Balcioglu, M.; Swenerton, R.; Zimmermann, B. Abstract 4542: Analytical validation of the SignateraTM RUO assay, a highly sensitive patient-specific multiplex PCR NGS-based noninvasive cancer recurrence detection and therapy monitoring assay. Cancer Res. 2018, 78, 4542. [Google Scholar] [CrossRef]
- Flach, S.; Howarth, K.; Hackinger, S.; Pipinikas, C.; Ellis, P.; McLay, K.; Marsico, G.; Forshew, T.; Walz, C.; Reichel, C.A.; et al. Liquid BIOpsy for MiNimal RESidual DiSease Detection in Head and Neck Squamous Cell Carcinoma (LIONESS)—A personalised circulating tumour DNA analysis in head and neck squamous cell carcinoma. Br. J. Cancer 2022, 126, 1186–1195. [Google Scholar] [CrossRef]
- Fakih, M.; Sandhu, J.; Wang, C.; Kim, J.; Chen, Y.-J.; Lai, L.; Melstrom, K.; Kaiser, A. Evaluation of Comparative Surveillance Strategies of Circulating Tumor DNA, Imaging, and Carcinoembryonic Antigen Levels in Patients with Resected Colorectal Cancer. JAMA Netw. Open 2022, 5, e221093. [Google Scholar] [CrossRef]
- Tan, A.C.; Saw, S.P.; Lai, G.G.; Chua, K.L.; Takano, A.; Ong, B.-H.; Koh, T.P.; Jain, A.; Tan, W.L.; Ng, Q.S.; et al. Abstract 5114: Ultra-sensitive detection of minimal residual disease (MRD) through whole genome sequencing (WGS) using an AI-based error suppression model in resected early-stage non-small cell lung cancer (NSCLC). Cancer Res. 2022, 82 (Suppl. 12), 5114. [Google Scholar] [CrossRef]
- Bauml, J.M.; Li, B.T.; Velcheti, V.; Govindan, R.; Curioni-Fontecedro, A.; Dooms, C.; Takahashi, T.; Duda, A.W.; Odegaard, J.I.; Cruz-Guilloty, F.; et al. Clinical validation of Guardant360 CDx as a blood-based companion diagnostic for sotorasib. Lung Cancer 2022, 166, 270–278. [Google Scholar] [CrossRef]
- Kurtz, D.M.; Soo, J.; Keh, L.C.T.; Alig, S.; Chabon, J.J.; Sworder, B.J.; Schultz, A.; Jin, M.C.; Scherer, F.; Garofalo, A.; et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat. Biotechnol. 2021, 39, 1537–1547. [Google Scholar] [CrossRef]
- Choi, J.; Dannebaum, R.; Singh, A.; Foley, R.; Dinis, J.; Choi, C.; Min, B.; Li, J.; Feng, L.; Casey, F.; et al. Abstract 3648: Performance of the AVENIO ctDNA assays across multiple high-throughput next-generation sequencing platforms. Cancer Res. 2018, 78 (Suppl. 13), 3648. [Google Scholar] [CrossRef]
- So, M.-K.; Park, J.-H.; Kim, J.-W.; Jang, J.-H. Analytical Validation of a Pan-Cancer Panel for Cell-Free Assay for the Detection of EGFR Mutations. Diagnostics 2021, 11, 1022. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE 2020, 15, e0237802. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Woo, H.I.; Kim, J.-W.; Kim, Y.; Lee, K.-A. Clinical Practice Guidelines for Pre-Analytical Procedures of Plasma Epidermal Growth Factor Receptor Variant Testing. Ann. Lab. Med. 2022, 42, 141–149. [Google Scholar] [CrossRef]
- Diaz, E.H.; Yachnin, J.; Grönberg, H.; Lindberg, J. The In Vitro Stability of Circulating Tumour DNA. PLoS ONE 2016, 11, e0168153. [Google Scholar] [CrossRef]
- Andersson, D.; Kristiansson, H.; Kubista, M.; Ståhlberg, A. Ultrasensitive circulating tumor DNA analysis enables precision medicine: Experimental workflow considerations. Expert Rev. Mol. Diagn. 2021, 21, 299–310. [Google Scholar] [CrossRef]
- Lam, N.Y.; Rainer, T.H.; Chiu, R.W.; Lo, Y.D. EDTA Is a Better Anticoagulant than Heparin or Citrate for Delayed Blood Processing for Plasma DNA Analysis. Clin. Chem. 2004, 50, 256–257. [Google Scholar] [CrossRef]
- Kang, Q.; Henry, N.L.; Paoletti, C.; Jiang, H.; Vats, P.; Chinnaiyan, A.M.; Hayes, D.F.; Merajver, S.D.; Rae, J.M.; Tewari, M. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes. Clin. Biochem. 2016, 49, 1354–1360. [Google Scholar] [CrossRef] [PubMed]
- Danesi, R.; Lo, Y.; Oellerich, M.; Beck, J.; Galbiati, S.; Del Re, M.; Lianidou, E.; Neumaier, M.; van Schaik, R. What do we need to obtain high quality circulating tumor DNA (ctDNA) for routine diagnostic test in oncology?—Considerations on pre-analytical aspects by the IFCC workgroup cfDNA. Clin. Chim. Acta 2021, 520, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Diaz, I.M.; Nocon, A.; Held, S.A.E.; Kobilay, M.; Skowasch, D.; Bronkhorst, A.J.; Ungerer, V.; Fredebohm, J.; Diehl, F.; Holdenrieder, S.; et al. Pre-Analytical Evaluation of Streck Cell-Free DNA Blood Collection Tubes for Liquid Profiling in Oncology. Diagnostics 2023, 13, 1288. [Google Scholar] [CrossRef] [PubMed]
- Martin-Alonso, C.; Tabrizi, S.; Xiong, K.; Blewett, T.; Sridhar, S.; Crnjac, A.; Patel, S.; An, Z.; Bekdemir, A.; Shea, D.; et al. Priming agents transiently reduce the clearance of cell-free DNA to improve liquid biopsies. Science 2024, 383, eadf2341. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Wang, Y.; Li, L.; Yao, W.; Song, Y.; Liu, B.; Chen, W.; Santarpia, M.; Rossi, E.; et al. Increased detection of circulating tumor DNA by short fragment enrichment. Transl. Lung Cancer Res. 2021, 10, 1501–1511. [Google Scholar] [CrossRef] [PubMed]
- Hudecova, I.; Smith, C.G.; Hänsel-Hertsch, R.; Chilamakuri, C.S.; Morris, J.A.; Vijayaraghavan, A.; Heider, K.; Chandrananda, D.; Cooper, W.N.; Gale, D.; et al. Characteristics, origin, and potential for cancer diagnostics of ultrashort plasma cell-free DNA. Genome Res. 2022, 32, 215–227. [Google Scholar] [CrossRef]
- Wang, F.; Li, X.; Li, M.; Liu, W.; Lu, L.; Li, Y.; Chen, X.; Yang, S.; Liu, T.; Cheng, W.; et al. Ultra-short cell-free DNA fragments enhance cancer early detection in a multi-analyte blood test combining mutation, protein and fragmentomics. cclm 2023, 62, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Lira, M.; Huang, D.; Wang, K.; Valdez, C.; Kinong, J.; Rejto, P.A.; Bienkowska, J.; Hardwick, J.; Xie, T. TNER: A novel background error suppression method for mutation detection in circulating tumor DNA. BMC Bioinform. 2018, 19, 387. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef]
- Wan, J.C.; Heider, K.; Gale, D.; Murphy, S.; Fisher, E.; Mouliere, F.; Ruiz-Valdepenas, A.; Santonja, A.; Morris, J.; Chandrananda, D.; et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 2020, 12, eaaz8084. [Google Scholar] [CrossRef]
- Wu, H.-T.; Kalashnikova, E.; Mehta, S.; Salari, R.; Sethi, H.; Zimmermann, B.; Billings, P.R.; Aleshin, A. Characterization of clonal hematopoiesis of indeterminate potential mutations from germline whole exome sequencing data. J. Clin. Oncol. 2020, 38, 1525. [Google Scholar] [CrossRef]
- Kamps-Hughes, N.; McUsic, A.; Kurihara, L.; Harkins, T.T.; Pal, P.; Ray, C.; Ionescu-Zanetti, C. ERASE-Seq: Leveraging replicate measurements to enhance ultralow frequency variant detection in NGS data. PLoS ONE 2018, 13, e0195272. [Google Scholar] [CrossRef] [PubMed]
- Campos-Carrillo, A.; Weitzel, J.N.; Sahoo, P.; Rockne, R.; Mokhnatkin, J.V.; Murtaza, M.; Gray, S.W.; Goetz, L.; Goel, A.; Schork, N.; et al. Circulating tumor DNA as an early cancer detection tool. Pharmacol. Ther. 2020, 207, 107458. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, Z.; Wang, S.; Zhu, Y.; Ma, D.; Mu, Y.; Ying, J.; Li, J.; Xing, P. Disease monitoring of epidermal growth factor receptor (EGFR)-mutated non-small-cell lung cancer patients treated with tyrosine kinase inhibitors via EGFR status in circulating tumor DNA. Thorac. Cancer 2022, 13, 2201–2209. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Mei, J.; Kang, R.; Deng, S.; Chen, Y.; Yang, Y.; Feng, G.; Deng, Y.; Gan, F.; Lin, Y.; et al. Perioperative ctDNA-Based Molecular Residual Disease Detection for Non-Small Cell Lung Cancer: A Prospective Multicenter Cohort Study (LUNGCA-1). Clin. Cancer Res. Off J. Am. Assoc. Cancer Res. 2022, 28, 3308–3317. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, X.; Cao, Y.; Mao, Y.; Zhu, Y.; Zhang, Q.; Zhang, T.; Chang, L.; Wang, C. Dynamic analysis of predictive biomarkers for radiation therapy efficacy in non-small cell lung cancer patients by next-generation sequencing based on blood specimens. Pathol.—Res. Pract. 2024, 253, 154972. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.; Zhang, H.; Ren, F.; Liu, J.; Li, Y.; Dong, M.; Zhao, H.; Xu, S.; Liu, H.; et al. Apatinib added when NSCLC patients get slow progression with EGFR-TKI: A prospective, single-arm study. Cancer Med. 2023, 12, 21735–21741. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhang, J.-T.; Gao, X.; Chen, Z.-Y.; Yan, B.; Tan, P.-X.; Yang, X.-R.; Gao, W.; Gong, Y.; Tian, Z.; et al. Dynamic circulating tumor DNA during chemoradiotherapy predicts clinical outcomes for locally advanced non-small cell lung cancer patients. Cancer Cell 2023, 41, 1763–1773.e4. [Google Scholar] [CrossRef] [PubMed]
- Raez, L.E.; Brice, K.; Dumais, K.; Lopez-Cohen, A.; Wietecha, D.; Izquierdo, P.A.; Santos, E.S.; Powery, H.W. Liquid Biopsy Versus Tissue Biopsy to Determine Front Line Therapy in Metastatic Non-Small Cell Lung Cancer (NSCLC). Clin. Lung Cancer 2023, 24, 120–129. [Google Scholar] [CrossRef]
- Stensgaard, S.; Thomsen, A.; Helstrup, S.; Meldgaard, P.; Sorensen, B.S. Plasma Immune Proteins and Circulating Tumor DNA Predict the Clinical Outcome for Non-Small-Cell Lung Cancer Treated with an Immune Checkpoint Inhibitor. Cancers 2023, 15, 5628. [Google Scholar] [CrossRef]
- Yamaguchi, O.; Kasahara, N.; Soda, H.; Imai, H.; Naruse, I.; Yamaguchi, H.; Itai, M.; Taguchi, K.; Uchida, M.; Sunaga, N.; et al. Predictive significance of circulating tumor DNA against patients with T790M-positive EGFR-mutant NSCLC receiving osimertinib. Sci. Rep. 2023, 13, 20848. [Google Scholar] [CrossRef] [PubMed]
- Waldeck, S.; Mitschke, J.; Wiesemann, S.; Rassner, M.; Andrieux, G.; Deuter, M.; Mutter, J.; Lüchtenborg, A.; Kottmann, D.; Titze, L.; et al. Early assessment of circulating tumor DNA after curative-intent resection predicts tumor recurrence in early-stage and locally advanced non-small-cell lung cancer. Mol. Oncol. 2022, 16, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Moding, E.J.; Liu, Y.; Nabet, B.Y.; Chabon, J.J.; Chaudhuri, A.A.; Hui, A.B.; Bonilla, R.F.; Ko, R.B.; Yoo, C.H.; Gojenola, L.; et al. Circulating Tumor DNA Dynamics Predict Benefit from Consolidation Immunotherapy in Locally Advanced Non-Small-Cell Lung Cancer. Nat. Cancer 2020, 1, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Su, M.; Zhou, D.; Zheng, F.; Zhang, B.; Qiang, L.; Ren, G.; Song, L.; Bu, B.; Fang, S.; et al. Author response: Dynamic analysis of circulating tumor DNA to predict the prognosis and monitor the treatment response of patients with metastatic triple-negative breast cancer: A prospective study. eLife 2023, 12, e90198. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.; Su, F.; Joshi, M.; Masuda, N.; Ishikawa, T.; Aruga, T.; Zarate, J.P.; Babbar, N.; Balbin, O.A.; Yap, Y.S. Potential value of ctDNA monitoring in metastatic HR + /HER2 - breast cancer: Longitudinal ctDNA analysis in the phase Ib MONALEESASIA trial. BMC Med. 2023, 21, 306. [Google Scholar] [CrossRef] [PubMed]
- Gerratana, L.; Davis, A.A.; Velimirovic, M.; Clifton, K.; Hensing, W.L.; Shah, A.N.; Dai, C.S.; Reduzzi, C.; D’amico, P.; Wehbe, F.; et al. Interplay between ESR1/PIK3CA codon variants, oncogenic pathway alterations and clinical phenotype in patients with metastatic breast cancer (MBC): Comprehensive circulating tumor DNA (ctDNA) analysis. Breast Cancer Res. 2023, 25, 112. [Google Scholar] [CrossRef]
- Lee, K.; Lee, J.; Choi, J.; Sim, S.H.; Kim, J.E.; Kim, M.H.; Park, Y.H.; Kim, J.H.; Koh, S.-J.; Park, K.H.; et al. Genomic analysis of plasma circulating tumor DNA in patients with heavily pretreated HER2 + metastatic breast cancer. Sci. Rep. 2023, 13, 9928. [Google Scholar] [CrossRef]
- Parsons, H.; Blewett, T.; Chu, X.; Sridhar, S.; Santos, K.; Xiong, K.; Abramson, V.; Patel, A.; Cheng, J.; Brufsky, A.; et al. Circulating tumor DNA association with residual cancer burden after neoadjuvant chemotherapy in triple-negative breast cancer in TBCRC 030. Ann. Oncol. 2023, 34, 899–906. [Google Scholar] [CrossRef]
- Turner, N.C.; Swift, C.; Jenkins, B.; Kilburn, L.; Coakley, M.; Beaney, M.; Fox, L.; Goddard, K.; Garcia-Murillas, I.; Proszek, P.; et al. Results of the c-TRAK TN trial: A clinical trial utilising ctDNA mutation tracking to detect molecular residual disease and trigger intervention in patients with moderate- and high-risk early-stage triple-negative breast cancer. Ann. Oncol. 2023, 34, 200–211. [Google Scholar] [CrossRef]
- Magbanua, M.; Swigart, L.; Wu, H.-T.; Hirst, G.; Yau, C.; Wolf, D.; Tin, A.; Salari, R.; Shchegrova, S.; Pawar, H.; et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann. Oncol. 2021, 32, 229–239. [Google Scholar] [CrossRef]
- Radovich, M.; Jiang, G.; Hancock, B.A.; Chitambar, C.; Nanda, R.; Falkson, C.; Lynce, F.C.; Gallagher, C.; Isaacs, C.; Blaya, M.; et al. Association of Circulating Tumor DNA and Circulating Tumor Cells After Neoadjuvant Chemotherapy with Disease Recurrence in Patients with Triple-Negative Breast Cancer: Preplanned Secondary Analysis of the BRE12-158 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1410–1415. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.C.; Kingston, B.; Kilburn, L.S.; Kernaghan, S.; Wardley, A.M.; Macpherson, I.R.; Baird, R.; Roylance, R.; Stephens, P.; Oikonomidou, O.; et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): A multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020, 21, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- McDonald, B.R.; Contente-Cuomo, T.; Sammut, S.-J.; Odenheimer-Bergman, A.; Ernst, B.; Perdigones, N.; Chin, S.-F.; Farooq, M.; Mejia, R.; Cronin, P.A.; et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 2019, 11, eaax7392. [Google Scholar] [CrossRef] [PubMed]
- Evrard, C.; Ingrand, P.; Rochelle, T.; Martel, M.; Tachon, G.; Flores, N.; Randrian, V.; Ferru, A.; Haineaux, P.-A.; Goujon, J.-M.; et al. Circulating tumor DNA in unresectable pancreatic cancer is a strong predictor of first-line treatment efficacy: The KRASCIPANC prospective study. Dig. Liver Dis. 2023, 55, 1562–1572. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.-H.; Yoon, H.; Kim, K.-P.; Ryoo, B.-Y.; Lee, S.S.; Park, D.H.; Song, T.J.; Hwang, D.W.; Lee, J.H.; Song, K.B.; et al. Analysis of Plasma Circulating Tumor DNA in Borderline Resectable Pancreatic Cancer Treated with Neoadjuvant Modified FOLFIRINOX: Clinical Relevance of DNA Damage Repair Gene Alteration Detection. Cancer Res. Treat. 2023, 55, 1313–1320. [Google Scholar] [CrossRef] [PubMed]
- Kitahata, Y.; Kawai, M.; Hirono, S.; Okada, K.I.; Miyazawa, M.; Motobayashi, H.; Ueno, M.; Hayami, S.; Miyamoto, A.; Yamaue, H. Circulating Tumor DNA as a Potential Prognostic Marker in Patients with Borderline-Resectable Pancreatic Cancer Undergoing Neoadjuvant Chemotherapy Followed by Pancreatectomy. Ann. Surg. Oncol. 2022, 29, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Hata, T.; Mizuma, M.; Iseki, M.; Takadate, T.; Ishida, M.; Nakagawa, K.; Hayashi, H.; Morikawa, T.; Motoi, F.; Unno, M. Circulating tumor DNA as a predictive marker for occult metastases in pancreatic cancer patients with radiographically non-metastatic disease. J. Hepato-Biliary-Pancreat. Sci. 2021, 28, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Sugimori, M.; Sugimori, K.; Tsuchiya, H.; Suzuki, Y.; Tsuyuki, S.; Kaneta, Y.; Hirotani, A.; Sanga, K.; Tozuka, Y.; Komiyama, S.; et al. Quantitative monitoring of circulating tumor DNA in patients with advanced pancreatic cancer undergoing chemotherapy. Cancer Sci. 2020, 111, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Uesato, Y.; Sasahira, N.; Ozaka, M.; Sasaki, T.; Takatsuki, M.; Zembutsu, H. Evaluation of circulating tumor DNA as a biomarker in pancreatic cancer with liver metastasis. PLoS ONE 2020, 15, e0235623. [Google Scholar] [CrossRef]
- Patel, H.; Okamura, R.; Fanta, P.; Patel, C.; Lanman, R.B.; Raymond, V.M.; Kato, S.; Kurzrock, R. Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J. Hematol. Oncol. 2019, 12, 130. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Ding, X.-Q.; Zhu, H.; Wang, R.-X.; Pan, X.-R.; Tong, J.-H. KRAS Mutant Allele Fraction in Circulating Cell-Free DNA Correlates with Clinical Stage in Pancreatic Cancer Patients. Front. Oncol. 2019, 9, 1295. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, F.; Suzuki, K.; Tamaki, S.; Abe, I.; Endo, Y.; Takayama, Y.; Ishikawa, H.; Kakizawa, N.; Saito, M.; Futsuhara, K.; et al. Longitudinal monitoring of KRAS-mutated circulating tumor DNA enables the prediction of prognosis and therapeutic responses in patients with pancreatic cancer. PLoS ONE 2019, 14, e0227366. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, Q.; Li, X.; Su, W.; Li, G.; Ma, T.; Gao, S.; Lou, J.; Que, R.; Zheng, L.; et al. Monitoring Tumor Burden in Response to FOLFIRINOX Chemotherapy Via Profiling Circulating Cell-Free DNA in Pancreatic Cancer. Mol. Cancer Ther. 2019, 18, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Liu, C.; Jiang, J.; Luo, G.; Lu, Y.; Jin, K.; Guo, M.; Zhang, Z.; Xu, J.; Liu, L.; et al. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients: Clinical value of ctDNA in metastatic pancreatic cancer. Int. J. Cancer 2017, 140, 2344–2350. [Google Scholar] [CrossRef]
- Brenne, S.S.; Madsen, P.H.; Pedersen, I.S.; Hveem, K.; Skorpen, F.; Krarup, H.B.; Giskeødegård, G.F.; Laugsand, E.A. Colorectal cancer detected by liquid biopsy 2 years prior to clinical diagnosis in the HUNT study. Br. J. Cancer 2023, 129, 861–868. [Google Scholar] [CrossRef]
- Gouda, M.A.; Duose, D.Y.; Lapin, M.; Zalles, S.; Huang, H.J.; Xi, Y.; Zheng, X.; I Aldesoky, A.; Alhanafy, A.M.; A Shehata, M.; et al. Mutation-Agnostic Detection of Colorectal Cancer Using Liquid Biopsy-Based Methylation-Specific Signatures. Oncologist 2022, 28, 368–372. [Google Scholar] [CrossRef]
- Mo, S.; Ye, L.; Wang, D.; Han, L.; Zhou, S.; Wang, H.; Dai, W.; Wang, Y.; Luo, W.; Wang, R.; et al. Early Detection of Molecular Residual Disease and Risk Stratification for Stage I to III Colorectal Cancer via Circulating Tumor DNA Methylation. JAMA Oncol. 2023, 9, 770. [Google Scholar] [CrossRef] [PubMed]
- Szász, I.; Kiss, T.; Mokánszki, A.; Koroknai, V.; Deák, J.; Patel, V.; Jámbor, K.; Ádány, R.; Balázs, M. Identification of liquid biopsy-based mutations in colorectal cancer by targeted sequencing assays. Mol. Cell. Probes 2023, 67, 101888. [Google Scholar] [CrossRef]
- Huang, Y.-S.; Wu, H.-X.; Wang, Z.-X.; Jin, Y.; Yao, Y.-C.; Chen, Y.-X.; Zhao, Q.; Chen, S.; He, M.-M.; Luo, H.-Y.; et al. Genomic temporal heterogeneity of circulating tumour DNA in unresectable metastatic colorectal cancer under first-line treatment. Gut 2021, 71, 1340–1349. [Google Scholar] [CrossRef]
- Procaccio, L.; Bergamo, F.; Daniel, F.; Rasola, C.; Munari, G.; Biason, P.; Crucitta, S.; Barsotti, G.; Zanella, G.; Angerilli, V.; et al. A Real-World Application of Liquid Biopsy in Metastatic Colorectal Cancer: The Poseidon Study. Cancers 2022, 13, 5128. [Google Scholar] [CrossRef]
- Wang, D.; O’rourke, D.; Sanchez-Garcia, J.F.; Cai, T.; Scheuenpflug, J.; Feng, Z. Development of a liquid biopsy based purely quantitative digital droplet PCR assay for detection of MLH1 promoter methylation in colorectal cancer patients. BMC Cancer 2021, 21, 797. [Google Scholar] [CrossRef]
- Cremolini, C.; Rossini, D.; Dell’Aquila, E.; Lonardi, S.; Conca, E.; Del Re, M.; Busico, A.; Pietrantonio, F.; Danesi, R.; Aprile, G.; et al. Rechallenge for Patients with RAS and BRAF Wild-Type Metastatic Colorectal Cancer with Acquired Resistance to First-line Cetuximab and Irinotecan: A Phase 2 Single-Arm Clinical Trial. JAMA Oncol. 2019, 5, 343–350. [Google Scholar] [CrossRef]
- van ’t Erve, I.; Greuter, M.J.E.; Bolhuis, K.; Vessies, D.C.L.; Leal, A.; Vink, G.R.; van den Broek, D.; Velculescu, V.E.; Punt, C.J.A.; Meijer, G.A.; et al. Diagnostic Strategies toward Clinical Implementation of Liquid Biopsy RAS/BRAF Circulating Tumor DNA Analyses in Patients with Metastatic Colorectal Cancer. J. Mol. Diagn. 2020, 22, 1430–1437. [Google Scholar] [CrossRef] [PubMed]
- Junca, A.; Tachon, G.; Evrard, C.; Villalva, C.; Frouin, E.; Karayan-Tapon, L.; Tougeron, D. Detection of Colorectal Cancer and Advanced Adenoma by Liquid Biopsy (Decalib Study): The ddPCR Challenge. Cancers 2020, 12, 1482. [Google Scholar] [CrossRef] [PubMed]
- Seremet, T.; Jansen, Y.; Planken, S.; Njimi, H.; Delaunoy, M.; El Housni, H.; Awada, G.; Schwarze, J.K.; Keyaerts, M.; Everaert, H.; et al. Undetectable circulating tumor DNA (ctDNA) levels correlate with favorable outcome in metastatic melanoma patients treated with anti-PD1 therapy. J. Transl. Med. 2019, 17, 303. [Google Scholar] [CrossRef]
- Marsavela, G.; McEvoy, A.C.; Pereira, M.R.; Reid, A.L.; Al-Ogaili, Z.; Warburton, L.; Khattak, M.A.; Abed, A.; Meniawy, T.M.; Millward, M.; et al. Detection of clinical progression through plasma ctDNA in metastatic melanoma patients: A comparison to radiological progression. Br. J. Cancer 2021, 126, 401–408. [Google Scholar] [CrossRef]
- Tan, L.; Sandhu, S.; Lee, R.; Li, J.; Callahan, J.; Ftouni, S.; Dhomen, N.; Middlehurst, P.; Wallace, A.; Raleigh, J.; et al. Prediction and monitoring of relapse in stage III melanoma using circulating tumor DNA. Ann. Oncol. 2019, 30, 804–814. [Google Scholar] [CrossRef]
- Eroglu, Z.; Krinshpun, S.; Kalashnikova, E.; Sudhaman, S.; Topcu, T.O.; Nichols, M.; Martin, J.; Bui, K.M.; Palsuledesai, C.C.; Malhotra, M.; et al. Circulating tumor DNA-based molecular residual disease detection for treatment monitoring in advanced melanoma patients. Cancer 2023, 129, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Saw, R.; Thompson, J.; Lo, S.; Spillane, A.; Shannon, K.; Stretch, J.; Howle, J.; Menzies, A.; Carlino, M.; et al. Pre-operative ctDNA predicts survival in high-risk stage III cutaneous melanoma patients. Ann. Oncol. 2019, 30, 815–822. [Google Scholar] [CrossRef]
- Gouda, M.; Polivka, J.; Huang, H.; Treskova, I.; Pivovarcikova, K.; Fikrle, T.; Woznica, V.; Dustin, D.; Call, S.; Meric-Bernstam, F.; et al. Ultrasensitive detection of BRAF mutations in circulating tumor DNA of non-metastatic melanoma. ESMO Open 2021, 7, 100357. [Google Scholar] [CrossRef]
- Lee, J.H.; Menzies, A.M.; Carlino, M.S.; McEvoy, A.C.; Sandhu, S.; Weppler, A.M.; Diefenbach, R.J.; Dawson, S.J.; Kefford, R.F.; Millward, M.J.; et al. Longitudinal Monitoring of ctDNA in Patients with Melanoma and Brain Metastases Treated with Immune Checkpoint Inhibitors. Clin. Cancer Res. 2020, 26, 4064–4071. [Google Scholar] [CrossRef]
- Váraljai, R.; Wistuba-Hamprecht, K.; Seremet, T.; Diaz, J.M.S.; Nsengimana, J.; Sucker, A.; Griewank, K.; Placke, J.-M.; Horn, P.A.; von Neuhoff, N.; et al. Application of Circulating Cell-Free Tumor DNA Profiles for Therapeutic Monitoring and Outcome Prediction in Genetically Heterogeneous Metastatic Melanoma. JCO Precis. Oncol. 2019, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lau, E.; McCoy, P.; Reeves, F.; Chow, K.; Clarkson, M.; Kwan, E.M.; Packwood, K.; Northen, H.; He, M.; Kingsbury, Z.; et al. Detection of ctDNA in plasma of patients with clinically localised prostate cancer is associated with rapid disease progression. Genome Med. 2020, 12, 72. [Google Scholar] [CrossRef]
- Sonpavde, G.; Agarwal, N.; Pond, G.R.; Nagy, R.J.; Nussenzveig, R.H.; Hahn, A.W.; Sartor, O.; Gourdin, T.S.; Nandagopal, L.; Ledet, E.M.; et al. Circulating tumor DNA alterations in patients with metastatic castration-resistant prostate cancer. Cancer 2019, 125, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Tukachinsky, H.; Madison, R.W.; Chung, J.H.; Gjoerup, O.V.; Severson, E.A.; Dennis, L.; Fendler, B.J.; Morley, S.; Zhong, L.; Graf, R.P.; et al. Genomic Analysis of Circulating Tumor DNA in 3334 Patients with Advanced Prostate Cancer Identifies Targetable BRCA Alterations and AR Resistance Mechanisms. Clin. Cancer Res. 2021, 27, 3094–3105. [Google Scholar] [CrossRef] [PubMed]
- Chi, K.N.; Barnicle, A.; Sibilla, C.; Lai, Z.; Corcoran, C.; Barrett, J.C.; Adelman, C.A.; Qiu, P.; Easter, A.; Dearden, S.; et al. Detection of BRCA1, BRCA2, and ATM Alterations in Matched Tumor Tissue and Circulating Tumor DNA in Patients with Prostate Cancer Screened in PROfound. Clin. Cancer Res. 2023, 29, 81–91. [Google Scholar] [CrossRef]
- Kohli, M.; Tan, W.; Zheng, T.; Wang, A.; Montesinos, C.; Wong, C.; Du, P.; Jia, S.; Yadav, S.; Horvath, L.G.; et al. Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer. EBioMedicine 2020, 54, 102728. [Google Scholar] [CrossRef]
- Mizuno, K.; Sumiyoshi, T.; Okegawa, T.; Terada, N.; Ishitoya, S.; Miyazaki, Y.; Kojima, T.; Katayama, H.; Fujimoto, N.; Hatakeyama, S.; et al. Clinical Impact of Detecting Low-Frequency Variants in Cell-Free DNA on Treatment of Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2021, 27, 6164–6173. [Google Scholar] [CrossRef]
- Fonseca, N.M.; Maurice-Dror, C.; Herberts, C.; Tu, W.; Fan, W.; Murtha, A.J.; Kwan, E.M.; Parekh, K.; Schönlau, E.; Bernales, C.Q.; et al. Prediction of plasma ctDNA fraction and prognostic implications of liquid biopsy in advanced prostate cancer. Nat. Commun. 2024, 15, 1828. [Google Scholar] [CrossRef]
- Porter, A.; Natsuhara, M.; Daniels, G.A.; Patel, S.P.; Sacco, A.G.; Bykowski, J.; Banks, K.C.; Cohen, E.E.W. Next generation sequencing of cell free circulating tumor DNA in blood samples of recurrent and metastatic head and neck cancer patients. Transl. Cancer Res. 2020, 9, 203. [Google Scholar] [CrossRef]
- Hanna, G.J.; Dennis, M.J.; Scarfo, N.; Mullin, M.S.; Sethi, R.K.; Sehgal, K.; Annino, D.J.; Goguen, L.A.; Haddad, R.I.; Tishler, R.B.; et al. Personalized circulating tumor DNA for monitoring disease status in head and neck squamous cell carcinoma. Clin. Cancer Res. 2024. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.-H.; Cheng, H.-W.; Liu, C.-J. Droplet digital polymerase chain reaction for detection and quantification of cell-free DNA TP53 target somatic mutations in oral cancer. Cancer Biomark. 2022, 33, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Economopoulou, P.; Spathis, A.; Kotsantis, I.; Maratou, E.; Anastasiou, M.; Moutafi, M.K.; Kirkasiadou, M.; Pantazopoulos, A.; Giannakakou, M.; Edelstein, D.L.; et al. Next-generation sequencing (NGS) profiling of matched tumor and circulating tumor DNA (ctDNA) in head and neck squamous cell carcinoma (HNSCC). Oral Oncol. 2023, 139, 106358. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.C.; Nguyen, T.H.; Doan, N.N.T.; Pham, T.M.Q.; Nguyen, G.T.H.; Nguyen, T.D.; Tran, T.T.T.; Vo, D.L.; Phan, T.H.; Jasmine, T.X.; et al. Multimodal analysis of methylomics and fragmentomics in plasma cell-free DNA for multi-cancer early detection and localization. eLife 2023, 12, RP89083. [Google Scholar] [CrossRef] [PubMed]
- Chabon, J.J.; Hamilton, E.G.; Kurtz, D.M.; Esfahani, M.S.; Moding, E.J.; Stehr, H.; Schroers-Martin, J.; Nabet, B.Y.; Chen, B.; Chaudhuri, A.A.; et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020, 580, 245–251. [Google Scholar] [CrossRef]
- Vitiello, P.P.; De Falco, V.; Giunta, E.F.; Ciardiello, D.; Cardone, C.; Vitale, P.; Zanaletti, N.; Borrelli, C.; Poliero, L.; Terminiello, M.; et al. Clinical Practice Use of Liquid Biopsy to Identify RAS/BRAF Mutations in Patients with Metastatic Colorectal Cancer (mCRC): A Single Institution Experience. Cancers 2019, 11, 1504. [Google Scholar] [CrossRef] [PubMed]
- Woolston, A.; Barber, L.J.; Griffiths, B.; Pich, O.; Lopez-Bigas, N.; Matthews, N.; Rao, S.; Watkins, D.; Chau, I.; Starling, N.; et al. Mutational signatures impact the evolution of anti-EGFR antibody resistance in colorectal cancer. Nat. Ecol. Evol. 2021, 5, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, B.; Liu, Z.; Gong, J.; Shao, L.; Ren, J.; Niu, Y.; Bo, S.; Li, Z.; Lai, Y.; et al. HER2 copy number of circulating tumour DNA functions as a biomarker to predict and monitor trastuzumab efficacy in advanced gastric cancer. Eur. J. Cancer 2018, 88, 92–100. [Google Scholar] [CrossRef]
- Wehrle, J.; Philipp, U.; Jolic, M.; Follo, M.; Hussung, S.; Waldeck, S.; Deuter, M.; Rassner, M.; Braune, J.; Rawluk, J.; et al. Personalized Treatment Selection and Disease Monitoring Using Circulating Tumor DNA Profiling in Real-World Cancer Patient Management. Diagnostics 2020, 10, 550. [Google Scholar] [CrossRef]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Kurata, T.; Chiappori, A.; Lee, K.H.; De Wit, M.; et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N. Engl. J. Med. 2018, 379, 2342–2350. [Google Scholar] [CrossRef]
- Sargent, D.; Sobrero, A.; Grothey, A.; O’Connell, M.J.; Buyse, M.; Andre, T.; Zheng, Y.; Green, E.; Labianca, R.; O‘Callaghan, C.; et al. Evidence for cure by adjuvant therapy in colon cancer: Observations based on individual patient data from 20,898 patients on 18 randomized trials. J. Clin. Oncol. Oncol Off J. Am. Soc. Clin. 2009, 27, 872–877. [Google Scholar] [CrossRef]
- Lee, J.-S.; Han, Y.; Yun, W.-G.; Kwon, W.; Kim, H.; Jeong, H.; Seo, M.-S.; Park, Y.; Cho, S.I.; Kim, H.; et al. Parallel Analysis of Pre- and Postoperative Circulating Tumor DNA and Matched Tumor Tissues in Resectable Pancreatic Ductal Adenocarcinoma: A Prospective Cohort Study. Clin. Chem. 2022, 68, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Pellini, B.; Pejovic, N.; Feng, W.; Earland, N.; Harris, P.K.; Usmani, A.; Szymanski, J.J.; Qaium, F.; Mudd, J.; Petty, M.; et al. ctDNA MRD Detection and Personalized Oncogenomic Analysis in Oligometastatic Colorectal Cancer From Plasma and Urine. JCO Precis. Oncol. 2021, 5, 378–388. [Google Scholar] [CrossRef]
- Lipsyc-Sharf, M.; de Bruin, E.C.; Santos, K.; McEwen, R.; Stetson, D.; Patel, A.; Kirkner, G.J.; Hughes, M.E.; Tolaney, S.M.; Partridge, A.H.; et al. Circulating Tumor DNA and Late Recurrence in High-Risk Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Breast Cancer. J. Clin. Oncol. Oncol Off J. Am. Soc. Clin. 2022, 40, 2408. [Google Scholar] [CrossRef] [PubMed]
- Jacob, S.; Davis, A.A.; Gerratana, L.; Velimirovic, M.; Shah, A.N.; Wehbe, F.; Katam, N.; Zhang, Q.; Flaum, L.; Siziopikou, K.P.; et al. The Use of Serial Circulating Tumor DNA to Detect Resistance Alterations in Progressive Metastatic Breast Cancer. Clin Cancer Res. 2021, 27, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Guibert, N.; Jones, G.; Beeler, J.F.; Plagnol, V.; Morris, C.; Mourlanette, J.; Delaunay, M.; Keller, L.; Rouquette, I.; Favre, G.; et al. Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer 2019, 137, 154. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, D.-L.; Yang, C.-P.; Chen, X.-X.; You, J.-Q.; Huang, J.-S.; Shao, Y.; Zhu, D.-Q.; Ouyang, Y.-M.; Luo, H.-Y.; et al. The predicting role of circulating tumor DNA landscape in gastric cancer patients treated with immune checkpoint inhibitors. Mol. Cancer 2020, 19, 1–6. [Google Scholar] [CrossRef]
- Ko, J.; Baldassano, S.N.; Loh, P.-L.; Kording, K.; Litt, B.; Issadore, D. Machine learning to detect signatures of disease in liquid biopsies—A user’s guide. Lab A Chip 2017, 18, 395–405. [Google Scholar] [CrossRef]
- Gillis, N.K.; Ball, M.; Zhang, Q.; Ma, Z.; Zhao, Y.; Yoder, S.J.; Balasis, M.E.; Mesa, T.E.; Sallman, D.A.; Lancet, J.E.; et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: A proof-of-concept, case-control study. Lancet Oncol. 2018, 18, 112–121. [Google Scholar] [CrossRef]
- Li, X.; Liu, T.; Bacchiocchi, A.; Li, M.; Cheng, W.; Wittkop, T.; Mendez, F.; Wang, Y.; Tang, P.; Yao, Q.; et al. Ultra-sensitive molecular residual disease detection through whole genome sequencing with single-read error correction. medRxiv 2024. [Google Scholar] [CrossRef]
- Hsiehchen, D.; Bucheit, L.; Yang, D.; Beg, M.S.; Lim, M.; Lee, S.S.; Kasi, P.M.; Kaseb, A.O.; Zhu, H. Genetic features and therapeutic relevance of emergent circulating tumor DNA alterations in refractory non-colorectal gastrointestinal cancers. Nat. Commun. 2022, 13, 7477. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Brasó-Maristany, F.; Martínez-Sáez, O.; Sanfeliu, E.; Xia, Y.; Bellet, M.; Galván, P.; Martínez, D.; Pascual, T.; Marín-Aguilera, M.; et al. Circulating tumor DNA reveals complex biological features with clinical relevance in metastatic breast cancer. Nat. Commun. 2023, 14, 1157. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.; Siu, L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lim, Y.; Kang, J.-K.; Kim, H.-P.; Roh, H.; Kim, S.Y.; Lee, D.; Bang, D.; Jeong, S.-Y.; Park, K.J.; et al. Dynamic changes in longitudinal circulating tumour DNA profile during metastatic colorectal cancer treatment. Br. J. Cancer 2022, 127, 898–907. [Google Scholar] [CrossRef]
- Haselmann, V.; Hedtke, M.; Neumaier, M. Liquid Profiling for Cancer Patient Stratification in Precision Medicine—Current Status and Challenges for Successful Implementation in Standard Care. Diagnostics 2022, 12, 748. [Google Scholar] [CrossRef]


| Digital PCR-Based ctDNA Detection | NGS-Based ctDNA Detection | |
|---|---|---|
| Advantages | Increased sensitivity (0.1–0.001%) | Enables comprehensive analysis—multiple targets, whole exome, and whole genome. |
| Provides absolute quantification of mutation load | Provides an unbiased discovery approach. | |
| Disadvantages | Targeted analysis—detection of known mutants only | Expensive with increased processing times and advanced analysis and data interpretation techniques. |
| Higher variant allele frequencies required for detection. |
| Name | Technology | Application | Sensitivity | Specificity | Current Clinical Trial ID(s) (Type; Cancer) | Reference |
|---|---|---|---|---|---|---|
| Personalized Cancer Monitoring (PCM™) | Anchored Multiplex PCR (AMP) (for target enrichment) for NGS | MRD | 99.8% | 99.9% | NCT05219734 (Observational; Pan-cancer) | [58] |
| TruSight Oncology 500 | Targeted NGS | Cancer recurrence detection, Tumor Profiling | >75% | 99.9% | NCT05763472 (Observational; Ovarian Cancer) NCT05111067 (Observational; TNBC) | [59] |
| RaDaR™ (Residual Disease and Recurrence) | Multiplex PCR-based NGS assay | MRD, Early detection of relapse | 95% | 100% | NCT05388149 (Phase 2; Breast Cancer | [60] |
| Signatera™ | Multiplex-based assay | MRD, Cancer recurrence detection, Therapy Monitoring | >65% | 99.9% | NCT04761783 (Observational; Melanoma, NSCLC, CRC) NCT05212779 (Observational; Epithelial Ovarian Cancer) NCT05757843 (Interventional; NSCLC) NCT04786600 (Interventional; CRC) NCT05174169 (Interventional; Colon Cancer) | [61] |
| MRDetect | WGS | MRD | 82–86% | 82–93% | - | [62] |
| Guardant360® CDx | NGS | Therapy Monitoring | 55.6% | 100% | NCT05935384 (Observational; NSCLC, CRC, Breast Cancer) | [63] |
| PhasED-seq (Phased Variant Enrichment and Detection Sequencing) | Hybrid capture-based NGS assays | MRD | 95% | 97% | NCT04417803 (Interventional; Lymphoma | [64] |
| AVENIO ctDNA Surveillance Kit | CAPP-seq | MRD, Monitoring tumor burden, Therapy Monitoring | 95–94% | 100% | NCT04585477 (Phase 2; NSCLC) NCT04585490 (Phase 3; NSLC) | [65] |
| Oncomine Pan-cancer cell-free assay | NGS | Mutation Detection | 90% | >98% | NCT04564079 (Observational; NSCLC) | [66] |
| FoundationOne® Liquid CDx | High throughput hybridization-based capture technology | Mutation Detection | 96.3% (PPA) * | 99.9% (NPA) ** | NCT05272423 (KRAS-driven cancers; Observational) NCT05032092 (Locally Advanced/Metastatic Cancers; Interventional) NCT05846594 (Lung & Gastrointestinal Cancer; Interventional) NCT04484636 (Multiple Cancers; Interventional) | [67] |
| Current Challenge | Technology | Application | Reference |
|---|---|---|---|
| Instability of ctDNA and cfDNA | Liposomal nanoparticle priming agents | Inhibits the uptake and degradation of cfDNA (including ctDNA) by liver macrophages and nucleases. | [75] |
| Background Noise (limiting analytical sensitivity) | Magnetic bead-based isolation, ssDNA library preparation | Enriches shorter ctDNA fragments to enable sensitive detection in low variant allele frequency samples. | [76,77,78] |
| Tri-nucleotide Error Reducer (TNER) | Background polishing algorithm that detects and eliminates background mutation errors from healthy subjects and sequencing artifacts from liquid biopsy data. | [79] | |
| Integrated Digital Error Suppression (iDES) | In silico background polishing to reduce common background artifacts and recover cfDNA molecules by molecular barcoding. | [80] | |
| INtegration of VAriant Reads (INVAR) | Molecular barcoding and locus-specific background polishing and detection. | [81] | |
| False Positives (non-tumor-derived genetic alterations) | Signatera™ Assay | Filters false positives due to clonal hematopoiesis of indeterminate potential (CHIP). | [82] |
| Elimination of Recurrent Artifacts and Stochastic Errors Sequencing (ERASE-seq) | Reduces false positives (10–100 fold) using deconvolution, iterative sequencing of background/negative DNA controls, and technical replicate analysis. | [83] |
| Cancer Type | Application | Technology | Total Patients | Reference |
|---|---|---|---|---|
| Lung Cancer | Therapy Response | NGS | 13 | [87] |
| Therapy Response | NGS | 12 | [88] | |
| MRD, Therapy Response | Targeted NGS | 139 (97.8% sensitivity) | [89] | |
| Therapy Selection | Guardant360™ NGS platform | 170 | [90] | |
| Therapy Response | Targeted NGS | 42 | [91] | |
| MRD, Therapy Response | dPCR | 40 | [92] | |
| MRD, Recurrence Monitoring | Multiplex PCR, NGS (RaDaR™ Assay) | 88 (86.7% sensitivity; 98.5% specificity) | [46] | |
| Prognosis | Real Time-Methylation-Specific PCR | 42 | [54] | |
| MRD | Targeted NGS | 33 (57% sensitivity) | [93] | |
| MRD, Recurrence Monitoring, Treatment Selection | NGS | 330 | [86] | |
| MRD, Therapy Response | CAPP-seq | 65 | [94] | |
| Breast Cancer | Prognosis, Therapy Response | Targeted capture-based NGS | 70 | [95] |
| Therapy Response | Targeted NGS, SNV detection (Mutect) | 88 | [96] | |
| Prognosis | Guardant360™ NGS platform | 703 | [97] | |
| Prognosis, Treatment Selection | Hybridization capture & targeted deep sequencing | 93 | [98] | |
| MRD, Recurrence Monitoring, Treatment Selection | WGS, Hybrid-capture duplex MRD Test | 139 | [99] | |
| Therapy Response, MRD, Metastasis Detection | dPCR | 208 (99.8% sensitivity) | [100] | |
| Therapy Response, MRD, Metastasis Marker | WES, multiplex PCR, NGS | 291 | [101] | |
| MRD, Recurrence Monitoring | Hybrid capture-based NGS | 142 | [102] | |
| Treatment Selection | ddPCR, Guardant360™ NGS platform | 1034 (93% sensitivity; 96–99% specificity) | [103] | |
| Therapy Response, MRD | TARDIS (Tumor-specific Analysis of Residual Disease in Solid Tumors) | 33 (19.6–94.6% sensitivity; 100% specificity) | [104] | |
| Pancreatic Cancer | Therapy Response, Treatment Selection | ddPCR | 69 | [105] |
| Prognosis | Guardant360™ NGS platform | 44 | [106] | |
| Prognosis | ddPCR | 55 | [107] | |
| Metastasis Marker | ddPCR | 172 | [108] | |
| Therapy Response | dPCR | 47 | [109] | |
| Therapy Response | Oncomine Colon cfDNA Assay targeted NGS | 106 | [110] | |
| Prognosis | NGS | 112 | [111] | |
| Mutation Detection | ddPCR | 162 | [112] | |
| Prognosis, Therapy Response | ddPCR | 67 (0.01–0.1% sensitivity) | [113] | |
| Therapy Response | Targeted NGS | 38 | [114] | |
| Prognosis, Therapy Response | NGS, ddPCR | 188 | [115] | |
| Colorectal Cancer | Cancer Detection | Methylation-specific PCR | 212 (43.1% sensitivity; >85.9% specificity) | [116] |
| Cancer Detection | Targeted Methylation Assay by NGS | 20 (85% sensitivity; 92% specificity) | [117] | |
| MRD | Multiplex QPCR | 299 (78% sensitivity; 90.2% specificity) | [118] | |
| Mutation Profiling | Targeted NGS | 23 | [119] | |
| Therapy Monitoring | WES, Targeted NGS | 171 | [120] | |
| Therapy Selection | ddPCR | 33 (80% sensitivity; 90% specificity) | [121] | |
| Prognosis | ddPCR | 48 (93% sensitivity; 95% specificity) | [122] | |
| Therapy Response | ddPCR, NGS | 28 | [123] | |
| Therapy Selection | ddPCR | 100 | [124] | |
| Cancer Detection, Mutation Detection | ddPCR | 155 (45% sensitivity; 100% specificity) | [125] | |
| Skin Cancer | Therapy Response | AS-PCR, RT-PCR, ddPCR | 85 | [126] |
| Disease Progression | ddPCR | 93 | [127] | |
| Recurrence Monitoring | ddPCR | 133 | [128] | |
| MRD | Signatera™ | 69 | [129] | |
| Prognosis | ddPCR | 174 | [130] | |
| Prognosis | ddPCR | 80 | [131] | |
| Therapy Response | ddPCR | 72 | [132] | |
| Therapy Response | ddPCR | 96 | [133] | |
| Prostate Cancer | Disease Progression | WGS; TAM-Seq | 10; 189 | [134] |
| Mutation Profiling, Therapy Response | Guardant360™ NGS platform | 514 | [135] | |
| Mutation Profiling | Hybrid capture–based comprehensive genomic profiling | 3334 | [136] | |
| Mutation Detection | FoundationOne® NGS | 619 | [137] | |
| Mutation Detection, Mutation Profiling | NGS | 279 | [138] | |
| Mutation Profiling | Targeted NGS sequencing | 100 | [139] | |
| Prognosis | Targeted NGS sequencing | 491 | [140] | |
| Head & Neck Cancer | Mutation Profiling | Guardant360™ NGS platform | 60 | [141] |
| Disease Progression | Signatera™ | 116 | [142] | |
| Mutation Detection | ddPCR | 107 | [143] | |
| Mutation Detection | Safe-Seqs | 62 | [144] | |
| Breast, Liver, Lung, Colorectal & Gastric Cancer | Early Detection, Localization | SPOT-MAS (tumor methylation screening), NGS | 738 (73.9–88.3% sensitivity; 97% specificity) | [145] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turabi, K.; Klute, K.; Radhakrishnan, P. Decoding the Dynamics of Circulating Tumor DNA in Liquid Biopsies. Cancers 2024, 16, 2432. https://doi.org/10.3390/cancers16132432
Turabi K, Klute K, Radhakrishnan P. Decoding the Dynamics of Circulating Tumor DNA in Liquid Biopsies. Cancers. 2024; 16(13):2432. https://doi.org/10.3390/cancers16132432
Chicago/Turabian StyleTurabi, Khadija, Kelsey Klute, and Prakash Radhakrishnan. 2024. "Decoding the Dynamics of Circulating Tumor DNA in Liquid Biopsies" Cancers 16, no. 13: 2432. https://doi.org/10.3390/cancers16132432
APA StyleTurabi, K., Klute, K., & Radhakrishnan, P. (2024). Decoding the Dynamics of Circulating Tumor DNA in Liquid Biopsies. Cancers, 16(13), 2432. https://doi.org/10.3390/cancers16132432







