Automated Deep Learning-Based Classification of Wilms Tumor Histopathology
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Image Data Sets
2.3. CNN Development and Design
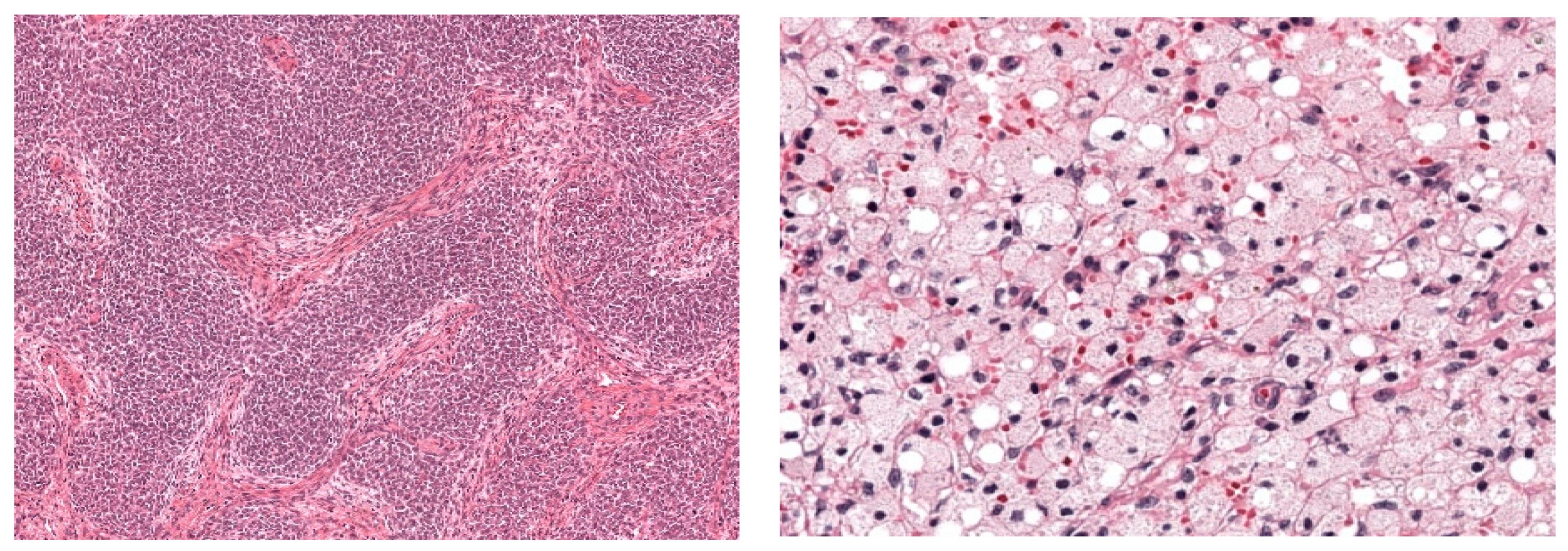
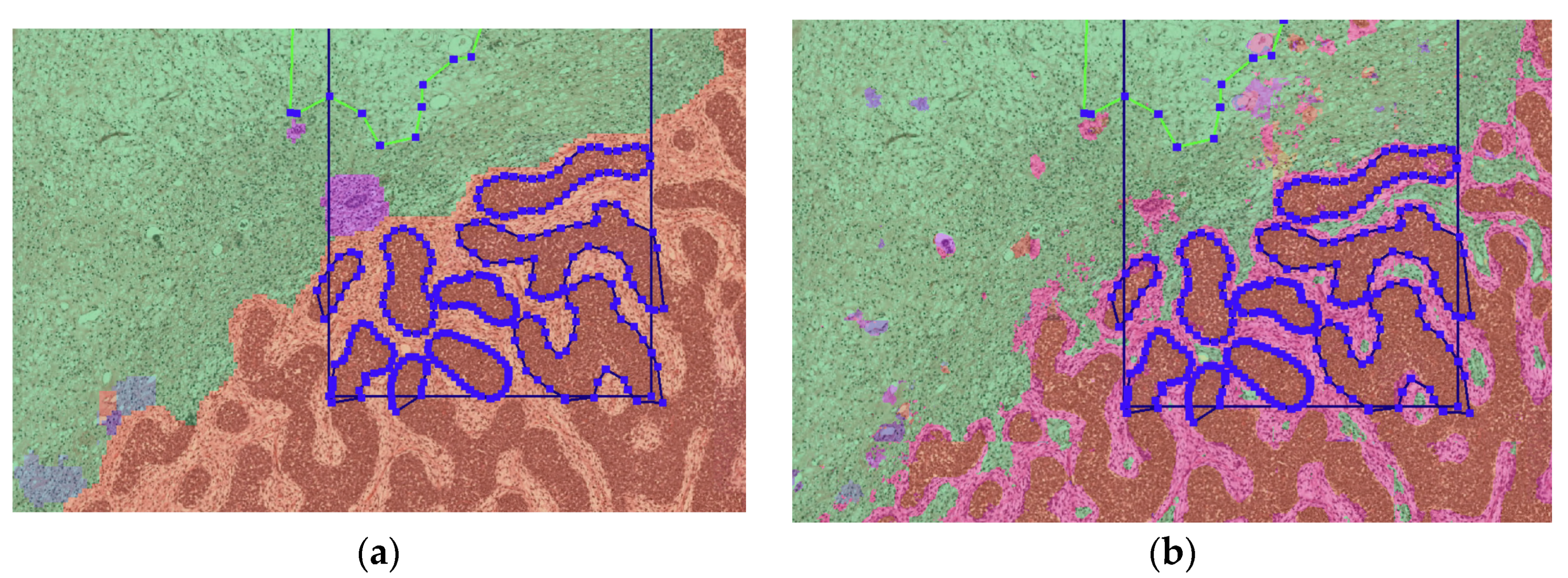
2.4. Histopathological Classification
2.5. Statistical Analysis
3. Results
3.1. Study Population
3.2. Algorithm Output
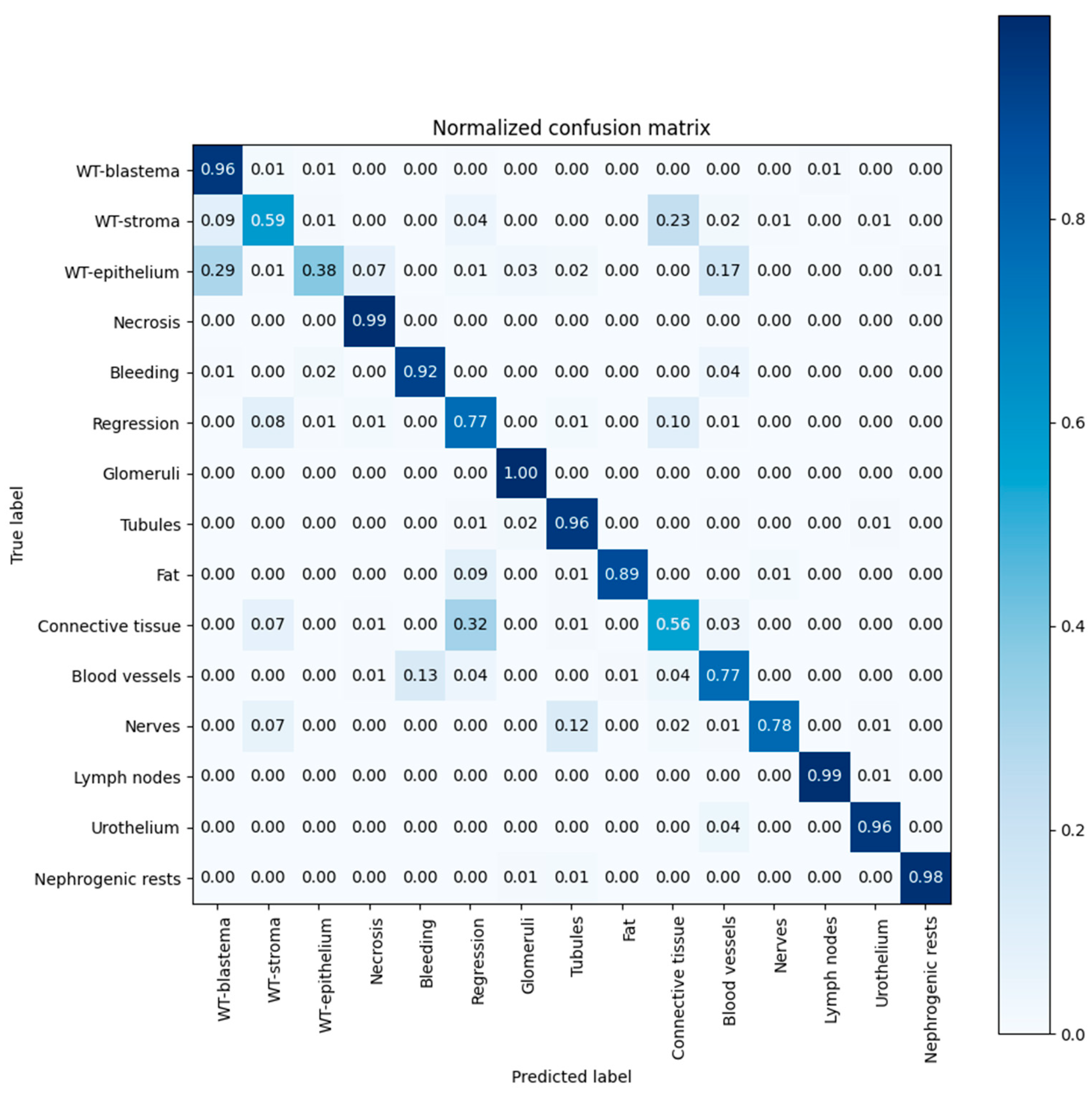
3.3. Algorithm Performance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Szychot, E.; Apps, J.; Pritchard-Jones, K. Wilms’ tumor: Biology, diagnosis and treatment. Transl. Pediatr. 2014, 3, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, P.F. Wilms tumor: Progress to date and future considerations. Expert Rev. Anticancer Ther. 2001, 1, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Malogolowkin, M.; Cotton, C.A.; Green, D.M.; Breslow, N.E.; Perlman, E.; Miser, J.; Ritchey, M.L.; Thomas, P.R.; Grundy, P.E.; D’Angio, G.J.; et al. Treatment of Wilms tumor relapsing after initial treatment with vincristine, actinomycin D, and doxorubicin. A report from the National Wilms Tumor Study Group. Pediatr. Blood Cancer 2008, 50, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.; Green, D.; Daw, N. Late effects of treatment for wilms tumor. Pediatr. Hematol. Oncol. 2009, 26, 407–413. [Google Scholar] [CrossRef] [PubMed]
- The SIOP Renal Tumour Study Group. Paediatric renal tumours: Perspectives from the SIOP–RTSG. Nat. Rev. Urol. 2017, 14, 3–4. [Google Scholar] [CrossRef]
- Wang, J.; Li, M.; Tang, D.; Gu, W.; Mao, J.; Shu, Q. Current treatment for Wilms tumor: COG and SIOP standards. World J. Pediatr. Surg. 2019, 2, 11–14. [Google Scholar] [CrossRef]
- Vujanić, G.M.; Gessler, M.; Ooms, A.H.A.G.; Collini, P.; Coulomb-l’Hermine, A.; D’Hooghe, E.; de Krijger, R.R.; Perotti, D.; Pritchard-Jones, K.; Vokuhl, C.; et al. The UMBRELLA SIOP-RTSG 2016 Wilms tumour pathology and molecular biology protocol. Nat. Rev. Urol. 2018, 15, 693–701. [Google Scholar] [CrossRef]
- Vujanić, G.M.; Sandstedt, B.; Kelsey, A.; Sebire, N.J. Central pathology review in multicenter trials and studies. Cancer 2009, 115, 1977–1983. [Google Scholar] [CrossRef]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69, S36–S40. [Google Scholar] [CrossRef]
- Chahal, A.; Gulia, P. Machine learning and deep learning. Int. J. Innov. Technol. Explor. Eng. 2019, 8, 4910–4914. [Google Scholar] [CrossRef]
- van der Kamp, A.; Waterlander, T.J.; de Bel, T.; van der Laak, J.; Heuvel-Eibrink, M.M.V.D.; Mavinkurve-Groothuis, A.M.C.; de Krijger, R.R. Artificial Intelligence in Pediatric Pathology: The Extinction of a Medical Profession or the Key to a Bright Future? Pediatr. Dev. Pathol. 2022, 25, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Tariq, A.; Purkayastha, S.; Padmanaban, G.P.; Krupinski, E.; Trivedi, H.; Banerjee, I.; Gichoya, J.W. Current Clinical Applications of Artificial Intelligence in Radiology and Their Best Supporting Evidence. J. Am. Coll. Radiol. 2020, 17, 1371–1381. [Google Scholar] [CrossRef]
- Lopez-Jimenez, F.; Attia, Z.; Arruda-Olson, A.M.; Carter, R.; Chareonthaitawee, P.; Jouni, H.; Kapa, S.; Lerman, A.; Luong, C.; Medina-Inojosa, J.R.; et al. Artificial Intelligence in Cardiology: Present and Future. Mayo Clin. Proc. 2020, 95, 1015–1039. [Google Scholar] [CrossRef]
- Hameed, B.; Dhavileswarapu, A.S.; Raza, S.; Karimi, H.; Khanuja, H.; Shetty, D.; Ibrahim, S.; Shah, M.; Naik, N.; Paul, R.; et al. Artificial Intelligence and Its Impact on Urological Diseases and Management: A Comprehensive Review of the Literature. J. Clin. Med. 2021, 10, 1864. [Google Scholar] [CrossRef]
- Yadav, A.S.; Kumar, S.; Karetla, G.R.; Cotrina-Aliaga, J.C.; Arias-Gonzáles, J.L.; Kumar, V.; Srivastava, S.; Gupta, R.; Ibrahim, S.; Paul, R.; et al. A Feature Extraction Using Probabilistic Neural Network and BTFSC-Net Model with Deep Learning for Brain Tumor Classification. J. Imaging 2022, 9, 10. [Google Scholar] [CrossRef]
- Khan, Y.F.; Kaushik, B.; Chowdhary, C.L.; Srivastava, G. Ensemble Model for Diagnostic Classification of Alzheimer’s Disease Based on Brain Anatomical Magnetic Resonance Imaging. Diagnostics 2022, 12, 3193. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Hara, S.; Haneda, E.; Kawakami, M.; Morita, K.; Nishioka, R.; Zoshima, T.; Kometani, M.; Yoneda, T.; Kawano, M.; Karashima, S.; et al. Evaluating tubulointerstitial compartments in renal biopsy specimens using a deep learning-based approach for classifying normal and abnormal tubules. PLoS ONE 2022, 17, e0271161. [Google Scholar] [CrossRef]
- Hermsen, M.; De Bel, T.; den Boer, M.; Steenbergen, E.J.; Kers, J.; Florquin, S.; Roelofs, J.; Stegall, M.D.; Alexander, M.P.; Smith, B.H.; et al. Deep Learning–Based Histopathologic Assessment of Kidney Tissue. J. Am. Soc. Nephrol. 2019, 30, 1968–1979. [Google Scholar] [CrossRef]
- Rutgers, J.J.; Bánki, T.; van der Kamp, A.; Waterlander, T.J.; Scheijde-Vermeulen, M.A.; Heuvel-Eibrink, M.M.v.D.; van der Laak, J.A.W.M.; Fiocco, M.; Mavinkurve-Groothuis, A.M.C.; de Krijger, R.R. Interobserver variability between experienced and inexperienced observers in the histopathological analysis of Wilms tumors: A pilot study for future algorithmic approach. Diagn. Pathol. 2021, 16, 77. [Google Scholar] [CrossRef]
- Roy, P.; van Peer, S.E.; de Witte, M.M.; Tytgat, G.A.M.; Karim-Kos, H.E.; van Grotel, M.; van de Ven, C.P.; Mavinkurve-Groothuis, A.M.C.; Merks, J.H.M.; Kuiper, R.P.; et al. Characteristics and outcome of children with renal tumors in the Netherlands: The first five-year’s experience of national centralization. PLoS ONE 2022, 17, e0261729. [Google Scholar] [CrossRef] [PubMed]
- van den Heuvel-Eibrink, M.M.; Hol, J.A.; Pritchard-Jones, K.; van Tinteren, H.; Furtwängler, R.; Verschuur, A.C.; Vujanic, G.M.; Leuschner, I.; Brok, J.; Rübe, C.; et al. Rationale for the treatment of Wilms tumour in the UMBRELLA SIOP–RTSG 2016 protocol. Nat. Rev. Urol. 2017, 14, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention 2015; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2016. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Popov, S.D.; Sebire, N.J.; Vujanic, G.M. Wilms’ Tumour–Histology and Differential Diagnosis. In Wilms Tumor; Codon Publications: Singapore, 2016; pp. 3–21. [Google Scholar] [CrossRef]
- Vujanic, G.M.; Sandstedt, B.; Vujanić, G.M.; Sandstedt, B. The pathology of Wilms’ tumour (nephroblastoma): The International Society of Paediatric Oncology approach. J. Clin. Pathol. 2010, 63, 102–109. [Google Scholar] [CrossRef]
| Vital Tumor Components | Chemotherapy-Induced Changes | Normal Renal Tissue |
|---|---|---|
| Blastema Stroma Epithelium | Necrosis Bleeding Regression | Glomeruli Tubules |
| Extra Renal Tissue | Adrenal Gland | Others |
| Fat Mesenchyme Vessels Nerves Lymph nodes | Adrenal cortex Adrenal medulla | Urothelium Anaplasia Nephrogenic rest Background |
| U-net Training Defaults | DenseNet Training Defaults | |
|---|---|---|
| Patch shape | (412, 412) | (128, 128) |
| Sampling spacing | 0.5 μm/pixel | 0.5 μm/pixel |
| Loss function | categorical cross-entropy | categorical cross-entropy |
| Optimization method | Adam [27] | Adam [27] |
| Epochs trained | 200 | 200 |
| Batch size | 4 | 16 |
| Initial learning rate | 0.0005 | 0.0005 |
| Learning rate decay | 0.5 after plateau of 5 epochs | 0.5 after plateau of 5 epochs |
| Augmentations used | rotation, flipping, Gaussian noise and color | rotation, flipping, Gaussian noise and color |
| Final layer | Softmax | Softmax |
| Patient Demographics (n = 72) | n (%) |
|---|---|
| Age in months at time of diagnosis, Mean (SD) | 51.4 (±41.3) |
| Female gender | 42 (58.3) |
| Left-sided WT localization | 41 (56.9) |
| Lymph node metastases | 11 (15.3) |
| Histology * (n = 72) | n (%) |
| Low risk | 2 (2.8) |
| Intermediate risk | 65 (90.2) |
| High risk | 5 (6.9) |
| Tumor histology * (n = 72) | n (%) |
| Completely necrotic1 | 2 (2.8) |
| Regressive 2 | 23 (31.9) |
| Epithelial 2 | 5 (6.9) |
| Stromal 2 | 12 (16.7) |
| Mixed 2 | 25 (33.8) |
| Blastemal 3 | 5 (6.9) |
| SIOP overall stage (n = 72) | n (%) |
| I | 25 (34.7) |
| II | 21 (29.2) |
| III | 26 (36.1) |
| Tumor Characteristics (n = 72) | n (%) |
|---|---|
| Chemotherapy-induced changes | |
| <66% | 47 (65.3) |
| >66% | 23 (31.9) |
| 100% | 2 (2.8) |
| Blastema | |
| <66% | 62 (86.1) |
| >66% | 8 (11.1) |
| Missing | 2 (2.8) |
| Epithelium | |
| <66% | 64 (88.9) |
| >66% | 7 (9.7) |
| Missing | 1 (1.4) |
| Stroma | |
| <66% | 57 (79.2) |
| >66% | 13 (18.1) |
| Missing | 2 (2.8) |
| Tissue Element | Precision | Recall | Dice Coef. |
|---|---|---|---|
| WT-blastema | 0.71 | 0.96 | 0.82 |
| WT-stroma | 0.77 | 0.59 | 0.67 |
| WT-epithelium | 0.65 | 0.38 | 0.48 |
| Necrosis | 0.98 | 0.99 | 0.98 |
| Bleeding | 0.23 | 0.92 | 0.37 |
| Regression | 0.62 | 0.77 | 0.69 |
| Glomeruli | 0.69 | 1.00 | 0.82 |
| Tubules | 0.98 | 0.96 | 0.97 |
| Fat | 1.00 | 0.89 | 0.94 |
| Mesenchyme | 0.57 | 0.67 | 0.62 |
| Vessels | 0.85 | 0.77 | 0.81 |
| Nerves | 0.85 | 0.77 | 0.81 |
| Lymph nodes | 0.99 | 0.99 | 0.99 |
| Urothelium | 0.46 | 0.96 | 0.62 |
| Nephrogenic rests | 0.82 | 0.98 | 0.89 |
| Chemotherapy-induced changes | 0.79 | 0.90 | 0.84 |
| Vital tumor components | 0.74 | 0.66 | 0.70 |
| Overall score | 0.85 | 0.85 | 0.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van der Kamp, A.; de Bel, T.; van Alst, L.; Rutgers, J.; van den Heuvel-Eibrink, M.M.; Mavinkurve-Groothuis, A.M.C.; van der Laak, J.; de Krijger, R.R. Automated Deep Learning-Based Classification of Wilms Tumor Histopathology. Cancers 2023, 15, 2656. https://doi.org/10.3390/cancers15092656
van der Kamp A, de Bel T, van Alst L, Rutgers J, van den Heuvel-Eibrink MM, Mavinkurve-Groothuis AMC, van der Laak J, de Krijger RR. Automated Deep Learning-Based Classification of Wilms Tumor Histopathology. Cancers. 2023; 15(9):2656. https://doi.org/10.3390/cancers15092656
Chicago/Turabian Stylevan der Kamp, Ananda, Thomas de Bel, Ludo van Alst, Jikke Rutgers, Marry M. van den Heuvel-Eibrink, Annelies M. C. Mavinkurve-Groothuis, Jeroen van der Laak, and Ronald R. de Krijger. 2023. "Automated Deep Learning-Based Classification of Wilms Tumor Histopathology" Cancers 15, no. 9: 2656. https://doi.org/10.3390/cancers15092656
APA Stylevan der Kamp, A., de Bel, T., van Alst, L., Rutgers, J., van den Heuvel-Eibrink, M. M., Mavinkurve-Groothuis, A. M. C., van der Laak, J., & de Krijger, R. R. (2023). Automated Deep Learning-Based Classification of Wilms Tumor Histopathology. Cancers, 15(9), 2656. https://doi.org/10.3390/cancers15092656










