Inhibition of PLK1 Destabilizes EGFR and Sensitizes EGFR-Mutated Lung Cancer Cells to Small Molecule Inhibitor Osimertinib
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
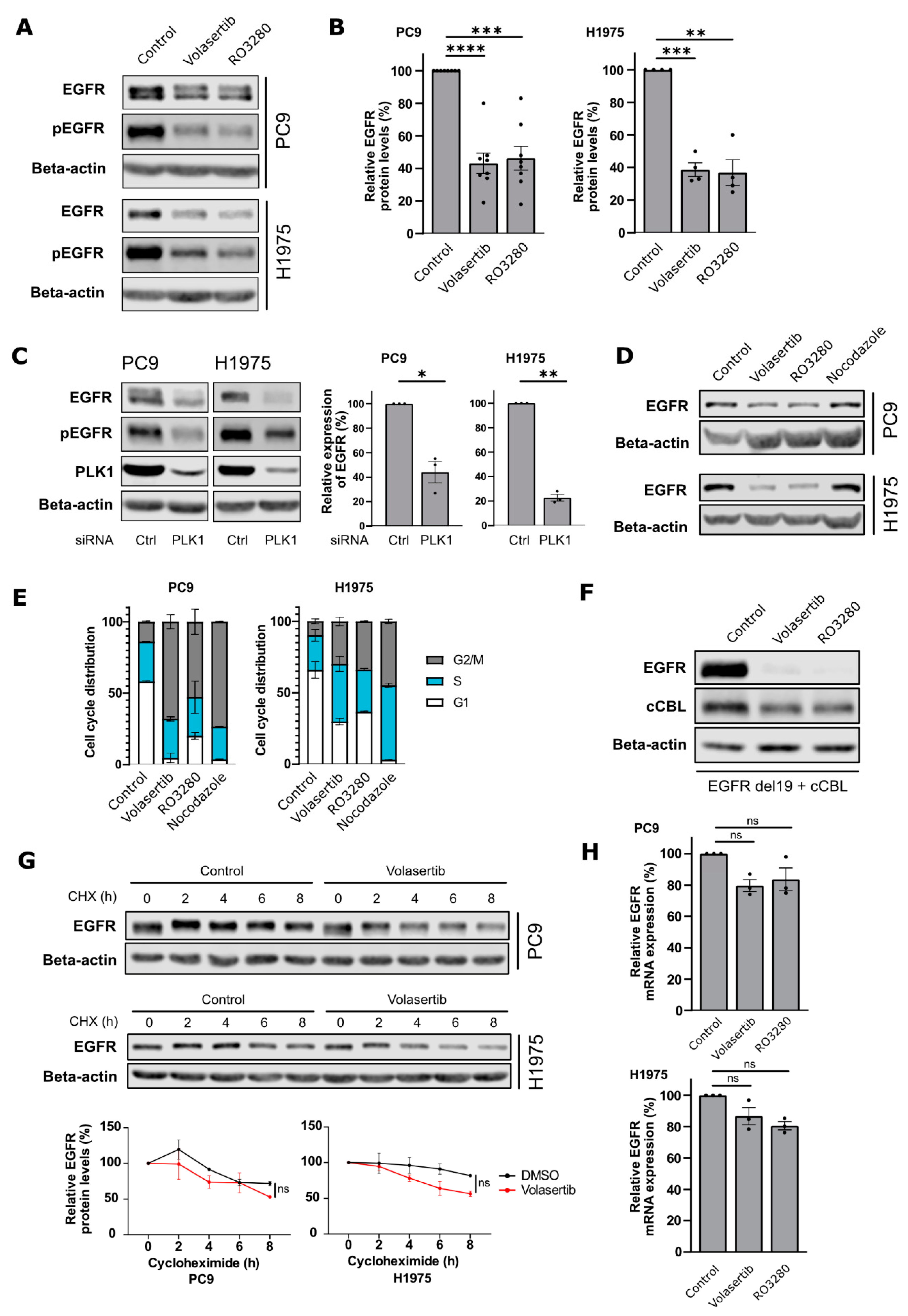
2.1. PLK1 Inhibition Decreases EGFR Protein Levels in NSCLC Cells
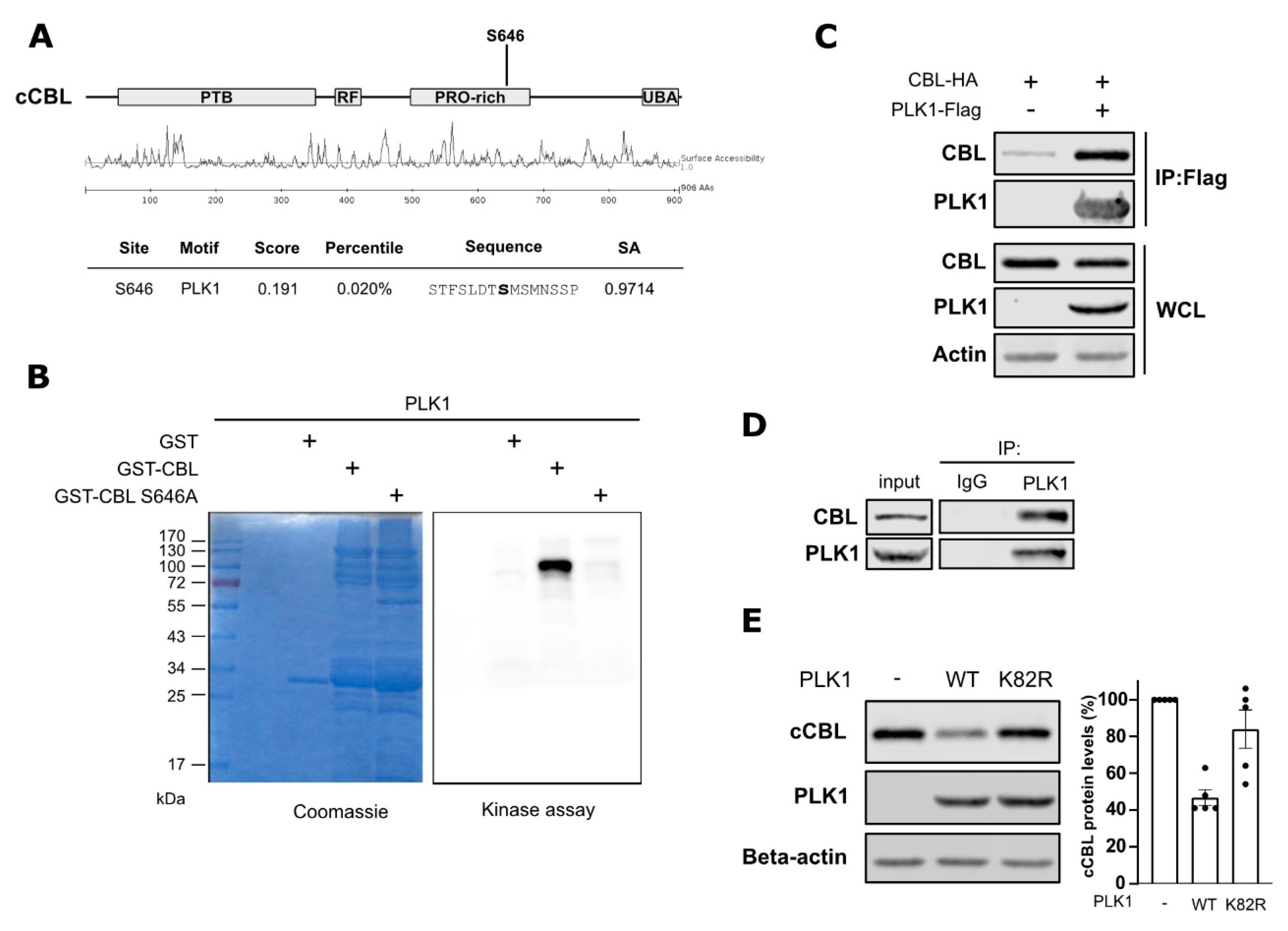
2.2. PLK1 Interacts with and Phosphorylates c-Cbl
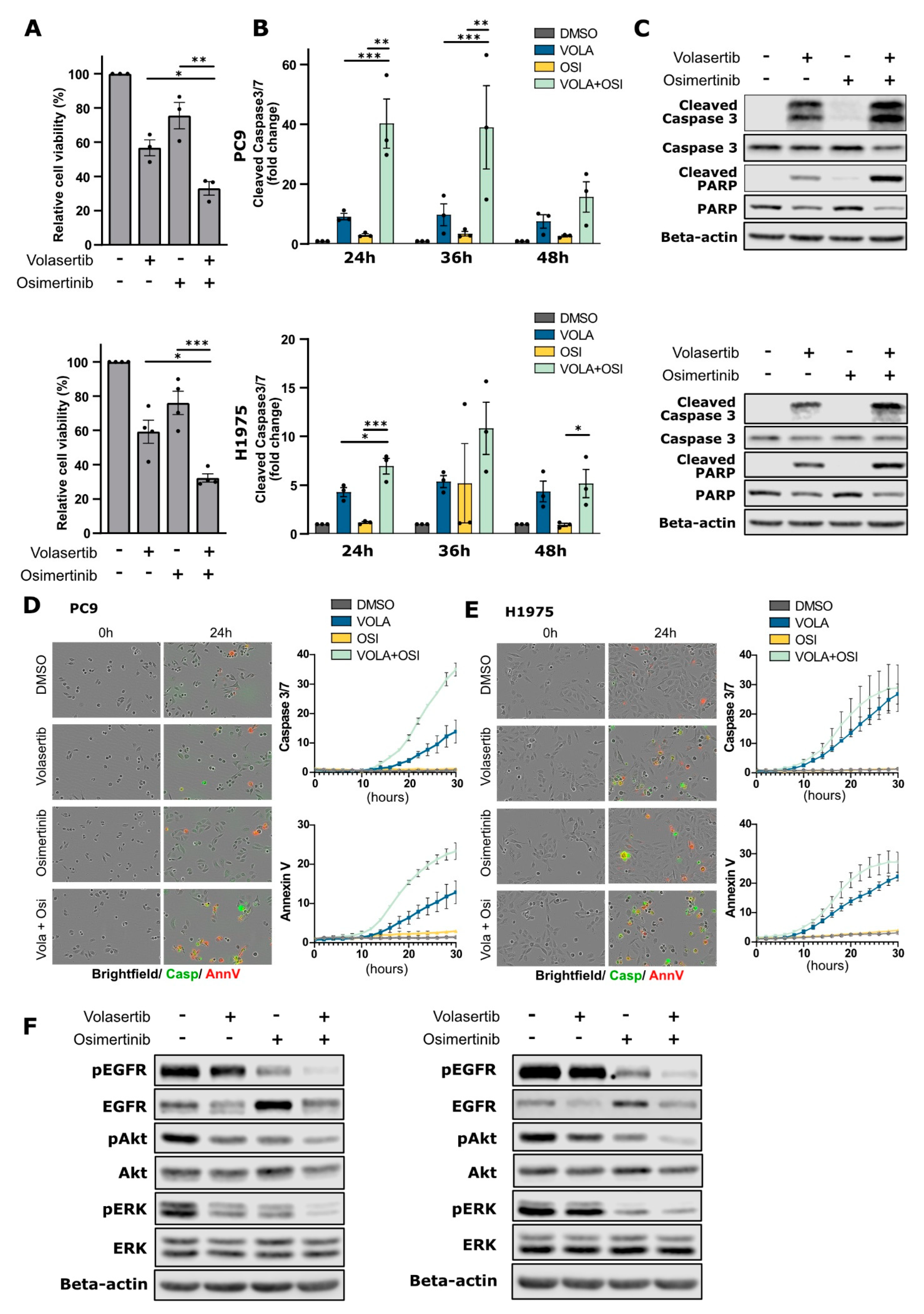
2.3. PLK1 Inhibition Enhances Cancer Cell Sensitivity to Osimertinib in EGFR-Mutant NSCLC Cells
3. Discussion
4. Materials & Methods
4.1. Cell Lines and Drugs
4.2. SiRNA and DNA Plasmids
4.3. Quantitative Real-Time PCR
4.4. Western Blotting
4.5. Co-Immunoprecipitation
4.6. Cell Viability
4.7. Apoptosis Assay Caspase-Glo 3/7
4.8. Apoptosis Assay Incucyte
4.9. Recombinant Proteins and In Vitro Kinase Assay
4.10. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non–Small-Cell Lung Cancer to Gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Suda, K.; Wiens, J.; Bunn, P.A. New and Emerging Targeted Treatments in Advanced Non-Small-Cell Lung Cancer. Lancet 2016, 388, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.S.; Vansteenkiste, J.; Planchard, D.; Cho, B.C.; Gray, J.E.; Ohe, Y.; Zhou, C.; Reungwetwattana, T.; Cheng, Y.; Chewaskulyong, B.; et al. Overall Survival with Osimertinib in Untreated, EGFR -Mutated Advanced NSCLC. N. Engl. J. Med. 2020, 382, 41–50. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance Mechanisms to Osimertinib in EGFR-Mutated Non-Small Cell Lung Cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]
- Suda, K.; Mitsudomi, T. Drug Tolerance to EGFR Tyrosine Kinase Inhibitors in Lung Cancers with EGFR Mutations. Cells 2021, 10, 1590. [Google Scholar] [CrossRef]
- Passaro, A.; Jänne, P.A.; Mok, T.; Peters, S. Overcoming Therapy Resistance in EGFR-Mutant Lung Cancer. Nat. Cancer 2021, 2, 377–391. [Google Scholar] [CrossRef]
- Zitouni, S.; Nabais, C.; Jana, S.C.; Guerrero, A.; Bettencourt-Dias, M. Polo-like Kinases: Structural Variations Lead to Multiple Functions. Nat. Rev. Mol. Cell Biol. 2014, 15, 433–452. [Google Scholar] [CrossRef]
- Wolf, G.; Elez, R.; Doermer, A.; Holtrich, U.; Ackermann, H.; Stutte, H.J.; Altmannsberger, H.-M.; Rübsamen-Waigmann, H.; Strebhardt, K. Prognostic Significance of Polo-like Kinase (PLK) Expression in Non-Small Cell Lung Cancer. Oncogene 1997, 14, 543–549. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Q.; Wang, X. PLK1, A Potential Target for Cancer Therapy. Transl. Oncol. 2017, 10, 22–32. [Google Scholar] [CrossRef]
- Shin, S.-B.; Kim, D.-H.; Kim, D.-E.; Aldonza, M.B.D.; Kim, Y.; Yim, H. Dual Targeting of EGFR with PLK1 Exerts Therapeutic Synergism in Taxane-Resistant Lung Adenocarcinoma by Suppressing ABC Transporters. Cancers 2021, 13, 4413. [Google Scholar] [CrossRef] [PubMed]
- Reda, M.; Ngamcherdtrakul, W.; Gu, S.; Bejan, D.S.; Siriwon, N.; Gray, J.W.; Yantasee, W. PLK1 and EGFR Targeted Nanoparticle as a Radiation Sensitizer for Non-Small Cell Lung Cancer. Cancer Lett. 2019, 467, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Noor, A.; Umelo, I.A.; Kronenberger, P.; Teugels, E.; de Greve, J. Targeting Polo-like Kinase 1 and TRAIL to Enhance Apoptosis in Non-Small Cell Lung Cancer (NSCLC) Cells. J. Clin. Oncol. 2017, 35, e14104. [Google Scholar] [CrossRef]
- Wang, Y.; Singh, R.; Wang, L.; Nilsson, M.; Goonatilake, R.; Tong, P.; Li, L.; Giri, U.; Villalobos, P.; Mino, B.; et al. Polo-like Kinase 1 Inhibition Diminishes Acquired Resistance to Epidermal Growth Factor Receptor Inhibition in Non-Small Cell Lung Cancer with T790M Mutations. Oncotarget 2016, 7, 47998–48010. [Google Scholar] [CrossRef]
- Takano, N.; Seike, M.; Sugano, T.; Matsuda, K.; Hisakane, K.; Yosshikawa, A.; Nakamichi, S.; Noro, R.; Gemma, A. A Novel Molecular Target in EGFR-Mutant Lung Cancer Treated with the Combination of Osimertinib and Pemetrexed. Anticancer Res. 2022, 42, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Obenauer, J.C. Scansite 2.0: Proteome-Wide Prediction of Cell Signaling Interactions Using Short Sequence Motifs. Nucleic Acids Res. 2003, 31, 3635–3641. [Google Scholar] [CrossRef]
- Giron, P.; Eggermont, C.; Noeparast, A.; Vandenplas, H.; Teugels, E.; Forsyth, R.; de Wever, O.; Aza-Blanc, P.; Gutierrez, G.J.; de Grève, J. Targeting USP13-mediated Drug Tolerance Increases the Efficacy of EGFR Inhibition of Mutant EGFR in Non-small Cell Lung Cancer. Int. J. Cancer 2021, 148, 2579–2593. [Google Scholar] [CrossRef]
- Schöffski, P.; Awada, A.; Dumez, H.; Gil, T.; Bartholomeus, S.; Wolter, P.; Taton, M.; Fritsch, H.; Glomb, P.; Munzert, G. A Phase I, Dose-Escalation Study of the Novel Polo-like Kinase Inhibitor Volasertib (BI 6727) in Patients with Advanced Solid Tumours. Eur. J. Cancer 2012, 48, 179–186. [Google Scholar] [CrossRef]
- Ellis, P.M.; Leighl, N.B.; Hirsh, V.; Reaume, M.N.; Blais, N.; Wierzbicki, R.; Sadrolhefazi, B.; Gu, Y.; Liu, D.; Pilz, K.; et al. A Randomized, Open-Label Phase II Trial of Volasertib as Monotherapy and in Combination With Standard-Dose Pemetrexed Compared With Pemetrexed Monotherapy in Second-Line Treatment for Non–Small-Cell Lung Cancer. Clin. Lung Cancer 2015, 16, 457–465. [Google Scholar] [CrossRef]
- Chen, G.; Kronenberger, P.; Teugels, E.; Umelo, I.; de Grève, J. Targeting the Epidermal Growth Factor Receptor in Non-Small Cell Lung Cancer Cells: The Effect of Combining RNA Interference with Tyrosine Kinase Inhibitors or Cetuximab. BMC Med. 2012, 10, 28. [Google Scholar] [CrossRef]
- Huang, K.Y.; Kao, S.H.; Wang, W.L.; Chen, C.Y.; Hsiao, T.H.; Salunke, S.B.; Chen, J.J.W.; Su, K.Y.; Yang, S.C.; Hong, T.M.; et al. Small Molecule T315 Promotes Casitas B-Lineage Lymphoma Dependent Degradation of Epidermal Growth Factor Receptor via Y1045 Autophosphorylation. Am. J. Respir. Crit. Care Med. 2016, 193, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Zhou, B.; Zhang, B.; Ren, H.; Zhu, L.; Zheng, G.; Ge, M.; Ge, J. Recent Advances in the Development of EGFR Degraders: PROTACs and LYTACs. Eur. J. Med. Chem. 2022, 239, 114533. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Weihua, Z. Rethink of EGFR in Cancer With Its Kinase Independent Function on Board. Front. Oncol. 2019, 9, 800. [Google Scholar] [CrossRef] [PubMed]
- Graves, L.M.; Duncan, J.S.; Whittle, M.C.; Johnson, G.L. The Dynamic Nature of the Kinome. Biochem. J. 2013, 450, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pedraza-Alva, G.; Sawasdikosol, S.; Liu, Y.C.; Mérida, L.B.; Cruz-Muñoz, M.E.; Oceguera-Yañez, F.; Burakoff, S.J.; Rosenstein, Y. Regulation of Cbl Molecular Interactions by the Co-Receptor Molecule CD43 in Human T Cells. J. Biol. Chem. 2001, 276, 729–737. [Google Scholar] [CrossRef]
- Kassenbrock, C.K.; Anderson, S.M. Regulation of Ubiquitin Protein Ligase Activity in C-Cbl by Phosphorylation-Induced Conformational Change and Constitutive Activation by Tyrosine to Glutamate Point Mutations. J. Biol. Chem. 2004, 279, 28017–28027. [Google Scholar] [CrossRef]
- Take, H.; Watanabe, S.; Takeda, K.; Yu, Z.X.; Iwata, N.; Kajigaya, S. Cloning and Characterization of a Novel Adaptor Protein, CIN85, That Interacts with c-Cbl. Biochem. Biophys. Res. Commun. 2000, 268, 321–328. [Google Scholar] [CrossRef]
- Soubeyran, P.; Kowanetz, K.; Szymkiewicz, I.; Langdon, W.Y.; Dikic, I. Cbl–CIN85–Endophilin Complex Mediates Ligand-Induced Downregulation of EGF Receptors. Nature 2002, 416, 183–187. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Chiang, Y.J.; Huang, F.; Li, Y.; Li, X.; Ning, Y.; Zhang, W.; Deng, H.; Chen, Y.-G. DNA Damage Activates TGF-β Signaling via ATM-c-Cbl-Mediated Stabilization of the Type II Receptor TβRII. Cell Rep. 2019, 28, 735–745.e4. [Google Scholar] [CrossRef]
- Ettenberg, S.A.; Magnifico, A.; Cuello, M.; Nau, M.M.; Rubinstein, Y.R.; Yarden, Y.; Weissman, A.M.; Lipkowitz, S. Cbl-b-Dependent Coordinated Degradation of the Epidermal Growth Factor Receptor Signaling Complex. J. Biol. Chem. 2001, 276, 27677–27684. [Google Scholar] [CrossRef]
- Tong, J.; Taylor, P.; Moran, M.F. Proteomic Analysis of the Epidermal Growth Factor Receptor (EGFR) Interactome and Post-Translational Modifications Associated with Receptor Endocytosis in Response to EGF and Stress. Mol. Cell Proteom. 2014, 13, 1644–1658. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.H.; Chou, C.C.; Lai, T.Y.; Hsu, J.E.; Lin, Y.S.; Liu, H.Y.; Chen, Y.K.; Ho, I.L.; Hsu, P.H.; Chuang, T.H.; et al. ZNRF1 Mediates Epidermal Growth Factor Receptor Ubiquitination to Control Receptor Lysosomal Trafficking and Degradation. Front. Cell Dev. Biol. 2021, 9, 642625. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Yoo, G.; Kim, T.; Lee, D.; Lee, C.S.; Cha, H.R.; Park, Y.H.; Moon, J.Y.; Jung, S.S.; Kim, J.O.; et al. The E3 Ubiquitin Ligase CHIP Selectively Regulates Mutant Epidermal Growth Factor Receptor by Ubiquitination and Degradation. Biochem. Biophys. Res. Commun. 2016, 479, 152–158. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eggermont, C.; Gutierrez, G.J.; De Grève, J.; Giron, P. Inhibition of PLK1 Destabilizes EGFR and Sensitizes EGFR-Mutated Lung Cancer Cells to Small Molecule Inhibitor Osimertinib. Cancers 2023, 15, 2589. https://doi.org/10.3390/cancers15092589
Eggermont C, Gutierrez GJ, De Grève J, Giron P. Inhibition of PLK1 Destabilizes EGFR and Sensitizes EGFR-Mutated Lung Cancer Cells to Small Molecule Inhibitor Osimertinib. Cancers. 2023; 15(9):2589. https://doi.org/10.3390/cancers15092589
Chicago/Turabian StyleEggermont, Carolien, Gustavo J. Gutierrez, Jacques De Grève, and Philippe Giron. 2023. "Inhibition of PLK1 Destabilizes EGFR and Sensitizes EGFR-Mutated Lung Cancer Cells to Small Molecule Inhibitor Osimertinib" Cancers 15, no. 9: 2589. https://doi.org/10.3390/cancers15092589
APA StyleEggermont, C., Gutierrez, G. J., De Grève, J., & Giron, P. (2023). Inhibition of PLK1 Destabilizes EGFR and Sensitizes EGFR-Mutated Lung Cancer Cells to Small Molecule Inhibitor Osimertinib. Cancers, 15(9), 2589. https://doi.org/10.3390/cancers15092589





