Detection of Ultra-Rare ESR1 Mutations in Primary Breast Cancer Using LNA-Clamp ddPCR
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and DNA/mRNA Extraction
2.2. Patients and Samples
2.3. mRNA Extraction from Primary Tumors and Reverse Transcription
2.4. LNA-Clamp ddPCR from Primary Tumors
2.5. Statistics
3. Results
3.1. Comparison of ESR1 Expression Levels between DNA and mRNA in Cell Lines
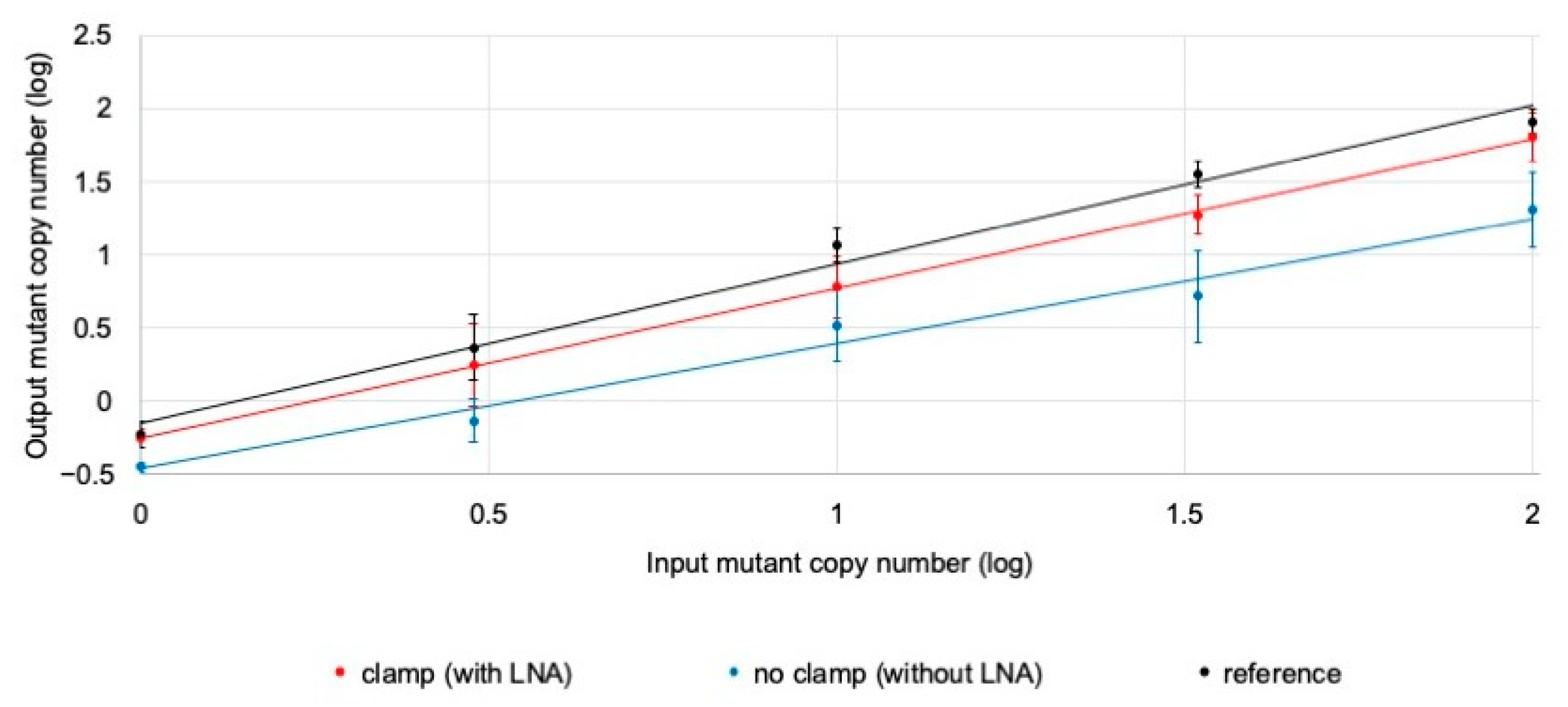
3.2. Confirmation of the Clamping Method Using LNA Oligo
3.3. Detection Sensitivity and Cutoff of the LNA-Clamp ddPCR
3.4. ESR1 Mutation Analysis of Primary Breast Cancer Using LNA-Clamp ddPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yamashita, H.; Iwase, H.; Toyama, T.; Takahashi, S.; Sugiura, H.; Yoshimoto, N.; Endo, Y.; Fujii, Y.; Kobayashi, S. Estrogen receptor-positive breast cancer in Japanese women: Trends in incidence, characteristics, and prognosis. Ann. Oncol. 2011, 22, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Hortobagyi, G.N. Treatment of Breast Cancer. N. Eng. J. Med. 1998, 339, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; Paluch-Shimon, S.; Senkus, E.; Curigliano, G.; Aapro, M.S.; Andre, F.; Barrios, C.H.; Bergh, J.; Bhattacharyya, G.S.; Biganzoli, L.; et al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5). Ann. Oncol. 2020, 31, 1623–1649. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.K.; Schiff, R. Mechanisms of endocrine resistance in breast cancer. Annu. Rev. Med. 2011, 62, 233–247. [Google Scholar] [CrossRef]
- Oesterreich, S.; Davidson, N.E. The search for ESR1 mutations in breast cancer. Nat. Genet. 2013, 45, 1415–1416. [Google Scholar] [CrossRef]
- Toy, W.; Shen, Y.; Won, H.; Green, B.; Sakr, R.A.; Will, M.; Li, Z.; Gala, K.; Fanning, S.; King, T.A.; et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 2013, 45, 1439–1445. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.M.; Vats, P.; Su, F.; Lonigro, R.J.; Cao, X.; Kalyana-Sundaram, S.; Wang, R.; Ning, Y.; Hodges, L.; et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 2013, 45, 1446–1451. [Google Scholar] [CrossRef]
- Jeselsohn, R.; Yelensky, R.; Buchwalter, G.; Frampton, G.; Meric-Bernstam, F.; Gonzalez-Angulo, A.M.; Ferrer-Lozano, J.; Perez-Fidalgo, J.A.; Cristofanilli, M.; Gomez, H.; et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin. Cancer Res. 2014, 20, 1757–1767. [Google Scholar] [CrossRef]
- Clatot, F.; Perdrix, A.; Augusto, L.; Beaussire, L.; Delacour, J.; Calbrix, C.; Sefrioui, D.; Viailly, P.J.; Bubenheim, M.; Moldovan, C.; et al. Kinetics, prognostic and predictive values of ESR1 circulating mutations in metastatic breast cancer patients progressing on aromatase inhibitor. Oncotarget 2016, 7, 74448–74459. [Google Scholar] [CrossRef]
- Zhang, K.; Hong, R.; Xu, F.; Xia, W.; Kaping, L.; Qin, G.; Zheng, Q.; Lu, Q.; Shi, Y.X.; Yuan, Z.Y.; et al. Clinical value of circulating ESR1 mutations for patients with metastatic breast cancer: A meta-analysis. Cancer Manag. Res. 2018, 10, 2573–2580. [Google Scholar] [CrossRef]
- Angus, L.; Beije, N.; Jager, A.; Martens, J.W.; Sleijfer, S. ESR1 mutations: Moving towards guiding treatment decision-making in metastatic breast cancer patients. Cancer Treat. Rev. 2017, 52, 33–40. [Google Scholar] [CrossRef] [PubMed]
- De Santo, I.; McCartney, A.; Migliaccio, I.; Di Leo, A.; Malorni, L. The Emerging Role of ESR1 Mutations in Luminal Breast Cancer as a Prognostic and Predictive Biomarker of Response to Endocrine Therapy. Cancers 2019, 11, 1894. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, T.; Kagara, N.; Miyake, T.; Tanei, T.; Naoi, Y.; Shimoda, M.; Shimazu, K.; Kim, S.J.; Noguchi, S. Detection of ESR1 mutations in plasma and tumors from metastatic breast cancer patients using next-generation sequencing. Breast Cancer Res. Treat. 2017, 163, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Brett, J.O.; Spring, L.M.; Bardia, A.; Wander, S.A. ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Res. 2021, 23, 85. [Google Scholar] [CrossRef]
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Inao, T.; Sueta, A.; Fujiwara, S.; Omoto, Y.; Iwase, H. Droplet digital polymerase chain reaction assay for screening of ESR1 mutations in 325 breast cancer specimens. Transl. Res. 2015, 166, 540–553.e2. [Google Scholar] [CrossRef]
- Toy, W.; Weir, H.; Razavi, P.; Lawson, M.; Goeppert, A.U.; Mazzola, A.M.; Smith, A.; Wilson, J.; Morrow, C.; Wong, W.L.; et al. Activating ESR1 Mutations Differentially Affect the Efficacy of ER Antagonists. Cancer Discov. 2017, 7, 277–287. [Google Scholar] [CrossRef]
- Razavi, P.; Chang, M.T.; Xu, G.; Bandlamudi, C.; Ross, D.S.; Vasan, N.; Cai, Y.; Bielski, C.M.; Donoghue, M.T.A.; Jonsson, P.; et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell. 2018, 34, 427–438.e6. [Google Scholar] [CrossRef]
- Quach, N.; Goodman, M.F.; Shibata, D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin. Pathol. 2004, 4, 1. [Google Scholar] [CrossRef]
- Bruskov, V.I.; Malakhova, L.V.; Masalimov, Z.K.; Chernikov, A.V. Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nucleic Acids Res. 2002, 30, 1354–1363. [Google Scholar] [CrossRef]
- Wang, P.; Bahreini, A.; Gyanchandani, R.; Lucas, P.C.; Hartmaier, R.J.; Watters, R.J.; Jonnalagadda, A.R.; Trejo Bittar, H.E.; Berg, A.; Hamilton, R.L.; et al. Sensitive Detection of Mono- and Polyclonal ESR1 Mutations in Primary Tumors, Metastatic Lesions, and Cell-Free DNA of Breast Cancer Patients. Clin. Cancer Res. 2016, 22, 1130–1137. [Google Scholar] [CrossRef]
- Gelsomino, L.; Gu, G.; Rechoum, Y.; Beyer, A.R.; Pejerrey, S.M.; Tsimelzon, A.; Wang, T.; Huffman, K.; Ludlow, A.; Ando, S.; et al. ESR1 mutations affect anti-proliferative responses to tamoxifen through enhanced cross-talk with IGF signaling. Breast Cancer Res. Treat. 2016, 157, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Glenn, T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011, 11, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Quail, M.A.; Kozarewa, I.; Smith, F.; Scally, A.; Stephens, P.J.; Durbin, R.; Swerdlow, H.; Turner, D.J. A large genome center’s improvements to the Illumina sequencing system. Nat. Methods 2008, 5, 1005–1010. [Google Scholar] [CrossRef]
- Masunaga, N.; Kagara, N.; Motooka, D.; Nakamura, S.; Miyake, T.; Tanei, T.; Naoi, Y.; Shimoda, M.; Shimazu, K.; Kim, S.J.; et al. Highly sensitive detection of ESR1 mutations in cell-free DNA from patients with metastatic breast cancer using molecular barcode sequencing. Breast Cancer Res. Treat. 2018, 167, 49–58. [Google Scholar] [CrossRef]
- Nagai, Y.; Miyazawa, H.; Huqun; Tanaka, T.; Udagawa, K.; Kato, M.; Fukuyama, S.; Yokote, A.; Kobayashi, K.; Kanazawa, M.; et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005, 65, 7276–7282. [Google Scholar] [CrossRef]
- Ikenaga, M.; Sakai, M. Application of Locked Nucleic Acid (LNA) oligonucleotide-PCR clamping technique to selectively PCR amplify the SSU rRNA genes of bacteria in investigating the plant-associated community structures. Microbes Environ. 2014, 29, 286–295. [Google Scholar] [CrossRef]
- Chung, J.H.; Pavlick, D.; Hartmaier, R.; Schrock, A.B.; Young, L.; Forcier, B.; Ye, P.; Levin, M.K.; Goldberg, M.; Burris, H.; et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with estrogen receptor-positive metastatic breast cancer. Ann. Oncol. 2017, 28, 2866–2873. [Google Scholar] [CrossRef]
- Allred, D.C.; Harvey, J.M.; Berardo, M.; Clark, G.M. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod. Pathol. 1998, 11, 155–168. [Google Scholar]
- Sun, Y.; Tian, H.; Liu, C.; Yang, D.; Li, Z. A Clamp-Based One-Step Droplet Digital Reverse Transcription PCR (ddRT-PCR) for Precise Quantitation of Messenger RNA Mutation in Single Cells. ACS Sens. 2018, 3, 1795–1801. [Google Scholar] [CrossRef] [PubMed]
- Thiede, C.; Creutzig, E.; Illmer, T.; Schaich, M.; Heise, V.; Ehninger, G.; Landt, O. Rapid and sensitive typing of NPM1 mutations using LNA-mediated PCR clamping. Leukemia 2006, 20, 1897–1899. [Google Scholar] [CrossRef] [PubMed]
- Fribbens, C.; O’Leary, B.; Kilburn, L.; Hrebien, S.; Garcia-Murillas, I.; Beaney, M.; Cristofanilli, M.; Andre, F.; Loi, S.; Loibl, S.; et al. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J. Clin. Oncol. 2016, 34, 2961–2968. [Google Scholar] [CrossRef] [PubMed]
- Chandarlapaty, S.; Chen, D.; He, W.; Sung, P.; Samoila, A.; You, D.; Bhatt, T.; Patel, P.; Voi, M.; Gnant, M.; et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016, 2, 1310–1315. [Google Scholar] [CrossRef]
- O’Leary, B.; Hrebien, S.; Morden, J.P.; Beaney, M.; Fribbens, C.; Huang, X.; Liu, Y.; Bartlett, C.H.; Koehler, M.; Cristofanilli, M.; et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 2018, 9, 896. [Google Scholar] [CrossRef]

| N | 212 | |
|---|---|---|
| Age | Median (range) | 60 (33–96) |
| Tumor size (cm) | ≤2 | 168 |
| >2 | 44 | |
| Lymph node metastasis | Negative | 178 |
| Positive | 34 | |
| Histological grade | 1/2 | 184 |
| 3 | 28 | |
| ER | Positive | 212 |
| Negative | 0 | |
| PgR | Positive | 177 |
| Negative | 35 | |
| HER2 | Positive | 26 |
| Negative | 186 | |
| Histology | IDC | 174 |
| ILC | 17 | |
| Other | 21 | |
| Prognosis | No recurrence | 199 |
| Recurrence | 13 |
| Case | AA Change | SNV | VAF (%) | Mutant Copy Number (per Well) | Total Copy Number (per Well) |
|---|---|---|---|---|---|
| 1 | Y537S | 1610A>C | 0.0025 | 2.8 | 111,150 |
| 38 | Y537S | 1610A>C | 0.0031 | 3.6 | 116,800 |
| 49 | Y537S | 1610A>C | 0.0022 | 2.8 | 129,000 |
| 59 | Y537S | 1610A>C | 0.0028 | 3 | 108,800 |
| 84 | Y537S | 1610A>C | 0.0018 | 2.8 | 158,000 |
| 117 | Y537S | 1610A>C | 0.0023 | 2.4 | 106,540 |
| 119 | Y537S | 1610A>C | 0.0021 | 2.6 | 121,200 |
| 120 | Y537S | 1610A>C | 0.0024 | 2.6 | 108,500 |
| 121 | Y537S | 1610A>C | 0.0037 | 3.8 | 102,200 |
| 139 | Y537S | 1610A>C | 0.1580 | 6.8 | 4304 |
| 165 | Y537S | 1610A>C | 0.0053 | 2.6 | 48,640 |
| 173 | Y537S | 1610A>C | 0.0106 | 3 | 28,320 |
| 177 | Y537S | 1610A>C | 0.0025 | 2.8 | 112,800 |
| 194 | Y537S | 1610A>C | 0.0026 | 2.6 | 101,700 |
| 202 | Y537S | 1610A>C | 0.0028 | 2.6 | 91,280 |
| 132 | Y537S | 1610A>C | 0.0034 | 2.8 | 81,280 |
| 132 | D538G | 1613A>G | 0.0034 | 2.8 | 81,280 |
| 2 | D538G | 1613A>G | 0.0041 | 2.6 | 63,840 |
| 7 | D538G | 1613A>G | 0.0119 | 2.8 | 23,520 |
| 68 | D538G | 1613A>G | 0.0110 | 4.8 | 43,520 |
| 144 | D538G | 1613A>G | 0.0422 | 2.8 | 6640 |
| 145 | D538G | 1613A>G | 0.0060 | 2.8 | 46,690 |
| 146 | D538G | 1613A>G | 0.0111 | 3 | 27,040 |
| 158 | D538G | 1613A>G | 0.0053 | 3 | 56,480 |
| 187 | D538G | 1613A>G | 0.0053 | 2.8 | 52,800 |
| 207 | D538G | 1613A>G | 0.0111 | 2.6 | 23,520 |
| 211 | D538G | 1613A>G | 0.0039 | 3.8 | 96,480 |
| 212 | D538G | 1613A>G | 1.6260 | 36 | 2178 |
| ESR1 | p | |||
|---|---|---|---|---|
| Positive | Negative | |||
| N | 27 | 185 | ||
| Age | Median (range) | 60 (36–83) | 60 (33–95) | 0.32 * |
| Tumor size (cm) | ≤2 | 23 | 145 | 0.61 ** |
| >2 | 4 | 40 | ||
| Lymph node metastasis | Positive | 4 | 30 | 1.00 ** |
| Negative | 23 | 155 | ||
| PgR | Positive | 23 | 154 | 1.00 ** |
| Negative | 4 | 31 | ||
| HER2 | Positive | 2 | 24 | 0.54 ** |
| Negative | 25 | 161 | ||
| Histological grade | 1 | 11 | 77 | 1.00 ** |
| 2/3 | 16 | 108 | ||
| Prognosis | No recurrence | 24 | 175 | 0.22 ** |
| Recurrence | 3 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashimoto, Y.; Masunaga, N.; Kagara, N.; Abe, K.; Yoshinami, T.; Tsukabe, M.; Sota, Y.; Miyake, T.; Tanei, T.; Shimoda, M.; et al. Detection of Ultra-Rare ESR1 Mutations in Primary Breast Cancer Using LNA-Clamp ddPCR. Cancers 2023, 15, 2632. https://doi.org/10.3390/cancers15092632
Hashimoto Y, Masunaga N, Kagara N, Abe K, Yoshinami T, Tsukabe M, Sota Y, Miyake T, Tanei T, Shimoda M, et al. Detection of Ultra-Rare ESR1 Mutations in Primary Breast Cancer Using LNA-Clamp ddPCR. Cancers. 2023; 15(9):2632. https://doi.org/10.3390/cancers15092632
Chicago/Turabian StyleHashimoto, Yoko, Nanae Masunaga, Naofumi Kagara, Kaori Abe, Tetsuhiro Yoshinami, Masami Tsukabe, Yoshiaki Sota, Tomohiro Miyake, Tomonori Tanei, Masafumi Shimoda, and et al. 2023. "Detection of Ultra-Rare ESR1 Mutations in Primary Breast Cancer Using LNA-Clamp ddPCR" Cancers 15, no. 9: 2632. https://doi.org/10.3390/cancers15092632
APA StyleHashimoto, Y., Masunaga, N., Kagara, N., Abe, K., Yoshinami, T., Tsukabe, M., Sota, Y., Miyake, T., Tanei, T., Shimoda, M., & Shimazu, K. (2023). Detection of Ultra-Rare ESR1 Mutations in Primary Breast Cancer Using LNA-Clamp ddPCR. Cancers, 15(9), 2632. https://doi.org/10.3390/cancers15092632






