Simple Summary
Lung cancer is the most common and lethal malignancy worldwide. Musashi-2 (MSI2) is an RNA-binding protein that is overexpressed in advanced NSCLC. VEGFR2 protein expression contributes to NSCLC progression and several FDA-approved drugs are used to target it in the clinic. Here, we show that MSI2 is a strong positive regulator of VEGFR2 protein levels in murine and human NSCLC cell lines. Furthermore, we found that MSI2 protein directly binds to VEGFR2 and PTEN mRNAs and impacts VEGFR2 downstream signaling, in part via PTEN regulation.
Abstract
Lung cancer is the most frequently diagnosed cancer type and the leading cause of cancer-related deaths worldwide. Non-small cell lung cancer (NSCLC) represents most of the diagnoses of lung cancer. Vascular endothelial growth factor receptor-2 (VEGFR2) is a member of the VEGF family of receptor tyrosine kinase proteins, which are expressed on both endothelial and tumor cells, are one of the key proteins contributing to cancer development, and are involved in drug resistance. We previously showed that Musashi-2 (MSI2) RNA-binding protein is associated with NSCLC progression by regulating several signaling pathways relevant to NSCLC. In this study, we performed Reverse Protein Phase Array (RPPA) analysis of murine lung cancer, which suggests that VEGFR2 protein is strongly positively regulated by MSI2. Next, we validated VEGFR2 protein regulation by MSI2 in several human lung adenocarcinoma cell line models. Additionally, we found that MSI2 affected AKT signaling via negative PTEN mRNA translation regulation. In silico prediction analysis suggested that both VEGFR2 and PTEN mRNAs have predicted binding sites for MSI2. We next performed RNA immunoprecipitation coupled with quantitative PCR, which confirmed that MSI2 directly binds to VEGFR2 and PTEN mRNAs, suggesting a direct regulation mechanism. Finally, MSI2 expression positively correlated with VEGFR2 and VEGF-A protein levels in human lung adenocarcinoma samples. We conclude that the MSI2/VEGFR2 axis contributes to lung adenocarcinoma progression and is worth further investigations and therapeutic targeting.
1. Introduction
Lung cancer is second in incidence and first in mortality among all cancer types in recent years based on World Health Organization and American Cancer Society reports [1,2]. Most lung cancer statistics include both small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). In general, NSCLC represents about 85% of all lung cancer and includes squamous (30%) and non-squamous (70%) subtypes [3,4,5]. Lung cancer typically has a poor prognosis if tumors have disseminated from the primary site [6]. Besides the most common oncogenic driver mutations, KRAS and TP53, NSCLC is associated with numerous genetic alterations such as EGFR mutations, ALK translocation, VEGFR2 and c-MET amplification, and others [7,8]. At this time, in addition to chemotherapy, immunotherapy, and targeted therapies, VEGFR2 and VEGF inhibitors have shown proven efficacy in NSCLC treatment in stage IV disease in combination with chemotherapy and also have also shown promise when combined with immunotherapy [3,9].
Vascular endothelial growth factor (VEGF-A) and its receptors (VEGFR1, VEGFR2, VEGFR3) are the main pro-angiogenic drivers of solid tumors [10]. In the canonical model, VEGF-A is expressed by tumors and recognizes and binds VEGFR2 on endothelial cells, thus leading to tumor vessel formation [11] and, as a result, a better supply of nutrients for tumor cells. Additionally, VEGF-A promotes an immunosuppressive tumor microenvironment via several mechanisms [12,13,14,15]. Published papers show that VEGFR2 and VEGFR1 are well expressed on tumor cells [16,17,18,19]. Moreover, VEGFR2 gain-of-function is associated with chemoresistance and poor survival in patients with lung cancer [20]. Currently, several drugs are used in the clinic, including VEGFR2 inhibitors (ramucirumab, cabozantinib, pazopanib), a VEGF-A inhibitor (bevacizumab) [3], and a VEGF-A/PIGF trap (aflibercept) [21]. Adding the VEGFR2 inhibitor nintedanib to second-line docetaxel showed improved progression-free survival (median 3.4 months with combination therapy versus 2.7 months with docetaxel alone; HR 0.79, p = 0.0019) [22]. Additionally, a randomized phase III study compared a combination of ramucirumab with docetaxel to docetaxel alone and reported a modest improvement in median overall survival from 9.1 months to 10.5 months (HR 0.86, p = 0.023) [23].
Furthermore, Musashi-2 (MSI2) protein expression is associated with advanced NSCLC [24,25]. MSI2 and its homolog, Musashi-1 (MSI1), are a family of RNA-binding proteins that regulate the stability and translation of target mRNAs through highly specific RNA binding of the core motif in the 3′-untranslated region of mRNAs (UAG) [26,27,28]. MSI2 regulates multiple critical biological processes in stem cells and cancer cells and contributes to cancer drug resistance [29,30]. It was previously indicated that MSI2 protein is increased in liquid and solid tumors, of which lung cancer is one of them [24,27,31,32,33,34]. Recently, we showed that MSI2 protein level correlates with NSCLC aggressiveness and has a specific role in promoting metastasis in these tumors via regulation of TGFβR1 and its target SMAD3 [25]. Furthermore, we have recently shown that MSI2 deficiency leads to higher sensitivity to Epidermal Growth Factor Receptor (EGFR) inhibitors in EGFR mutant NSCLC due to EGFR protein downregulation resulting from MSI2-mediated direct regulation of EGFR mRNA [24].
In the light of published papers, we have validated proteomic analysis data that suggested VEGFR2 regulation upon MSI2 depletion in murine lung cancer. We found that MSI2 depletion leads to a strong decrease in VEGFR2 protein levels in murine and human lung adenocarcinoma cell lines. Furthermore, we found that MSI2 protein directly binds to VEGFR2 and PTEN mRNAs and impacts VEGFR2 downstream signaling via PTEN regulation. Finally, MSI2 protein expression correlated with VEGFR2 and VEGF-A protein levels in NSCLC patient samples.
2. Materials and Methods
2.1. Cell Culture
Human lung adenocarcinoma cell lines A549 (KRAS34G>A, CDKNA1_471del471), Calu-1 (KRAS34G>T, TP53del), H23 (KRAS34G>T, TP53738G>C), and H441 (KRAS35G>T, TP53473G>T) were obtained from the American Type Culture Collection (ATCC). Human lung cancer cell lines Hcc1171 (KRAS34G>T, TP53740A>T) and Hcc461 (KRAS35G>A, TP53445delT) were obtained from UT Southwestern Medical Center. The murine NSCLC cell line (344SQ) from Trp53R172HΔG/+ KrasLA1/+ mice was previously described [35]. Initial stocks were cryopreserved and, at every 6-month interval, a fresh aliquot of frozen cells was used for the experiments. No additional authentication was performed. All cells were cultured in RPMI 1640 (Gibco, Gaithersburg, MD, USA) supplemented with 10% FBS (Hyclone, Logan, UT, USA), penicillin (100 U/mL), streptomycin (100 µg/mL), sodium pyruvate (1 mM), and non-essential amino acids (0.1 mM) under the conditions indicated in the figure legends. Hypoxic incubations were performed with 1% O2, 94% N2, and 5% CO2.
2.2. Antibodies and Drugs
Anti-MSI2 (#ab76148) and anti-VEGF-A (#ab46154) were obtained from Abcam (Cambridge, UK). Anti-VEGFR2 (#2479), anti-phVEGFR2 (#3817), anti-β-actin (#3700), anti-phAKT (T308) (#13038), anti-phAKT (S473) (#4060), anti-AKT total (#2920), anti-phGSKαβ (#9331), anti-GSKαβ total (#5676), anti-PTEN (#14642), anti-phERK (#4370), anti-ERK total (4695), anti-ph4EBP(#2855), anti-phP70SK (#9206), anti-P70S6K total (#2708), anti-rabbit HRP-linked (#7074), and anti-mouse HRP-linked (#7076) were obtained from Cell Signaling (Danvers, MA, USA). Doxycycline (#HY-N0565), hygromycin (HY-B0490), and puromycin (HY-B1743A) were obtained from MedChemExpress (Monmouth Junction, NJ, USA).
2.3. Vector Construction and Lentivirus Production
To generate cell lines with inducible knockdown of MSI2, we used self-complementary single-stranded DNA oligos from Supplementary Table S1 as shown in our published paper [24]. To generate stable cell lines with KDR (VEGFR2) overexpression, we used pHage-KDR-hygro vector and pHage-hygro vector as negative control. We used plasmids pHage-KDR (#116754) and pHage-puro (#118692) from Addgene with a changed resistance gene to hygromycin using a cDNA containing a Hygro gene and cloned into ClaI/NdeI and BfuAI/ClaI sites to generate overexpression constructs. The pLV-CMV-hygro vector used as a template for the Hygro gene. All generated cell lines used in the study are noted in Supplementary Table S2.
2.4. SiRNA Transfections
SiRNA transfection was performed using nonspecific control pool siRNAs and anti-human MSI2 SiRNAs (Supplementary Table S3). Details of the procedure were previously described in our published paper [24].
2.5. Western Blot Analysis
Cell lysate preparation and Western blot analysis were performed using standard methods as previously described [1]. Band signals were detected using X-ray film, and the film was digitized using a photo scanner. Image analysis was conducted using ImageJ (version 1.53e, National Institutes of Health, Bethesda, MD, USA) with signal intensity normalized to β-actin, and 3–4 repeats were used for each experiment’s quantitative analysis. Final data were analyzed in GraphPad Prism using an unpaired t-test or ANOVA to determine statistical significance.
2.6. Cell Viability Assay
To evaluate the effects of MSI2 protein level on cell proliferation, we plated cells at a concentration of 500 cells/well in 96-well cell culture plates in triplicate. After 72 h of incubation to allow cells to grow, we added reagents for the CellTiter-Blue® assay (Promega, Fitchburg, WI, USA) based on the manufacturer’s protocol to measure OD 562 nm.
2.7. Reverse Transcription and qPCR
RNA was extracted using a phenol–chloroform-based method. RNA concentration and quantity were measured using NanoDrop Lite (cat# ND-LITE ThermoFisher Scientific, Waltham, MA, USA). First-strand cDNA synthesis was performed using the iScript cDNA synthesis kit (cat#1708841, Biorad, CA, USA) according to the manufacturer’s instructions. The generated cDNA was diluted tenfold and used as a template for qPCR, which was performed with an Applied Biosystems QuantStudio 3 system using PowerTrack™ SYBR Green Master Mix (Applied Biosystems, Waltham, MA, USA). Relative quantification of gene expression was performed using the 2−ΔΔCt method using the primers indicated in Supplementary Table S4.
2.8. ELISA
The concentration of VEGF-A in human lung cancer cell lines was determined by a VEGF Human ELISA kit (Abcam, #ab100662). ELISA assays were performed according to the manufacturer’s instructions. Briefly, the collected condition media from cells were added to a well coated with primary antibody and then immunosorbented using a biotinylated primary antibody at room temperature for 2.5 h. The color development catalyzed by horseradish peroxidase was terminated with 2.5 mol/L sulfuric acid and the absorption was measured at 450 nm. The protein concentration was determined by comparing the relative absorbance of the samples with the standards.
2.9. RNA-IP Assays
RNA-IP was performed using the Magna RIP RNA-binding Protein Immunoprecipitation kit (cat#17-700, Millipore, Burlington, MA, USA) according to the manufacturer’s protocol. Immunoprecipitated RNAs were quantified by quantitative PCR (qPCR) using the primers indicated in Supplementary Table S4, with PTP4A1 as a normalization (positive) control and GAPDH as a negative control.
2.10. RPPA
The RPPA analysis procedure of 344SQ-SCR, 344SQ-m1, and 344SQ-m2 mouse cells was previously described [25].
2.11. Immunohistochemistry of Human NSCLC
Human non-squamous NSCLC patient samples from the Rоstov Research Institute’s Human Tissue Repository Facility (HTRF) and from the Republican Clinical Oncology Dispensary named after Prof. M. Z. Sigal (RCOD at Kazan) were used. At the time of tissue acquisition, patients provided Institutional Review Board (IRB)–approved informed consent for the storage of tissues and the review of deidentified clinical data. Clinical information (Supplementary Table S5) from the repository database was abstracted in an anonymized fashion. Tissue samples were stained for VEGFR2 and MSI2 proteins via an immunohistochemical (IHC) approach, and hematoxylin and eosin (H&E) stained sections were used for morphological evaluation purposes, with unstained sections used for IHC staining using standard methods. The sections were incubated overnight with primary antibodies to MSI2 (EP1305Y, Rabbit, 1:100, Abcam #ab76148), VEGF-A (VG-1, Mouse, 1:50, Abcam #ab1316), and VEGFR2 (55B11, Rabbit, 1:50, Cell signaling, Cat #2479) at 4 °C in a humidified slide chamber. As a negative control, the primary antibody was replaced with normal mouse/rabbit IgG to confirm the absence of specific staining. Immunodetection was performed using the Dako Envision+ polymer system and immunostaining was visualized with the chromogen 3,3′-diaminobenzidine. All slides were viewed with a Nikon Eclipse 50i microscope and photomicrographs were taken with an attached Nikon DS-Fi1 camera (Melville, NY, USA).
2.12. In Silico Evaluation of MSI2 Binding to VEGFR2, VEGF-A, and PTEN mRNAs
Human and murine genome sequences for EGFR were obtained from the UCSC Human Gene Sorter December 2013 (GRCh38/hg38) assembly and scanned for the Musashi binding motifs previously defined by Bennett et al. [26] (15 motifs with the highest p values) and Wang et al. [27] (8 motifs with the highest p values; Supplementary Tables S6 and S7).
2.13. Statistical Analysis
All statistical analyses, including unpaired two-tailed t-tests, ANOVA analysis, and Spearman correlations, were performed in GraphPad Prism 9 (San Diego, CA, USA).
3. Results
3.1. Musashi-2 Regulates VEGFR2 mRNA and Protein Levels and Directly Binds VEGFR2 mRNA in Human NSCLC Cell Lines
Our published paper suggested that MSI2 depletion in murine lung cancer cell line 344SQ may decrease VEGFR2 protein levels based on RPPA data (Supplementary Figure S1A) [25]. To verify this result, we used our previously established KRAS/P53-driven 344SQ cell line with doxycycline (DOX)-inducible knockdown (KD) of MSI2 (sh1, sh2) and with constant MSI2 overexpression (OE) (MSI2) (Supplementary Figure S1B) and evaluated VEGFR2 and VEGF-A protein and mRNA levels (Supplementary Figure S1C,D). We found that MSI2 KD leads to a significant decrease in VEGFR2 protein levels in mouse cells, though not a decrease in mRNA levels. In addition, MSI2 OE leads to a mild increase in VEGF-A mRNA and protein levels, but not VEGFR2 protein and mRNA levels. Thus, we conclude that MSI2 may regulate VEGFR2 and VEGF-A in mouse cells. Next, based on previously published papers [19,36,37], we selected several NSCLC cell lines with significant VEGFR2 expression and examined VEGFR2 and VEGF-A protein levels using Western blot analysis (Figure 1A) and VEGFR2 mRNA levels (Figure 1B). We selected cell lines with higher expression of VEGFR2 (H441, Hcc1171, and Calu-1) and the A549 cell line as a negative control/low VEGFR2 expression sample for further analysis. Previously, Bennett et al. [26], Wang et al. [27], and Nguyen et al. [28] in their papers showed that MSI2 recognizes and directly binds consensus sequences with the core motif UAG in the 3′-untranslated region (3′-UTR) of mRNAs, and as a result regulates the stability and/or translation of target mRNAs. Based on this published data, we performed in silico analysis (using Bennett and Wang data) and found that VEGFR2 and VEGF-A mRNAs have predicted binding sites for MSI2 binding (Figure 1C, Supplementary Tables S6 and S7). To confirm the in silico results, we performed RNA immunoprecipitation (RIP) assays with an MSI2 antibody pulldown coupled with RT-qPCR in three lung adenocarcinoma cell lines: A549 (Supplementary Figure S2A), Hcc1171, and H441 (Figure 1D), using the previously defined MSI2 target mRNAs (PTP4A, TGFβR1, SMAD3) [25,38] as positive controls and ACTB and GAPDH as negative controls. Antibodies to MSI2 specifically immunoprecipitated the VEGFR2 mRNA as efficiently as they did for the positive controls (Figure 1D). Taken together, we conclude that MSI2 directly regulates VEGFR2 mRNA translation in human lung adenocarcinoma.
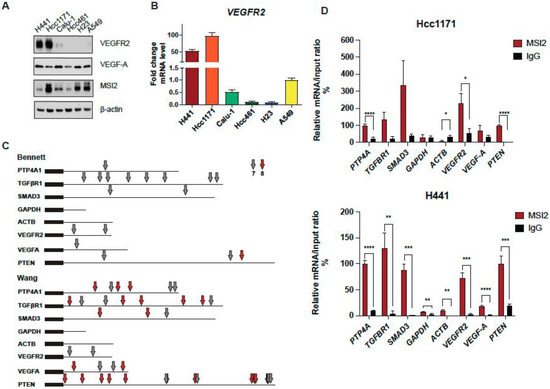
Figure 1.
MSI2 directly binds VEGFR2 mRNA in human NSCLC. (A) Western blot representing images of human NSCLC cell lines. (B) VEGFR2 mRNA level in human NSCLC cell lines measured through RT-qPCR analysis. Data from at least three independent experiments normalized to 18S rRNA and to A549. (C) Predicted MSI2 binding sites in human mRNAs. Location of consensus binding sites for Musashi proteins in the noted human genes, as defined from studies by Bennett et al. and Wang et al. Coding sequences are thick lines and 3′-untranslated regions are thin lines. The 7- or 8-bp consensus sequences are indicated by arrows. VEGFR2 reference sequence–NCBI Reference Sequence: NM_002253.4; PTEN reference sequence–NCBI Reference Sequence: NM_000314.8. (D) mRNA immunoprecipitation (RIP) analysis of indicated cell lines lysates using antibodies to MSI2 or IgG (negative control, followed by qRT-PCR). Data are normalized to positive control PTP4A1, TGFBR1 and SMAD3 are additional positive controls, and GAPDH and ACTB are negative controls. The data shown reflect the average of three independent RIP experiments. Error bars represented by SEM. Statistical analysis was performed using an unpaired two-tailed t-test. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 for all graphs.
3.2. Musashi-2 Regulation of VEGFR2 and VEGF-A Protein Levels in NSCLC
Since MSI2 directly binds VEGFR2 mRNA in human lung adenocarcinoma cell lines, we established cell lines with MSI2 depletion and overexpression to evaluate the effect of MSI2 expression on VEGFR2 and VEGF-A protein levels. Depletion of MSI2 in human NSCLC cell lines led to a significant decrease in VEGFR2 protein levels and had a mixed effect on intracellular VEGF-A protein levels (Figure 2A,B, Supplementary Figure S2B). Additionally, MSI2 OE leads to slight increase in VEGFR2 protein levels in the A549 and H441 cell lines and a significant decrease in VEFR2 protein levels in the Hcc1171 and Calu-1 cell lines (Supplementary Figure S2C,D). In addition, we performed ELISA analysis of extracellular VEGF-A concentration after MSI2 depletion and overexpression (Figure 2C). ELISA analysis indicated that MSI2 depletion only leads to a decrease in VEGF-A in the A549 cell line, while MSI2 OE leads to a significant increase in VEGF-A concentration in cell lines that have moderate to high expression of VEGFR2 (H441, Hcc1171, and Calu-1). Moreover, RT-qPCR analysis of VEGFR2 and VEGF-A with MSI2 depletion and overexpression showed that VEGFR2 mRNA is significantly decreased in human lung adenocarcinoma cell lines with MSI2 depletion and increased with MSI2 OE in cell lines with moderate to high VEGFR2 expression (H441, Hcc1171, and Calu-1) (Supplementary Figure S3). In parallel, evaluation of the effect of MSI2 depletion on cell growth in these cell lines showed that MSI2 level does not affect NSCLC cell growth (Supplementary Figure S4A). Therefore, we conclude that MSI2 positively regulates both VEGFR2 mRNA and protein levels and also regulates VEGF-A mRNA translation in human lung cancer cells.
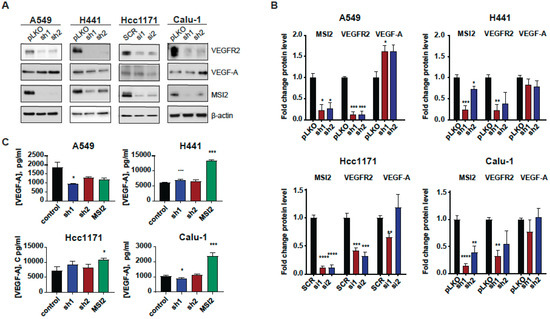
Figure 2.
MSI2 regulation of VEGFR2 and VEGF-A protein levels in human NSCLC cell lines. (A) Representative Western blot images of NSCLC cell lines after MSI2 depletion by shRNA (sh1, sh2) and siRNA (si1, si2). Negative controls include pLKO and SCR. (B) Western blot quantifications of data from (A) were performed from at least three independent experiments using Image J software (version 1.53e), with values normalized to negative controls and β-actin. (C) The concentration of VEGF-A in cell culture media from the indicated cell lines following depletion by shRNA (sh1, sh2) and siRNA (si1, si2) and overexpression (MSI2) of MSI2. The ELISA data shown reflect the average of three independent experiments. shRNA KD of MSI2 was induced by the addition of 1 μg/mL of doxycycline for 48 h. Error bars represented by SEM. Statistical analysis was performed using an unpaired two-tailed t-test. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 for all graphs.
3.3. Musashi-2 Regulates AKT Signaling via PTEN mRNA Binding Independent of VEGFR2
Since MSI2 depletion leads to a reduction in VEGFR2 protein levels, we evaluated VEGFR2 downstream signaling in human NSCLC cell lines. Western blot analysis showed that MSI2 depletion reduces VEGFR2 and phAKT protein levels (Figure 3A,B, Supplementary Figure S4B). To assess the role of VEGFR2 in the downregulation of phAKT with MSI2 KD, we established A549 and H441 cell lines with inducible MSI2 KD and constant VEGFR2 overexpression (OE). Analysis of AKT signaling indicated that MSI2 KD leads to a decrease in phAKT and its downstream target phGSK3⍺/β with and without VEGFR2 OE of VEGFR2 (Figure 3C,D). These findings suggest that a decrease in VEGFR2 protein level does not affect AKT signaling.
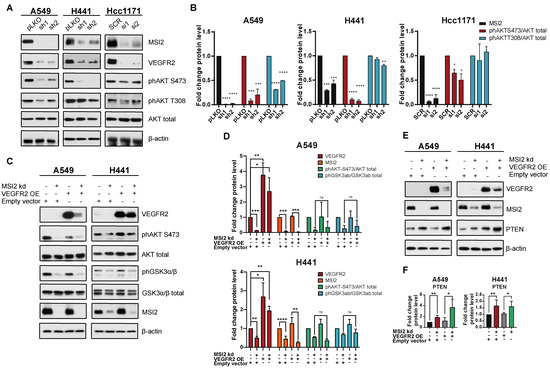
Figure 3.
VEGFR2-independent effects of MSI2 KD on AKT signaling. (A) Representative Western blot images of NSCLC cell lines after MSI2 depletion by shRNA (sh1, sh2) and siRNA (si1, si2). Negative controls include pLKO and SCR. Negative controls include pLKO and SCR. (B) Quantification of Western blot data from (A) with values normalized to negative controls (pLKO, SCR) and β-actin. (C) Rescue experiment: representative Western blot images of A549 and H441 cell lines with MSI2 depletion by shRNA (sh1) and VEGFR2 overexpression (VEGFR2 OE). Negative control is an empty vector (pHAGE). (D) Quantification of Western blot data from (C) with values normalized to the empty vector and β-actin. (E) Western blot of indicated cell lines following depletion by shRNA (sh1) of MSI2 and VEGFR2 overexpression (VEGFR2 OE). (F) Quantification of Western blot data from (E) with values normalized to the empty vector and β-actin. Data from (B–D) were quantified from at least three independent experiments using Image J software. Error bars represented by SEM. Statistical analysis was performed using an unpaired two-tailed t-test. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Wang et al. [27] showed that MSI2 inhibits PTEN (tumor suppressor and inhibitor of AKT signaling) protein level in murine intestinal epithelia. Therefore, we evaluated PTEN protein level in human lung adenocarcinoma cell lines upon MSI2 depletion (Figure 3E,F). Western blot analysis showed significant upregulation of PTEN with MSI2 KD regardless of VEGFR2 level. This suggests that MSI2 may directly regulate PTEN mRNA levels and affect AKT signaling in lung adenocarcinoma. To test this, we performed in silico analysis and found that the 3′ UTR of PTEN mRNA has predicted binding sites for MSI2 (Figure 1D, Supplementary Figure S2A). Next, we performed RIP-qPCR analysis using an MSI2 antibody, which supported the in silico predictions and showed that MSI2 directly binds PTEN mRNA. Taken together, we conclude that MSI2 affects AKT signaling via direct PTEN mRNA binding in lung adenocarcinoma, independent of VEGFR2 mRNA regulation.
3.4. Correlation of MSI2 with VEGFR2 and VEGF-A Expression in Human NSCLC
To evaluate the relationship between MSI2, VEGFR2, and VEGF-A in the subset of lung adenocarcinoma, we performed IHC analysis of MSI2, VEGFR2, and VEGF-A expression in an independent group of 116 non-squamous clinical tumor samples (Figure 4A,B, Supplementary Table S8). Spearman’s analysis of H scores showed a strong positive correlation for MSI2 vs. VEGFR2 (r = 0.673) and a moderate positive correlation for MSI2 vs. VEGF-A (r = 0.533).
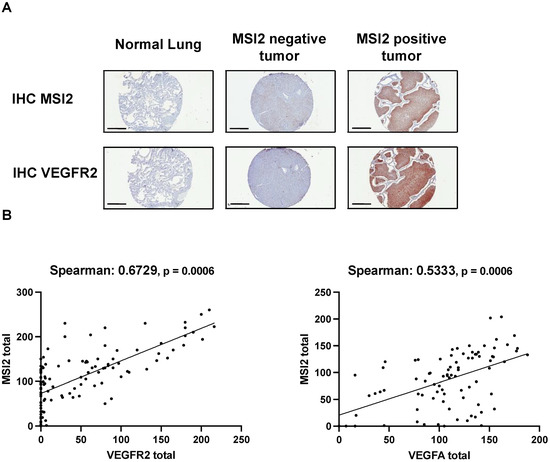
Figure 4.
Expression of MSI2, VEGFR2, and VEGF-A proteins in human non-squamous NSCLC patient samples. (A) Representative IHC images of MSI2 and VEGFR2 expression in human normal lungs and lung tumors. (B) MSI2 and VEGF-A (n = 94) and MSI2 and VEGFR2 (n = 116) H score correlation in human NSCLC TMAs.
4. Discussion
Published papers show that VEGFR2 and VEGF-A proteins are widely expressed by various tumor types, including lung tumors [16,17,18,19]. Additionally, VEGFR2 overexpression is associated with chemoresistance and poor survival in patients with lung cancer [20]. Our study shows for the first time that MSI2 positively regulates VEGFR2 protein levels in NSCLC.
First, we selected human lung adenocarcinoma cell lines with high levels of VEGFR2 expression (H441, Hcc1171, and Calu-1) and A549 as a control with a low level of VEGFR2 expression (Figure 1A,B). MSI2 typically regulates its targets by directly binding to specific motifs in the 3′-UTR fragments of mRNAs [26,27,28]. Therefore, we performed in silico analysis and then validated it through RNA-IP analysis (Figure 1C,D). In silico analysis and RNA-IP validation showed that MSI2 directly binds VEGFR2 but not VEGF-A mRNA. These results expand our knowledge about the role of Musashi-2 in NSCLC progression.
We then established human NSCLC cell lines with MSI2 KD and MSI2 OE to see how MSI2 levels affect VEGFR2 and VEGF-A protein and mRNA levels (Figure 2, Supplementary Figure S3). We found that MSI2 depletion leads not only to a decrease in VEGFR2 protein levels, but also a decrease in mRNA levels. That result can be interpreted in such a way that MSI2 regulates not only protein translation, but also affects mRNA transcript stability [29]. Moreover, we found that MSI2 overexpression leads to a modest increase in extracellular VEGF-A from cells with VEGFR2 overexpression. This effect may be interpreted as the indirect positive regulation of VEGF-A by MSI2 [39,40].
As MSI2 regulates VEGFR2 protein levels, we evaluated cell proliferation along with VEGFR2 downstream signaling in human lung adenocarcinoma cell lines upon MSI2 depletion. The viability assay showed that MSI2 does not affect cell growth in these KRAS-driven cells (Supplementary Figure S4A). Signaling evaluations indicated a decrease in VEGFR2 and phAKT protein levels with MSI2 KD (Figure 3A,B), while phERK protein levels were not changed (Supplementary Figure S4B). This result is expected because our models involve KRASmut, and published papers show that KRASmut results in constitutive activation of ERK [41,42]. Next, we established cell lines with VEGFR2 OE to evaluate the effects of MSI2 on AKT signaling via VEGFR2 regulation. Our results indicated that MSI2 KD induces a decrease in phAKT and its downstream target GSK3⍺/β regardless of the VEGFR2 level (Figure 3C,D).
Wang et al. [27] previously showed that MSI2 inhibits PTEN protein levels in murine intestinal epithelia. Based on that, the evaluation of PTEN levels with MSI2 KD and VEGFR2 OE indicated that PTEN protein levels are increased upon MSI2 KD regardless of the VEGFR2 level (Figure 3E,F). In addition, we showed that MSI2 negatively directly regulates PTEN protein levels (Figure 1C,D, Supplementary Figure S2A). We next performed in silico analysis, which predicted MSI2 binding sites in PTEN mRNA, and such binding was validated using RNA IP/QPCR analysis. Taken together, our data suggest that PTEN is a direct MSI2 target in NSCLC. Therefore, MSI2 affects AKT signaling via direct PTEN mRNA binding, independent of VEGFR2 mRNA regulation.
In addition, TMA analysis of human lung adenocarcinoma samples showed a positive correlation between MSI2 and VEGFR2 and VEGF-A protein levels (Figure 4). Taken together, we conclude that the previously unsuspected novel Musashi-2/VEGFR2 signaling axis is worth additional investigations and could be targeted for better NSCLC control in the future.
5. Conclusions
MSI2 protein directly regulates VEGFR2 and PTEN protein levels via VEGFR2 and PTEN mRNA binding in lung adenocarcinoma. MSI2 protein expression was correlated with VEGFR2 and VEGF-A protein levels in non-squamous NSCLC patient samples. MSI2 is a promising therapeutic target for better control of NSCLC.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15092529/s1, Figure S1: MSI2 regulation of VEGFR2 protein level in a mouse cell line; Figure S2: Effect of MSI2 OE in human NSCLC cell lines; Figure S3: Consequences of MSI2 depletion (left) and overexpression (right) on mRNA expression levels of VEGFR2 and VEGF-A; Figure S4: Effect of MSI2 KD on cell growth and signaling; Table S1: DNA oligonucleotides used for construction of shRNA vectors. Table lists single stranded DNA (ssDNA) oligos used for generation of Tet-pLKO vectors expressing specific shRNAs, used for lentiviral infection and selection of stable cell lines. MSI2 targeting sequences are underscored; Table S2: List of cell line derivatives used in the study; Table S3: siRNAs targeting human and murine MSI2; Table S4: Primers used for RT-PCR to quantify gene expression; Table S5: Clinical characteristics of NSCLC specimens; Table S6: Consensus MSI2 binding site sequences, defined by Wang et al.; Table S7: Consensus MSI2 binding site sequences, defined by Bennett et al.; Table S8: Clinical characteristics of NSCLC specimens.
Author Contributions
Conceptualization, I.B., A.K. and Y.B.; methodology, I.B. and I.T.; validation, I.B. and I.T.; investigation, Y.B.; resources, Y.B.; data curation, P.M.; writing—original draft preparation, I.B.; writing—review and editing, Y.B., P.M., I.T. and J.D.P.; visualization, I.B.; supervision, Y.B. and J.D.P.; project administration, Y.B.; funding acquisition, Y.B. and P.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work and the authors were supported by NIH R01 grant CA218802 (to Y.B.); a Translational Bridge Award from Northwestern University, number 2022-001 (to Y.B.); NCI Core Grant P30 CA060553 (to Robert H Lurie, Comprehensive Cancer Center at Northwestern University); NCI Core Grant P30 CA006927 (to the Fox Chase Cancer Center), and NCI R21 grant CA263362 (to P.M.).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Fox Chase Cancer Center (protocol code 17-9029, approved on 15 June 2017.
Informed Consent Statement
All clinical samples were collected under IRB approved consents at Rostov National Research Cancer Institute Human Tissue Repository Facility (Rostov-on-Don, Russia), Republican Clinical Oncology Dispensary named after Prof. M.Z. Sigal (Kazan, Russia) and Fox Chase Cancer Center (Philadelphia, PA, USA).
Data Availability Statement
The data presented in this study are available on request from the corresponding author and available to public.
Acknowledgments
We acknowledge assistance from the Fox Chase Cancer Center Cell Culture Facility, Histopathology Facility, Biostatistics Facility, and Biosample Repository.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Source: Globocan 2020. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/15-Lung-fact-sheet.pdf (accessed on 20 December 2022).
- Society, A.C. Cancer Facts & Figures. Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2022/2022-cancer-facts-and-figures.pdf (accessed on 15 February 2023).
- Gridelli, C.; Rossi, A.; Carbone, D.P.; Guarize, J.; Karachaliou, N.; Mok, T.; Petrella, F.; Spaggiari, L.; Rosell, R. Non-small cell lung cancer. Nat. Rev. Dis. Prim. 2015, 1, 15009. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Riely, G.J. New pathologic classification of lung cancer: Relevance for clinical practice and clinical trials. J. Clin. Oncol. 2013, 31, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Govindan, R.; Page, N.; Morgensztern, D.; Read, W.; Tierney, R.; Vlahiotis, A.; Spitznagel, E.L.; Piccirillo, J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: Analysis of the surveillance, epidemiologic, and end results database. J. Clin. Oncol. 2006, 24, 4539–4544. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Goel, H.L.; Mercurio, A.M. VEGF targets the tumour cell. Nat. Rev. Cancer 2013, 13, 871–882. [Google Scholar] [CrossRef]
- Minguet, J.; Smith, K.H.; Bramlage, P. Targeted therapies for treatment of non-small cell lung cancer—Recent advances and future perspectives. Int. J. Cancer 2016, 138, 2549–2561. [Google Scholar] [CrossRef]
- Reckamp, K.L.; Redman, M.W.; Dragnev, K.H.; Minichiello, K.; Villaruz, L.C.; Faller, B.; Baghdadi, T.A.; Hines, S.; Everhart, L.; Highleyman, L.; et al. Phase II Randomized Study of Ramucirumab and Pembrolizumab Versus Standard of Care in Advanced Non–Small-Cell Lung Cancer Previously Treated With Immunotherapy—Lung-MAP S1800A. J. Clin. Oncol. 2022, 40, 2295–2307. [Google Scholar] [CrossRef]
- Moens, S.; Goveia, J.; Stapor, P.C.; Cantelmo, A.R.; Carmeliet, P. The multifaceted activity of VEGF in angiogenesis—Implications for therapy responses. Cytokine Growth Factor Rev 2014, 25, 473–482. [Google Scholar] [CrossRef]
- Carmeliet, P. Mechanisms of angiogenesis and arteriogenesis. Nat. Med. 2000, 6, 389–395. [Google Scholar] [CrossRef]
- Ziogas, A.C.; Gavalas, N.G.; Tsiatas, M.; Tsitsilonis, O.; Politi, E.; Terpos, E.; Rodolakis, A.; Vlahos, G.; Thomakos, N.; Haidopoulos, D.; et al. VEGF directly suppresses activation of T cells from ovarian cancer patients and healthy individuals via VEGF receptor Type 2. Int. J. Cancer 2012, 130, 857–864. [Google Scholar] [CrossRef]
- Voron, T.; Marcheteau, E.; Pernot, S.; Colussi, O.; Tartour, E.; Taieb, J.; Terme, M. Control of the Immune Response by Pro-Angiogenic Factors. Front. Oncol. 2014, 4, 70. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Pan, P.-Y.; Li, Q.; Sato, A.I.; Levy, D.E.; Bromberg, J.; Divino, C.M.; Chen, S.-H. Gr-1+CD115+ Immature Myeloid Suppressor Cells Mediate the Development of Tumor-Induced T Regulatory Cells and T-Cell Anergy in Tumor-Bearing Host. Cancer Res. 2006, 66, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Gavalas, N.G.; Tsiatas, M.; Tsitsilonis, O.; Politi, E.; Ioannou, K.; Ziogas, A.C.; Rodolakis, A.; Vlahos, G.; Thomakos, N.; Haidopoulos, D.; et al. VEGF directly suppresses activation of T cells from ascites secondary to ovarian cancer via VEGF receptor type 2. Br. J. Cancer 2012, 107, 1869–1875. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.R.; Bowen, K.A.; Rychahou, P.G.; Jackson, L.N.; Weiss, H.L.; Lee, E.Y.; Townsend, C.M., Jr.; Evers, B.M. VEGFR-2 expression in carcinoid cancer cells and its role in tumor growth and metastasis. Int. J. Cancer 2011, 128, 1045–1056. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Seng, S.; Sekine, M.; Hinton, C.; Fu, Y.; Avraham, H.K.; Avraham, S. Vascular endothelial growth factor mediates intracrine survival in human breast carcinoma cells through internally expressed VEGFR1/FLT1. PLoS Med. 2007, 4, e186. [Google Scholar] [CrossRef]
- Chung, G.G.; Yoon, H.H.; Zerkowski, M.P.; Ghosh, S.; Thomas, L.; Harigopal, M.; Charette, L.A.; Salem, R.R.; Camp, R.L.; Rimm, D.L.; et al. Vascular endothelial growth factor, FLT-1, and FLK-1 analysis in a pancreatic cancer tissue microarray. Cancer 2006, 106, 1677–1684. [Google Scholar] [CrossRef]
- Sampurna, C.; Heukamp, L.C.; Siobal, M.; Schöttle, J.; Wieczorek, C.; Peifer, M.; Frasca, D.; Koker, M.; König, K.; Meder, L.; et al. Tumor vegf:vegfr2 autocrine feed-forward loop triggers angiogenesis in lung cancer. J. Clin. Investig. 2013, 123, 1732–1740. [Google Scholar] [CrossRef]
- Yang, F.; Tang, X.; Riquelme, E.; Behrens, C.; Nilsson, M.B.; Giri, U.; Varella-Garcia, M.; Byers, L.A.; Lin, H.Y.; Wang, J.; et al. Increased VEGFR-2 gene copy is associated with chemoresistance and shorter survival in patients with non-small-cell lung carcinoma who receive adjuvant chemotherapy. Cancer Res. 2011, 71, 5512–5521. [Google Scholar] [CrossRef]
- Ferrara, N.; Adamis, A.P. Ten years of anti-vascular endothelial growth factor therapy. Nat. Rev. Drug Discov. 2016, 15, 385–403. [Google Scholar] [CrossRef]
- Reck, M.; Kaiser, R.; Mellemgaard, A.; Douillard, J.Y.; Orlov, S.; Krzakowski, M.; von Pawel, J.; Gottfried, M.; Bondarenko, I.; Liao, M.; et al. Docetaxel plus nintedanib versus docetaxel plus placebo in patients with previously treated non-small-cell lung cancer (LUME-Lung 1): A phase 3, double-blind, randomised controlled trial. Lancet Oncol. 2014, 15, 143–155. [Google Scholar] [CrossRef]
- Garon, E.B.; Ciuleanu, T.E.; Arrieta, O.; Prabhash, K.; Syrigos, K.N.; Goksel, T.; Park, K.; Gorbunova, V.; Kowalyszyn, R.D.; Pikiel, J.; et al. Ramucirumab plus docetaxel versus placebo plus docetaxel for second-line treatment of stage IV non-small-cell lung cancer after disease progression on platinum-based therapy (REVEL): A multicentre, double-blind, randomised phase 3 trial. Lancet 2014, 384, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Makhov, P.; Bychkov, I.; Faezov, B.; Deneka, A.; Kudinov, A.; Nicolas, E.; Brebion, R.; Avril, E.; Cai, K.Q.; Kharin, L.V.; et al. Musashi-2 (MSI2) regulates epidermal growth factor receptor (EGFR) expression and response to EGFR inhibitors in EGFR-mutated non-small cell lung cancer (NSCLC). Oncogenesis 2021, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Kudinov, A.E.; Deneka, A.; Nikonova, A.S.; Beck, T.N.; Ahn, Y.-H.; Liu, X.; Martinez, C.F.; Schultz, F.A.; Reynolds, S.; Yang, D.-H.; et al. Musashi-2 (MSI2) supports TGF-β signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, 6955–6960. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.G.; Riemondy, K.; Chapnick, D.A.; Bunker, E.; Liu, X.; Kuersten, S.; Yi, R. Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration. Nucleic Acids Res. 2016, 44, 3788–3800. [Google Scholar] [CrossRef]
- Wang, S.; Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Parada, K.; Rao, S.; Minuesa, G.; Katz, Y.; Gregory, B.D.; et al. Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat. Commun. 2015, 6, 6517. [Google Scholar] [CrossRef]
- Nguyen, D.T.T.; Lu, Y.; Chu, K.L.; Yang, X.; Park, S.-M.; Choo, Z.-N.; Chin, C.R.; Prieto, C.; Schurer, A.; Barin, E.; et al. HyperTRIBE uncovers increased MUSASHI-2 RNA binding activity and differential regulation in leukemic stem cells. Nat. Commun. 2020, 11, 2026. [Google Scholar] [CrossRef]
- Fox, R.G.; Park, F.D.; Koechlein, C.S.; Kritzik, M.; Reya, T. Musashi Signaling in Stem Cells and Cancer. Annu. Rev. Cell Dev. Biol. 2015, 31, 249–267. [Google Scholar] [CrossRef]
- Kudinov, A.E.; Karanicolas, J.; Golemis, E.A.; Boumber, Y. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clin. Cancer Res. 2017, 23, 2143–2153. [Google Scholar] [CrossRef]
- Vu, L.P.; Prieto, C.; Amin, E.M.; Chhangawala, S.; Krivtsov, A.; Calvo-Vidal, M.N.; Chou, T.; Chow, A.; Minuesa, G.; Park, S.M.; et al. Functional screen of MSI2 interactors identifies an essential role for SYNCRIP in myeloid leukemia stem cells. Nat. Genet. 2017, 49, 866–875. [Google Scholar] [CrossRef]
- Palacios, F.; Yan, X.J.; Ferrer, G.; Chen, S.S.; Vergani, S.; Yang, X.; Gardner, J.; Barrientos, J.C.; Rock, P.; Burack, R.; et al. Musashi 2 influences chronic lymphocytic leukemia cell survival and growth making it a potential therapeutic target. Leukemia 2021, 35, 1037–1052. [Google Scholar] [CrossRef]
- Kharas, M.G.; Lengner, C.J.; Al-Shahrour, F.; Bullinger, L.; Ball, B.; Zaidi, S.; Morgan, K.; Tam, W.; Paktinat, M.; Okabe, R.; et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010, 16, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Kharin, L.; Bychkov, I.; Karnaukhov, N.; Voloshin, M.; Fazliyeva, R.; Deneka, A.; Frantsiyants, E.; Kit, O.; Golemis, E.; Boumber, Y. Prognostic role and biologic features of Musashi-2 expression in colon polyps and during colorectal cancer progression. PLoS ONE 2021, 16, e0252132. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, D.L.; Lin, W.; Creighton, C.J.; Rizvi, Z.H.; Gregory, P.A.; Goodall, G.J.; Thilaganathan, N.; Du, L.; Zhang, Y.; Pertsemlidis, A.; et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009, 23, 2140–2151. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, E.; Suraokar, M.; Behrens, C.; Lin, H.Y.; Girard, L.; Nilsson, M.B.; Simon, G.; Wang, J.; Coombes, K.R.; Lee, J.J.; et al. VEGF/VEGFR-2 upregulates EZH2 expression in lung adenocarcinoma cells and EZH2 depletion enhances the response to platinum-based and VEGFR-2-targeted therapy. Clin. Cancer Res. 2014, 20, 3849–3861. [Google Scholar] [CrossRef]
- Chen, T.T.; Filvaroff, E.; Peng, J.; Marsters, S.; Jubb, A.; Koeppen, H.; Merchant, M.; Ashkenazi, A. MET Suppresses Epithelial VEGFR2 via Intracrine VEGF-induced Endoplasmic Reticulum-associated Degradation. EBioMedicine 2015, 2, 406–420. [Google Scholar] [CrossRef]
- Park, S.-M.; Deering, R.P.; Lu, Y.; Tivnan, P.; Lianoglou, S.; Al-Shahrour, F.; Ebert, B.L.; Hacohen, N.; Leslie, C.; Daley, G.Q.; et al. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J. Exp. Med. 2014, 211, 71–87. [Google Scholar] [CrossRef]
- Sheng, W.; Shi, X.; Lin, Y.; Tang, J.; Jia, C.; Cao, R.; Sun, J.; Wang, G.; Zhou, L.; Dong, M. Musashi2 promotes EGF-induced EMT in pancreatic cancer via ZEB1-ERK/MAPK signaling. J. Exp. Clin. Cancer Res. 2020, 39, 16. [Google Scholar] [CrossRef]
- Park, S.M.; Gönen, M.; Vu, L.; Minuesa, G.; Tivnan, P.; Barlowe, T.S.; Taggart, J.; Lu, Y.; Deering, R.P.; Hacohen, N.; et al. Musashi2 sustains the mixed-lineage leukemia-driven stem cell regulatory program. J. Clin. Investig. 2015, 125, 1286–1298. [Google Scholar] [CrossRef]
- Suzawa, K.; Offin, M.; Lu, D.; Kurzatkowski, C.; Vojnic, M.; Smith, R.S.; Sabari, J.K.; Tai, H.; Mattar, M.; Khodos, I.; et al. Activation of KRAS Mediates Resistance to Targeted Therapy in MET Exon 14–mutant Non–small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 1248–1260. [Google Scholar] [CrossRef]
- Brandt, R.; Sell, T.; Lüthen, M.; Uhlitz, F.; Klinger, B.; Riemer, P.; Giesecke-Thiel, C.; Schulze, S.; El-Shimy, I.A.; Kunkel, D.; et al. Cell type-dependent differential activation of ERK by oncogenic KRAS in colon cancer and intestinal epithelium. Nat. Commun. 2019, 10, 2919. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).