The Role of MicroRNAs in Uterine Leiomyosarcoma Diagnosis and Treatment
Abstract
Simple Summary
Abstract
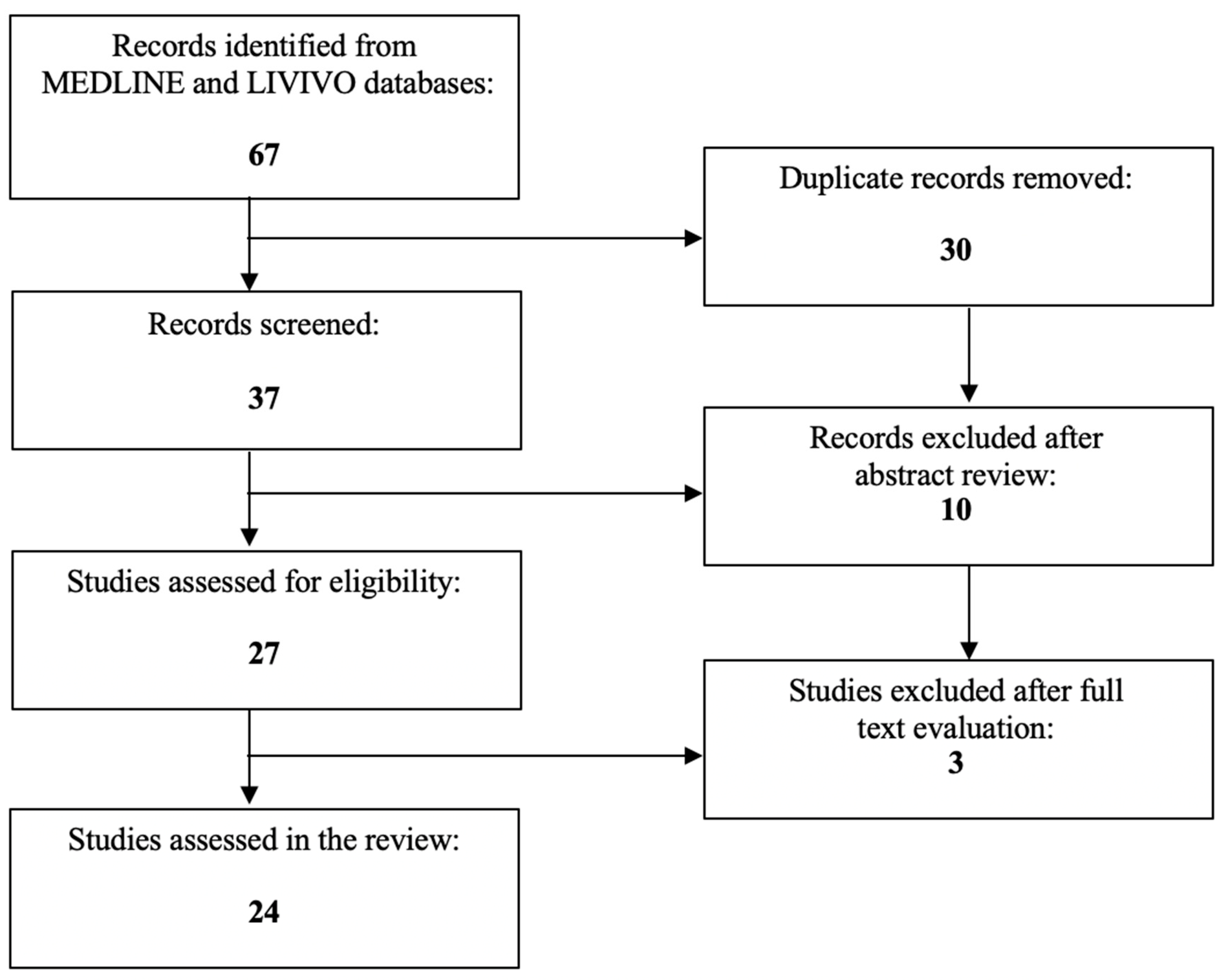
1. Introduction
2. The Expression of miRNAs in Uterine Sarcoma
2.1. Differential Expression of miRNAs in Uterine Leiomyosarcoma Cell Lines
2.2. Different miRNA Profiles between Uterine Leiomyosarcomas versus Normal Uteri and Benign Uterine Tumors
2.3. Other Smooth Muscle Neoplasms versus Leiomyosarcoma
2.4. Endometrial Stromal Sarcoma versus Leiomyosarcoma
2.5. Undifferentiated Pleomorphic Sarcoma versus Leiomyosarcoma
2.6. miRNAs Correlate with Prognosis of Uterine Leiomyosarcomas and Could Predict Response to Treatment
3. Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- American Cancer Society. What Is Uterine Sarcoma? American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- American Cancer Society. Key Statistics for Uterine Sarcoma; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- American Cancer Society. Risk Factors for Uterine Sarcoma; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Trope, C.G.; Abeler, V.M.; Kristensen, G.B. Diagnosis and treatment of sarcoma of the uterus. A review. Acta Oncol. 2012, 51, 694–705. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Signs and Symptoms of Uterine Sarcomas; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- American Cancer Society. Tests for Uterine Sarcoma; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Liu, J.; Wang, Z. Advances in the Preoperative Identification of Uterine Sarcoma. Cancers 2022, 14, 3517. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Treatment for Uterine Sarcoma, by Type and Stage; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Bose, S.; Schwartz, G.K.; Ingham, M. Novel Therapeutics in the Treatment of Uterine Sarcoma. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Holmes, B. Identifying New Biomarkers and Targets in Uterine Sarcomas. Target. Ther. Oncol. 2022, 11. Available online: https://www.targetedonc.com/view/identifying-new-biomarkers-and-targets-in-uterine-sarcomas (accessed on 19 April 2023).
- Zhang, X.; Wei, C.; Liang, H.; Han, L. Polo-Like Kinase 4’s Critical Role in Cancer Development and Strategies for Plk4-Targeted Therapy. Front. Oncol. 2021, 11, 587554. [Google Scholar] [CrossRef]
- Gutierrez, A.; Demond, H.; Brebi, P.; Ili, C.G. Novel Methylation Biomarkers for Colorectal Cancer Prognosis. Biomolecules 2021, 11, 1722. [Google Scholar] [CrossRef]
- Roy, S.K.; Shrivastava, A.; Srivastav, S.; Shankar, S.; Srivastava, R.K. SATB2 is a novel biomarker and therapeutic target for cancer. J. Cell. Mol. Med. 2020, 24, 11064–11069. [Google Scholar] [CrossRef] [PubMed]
- Shang, B.; Liu, Y.; Jiang, S.J.; Liu, Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: A systematic review and meta-analysis. Sci. Rep. 2015, 5, 15179. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Iftikhar, H.; Carney, G.E. Evidence and potential in vivo functions for biofluid miRNAs: From expression profiling to functional testing: Potential roles of extracellular miRNAs as indicators of physiological change and as agents of intercellular information exchange. Bioessays 2016, 38, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef]
- Hu, Y.; Rao, S.S.; Wang, Z.X.; Cao, J.; Tan, Y.J.; Luo, J.; Li, H.M.; Zhang, W.S.; Chen, C.Y.; Xie, H. Exosomes from human umbilical cord blood accelerate cutaneous wound healing through miR-21-3p-mediated promotion of angiogenesis and fibroblast function. Theranostics 2018, 8, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M. MicroRNAs and miRceptors: A new mechanism of action for intercellular communication. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 1737. [Google Scholar] [CrossRef]
- Lehmann, S.M.; Kruger, C.; Park, B.; Derkow, K.; Rosenberger, K.; Baumgart, J.; Trimbuch, T.; Eom, G.; Hinz, M.; Kaul, D.; et al. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012, 15, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef]
- Zhu, J.J.; Liu, Y.F.; Zhang, Y.P.; Zhao, C.R.; Yao, W.J.; Li, Y.S.; Wang, K.C.; Huang, T.S.; Pang, W.; Wang, X.F.; et al. VAMP3 and SNAP23 mediate the disturbed flow-induced endothelial microRNA secretion and smooth muscle hyperplasia. Proc. Natl. Acad. Sci. USA 2017, 114, 8271–8276. [Google Scholar] [CrossRef]
- Gumurdu, A.; Yildiz, R.; Eren, E.; Karakulah, G.; Unver, T.; Genc, S.; Park, Y. MicroRNA exocytosis by large dense-core vesicle fusion. Sci. Rep. 2017, 7, 45661. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Carpenter, K.J.; Berry, W.L.; Janknecht, R.; Dooley, W.C.; Ding, W.Q. Exosome-mediated microRNA signaling from breast cancer cells is altered by the anti-angiogenesis agent docosahexaenoic acid (DHA). Mol. Cancer 2015, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Liu, Y.; Huang, J.D.; Song, E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Tufekci, K.U.; Meuwissen, R.L.; Genc, S. The role of microRNAs in biological processes. Methods Mol. Biol. 2014, 1107, 15–31. [Google Scholar] [CrossRef]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.S.; Malakar, A.K.; Chakraborty, S. Interplay between miRNAs and human diseases. J. Cell. Physiol. 2018, 233, 2007–2018. [Google Scholar] [CrossRef] [PubMed]
- Filipow, S.; Laczmanski, L. Blood Circulating miRNAs as Cancer Biomarkers for Diagnosis and Surgical Treatment Response. Front. Genet. 2019, 10, 169. [Google Scholar] [CrossRef] [PubMed]
- Duica, F.; Condrat, C.E.; Danila, C.A.; Boboc, A.E.; Radu, M.R.; Xiao, J.; Li, X.; Cretoiu, S.M.; Suciu, N.; Cretoiu, D.; et al. MiRNAs: A Powerful Tool in Deciphering Gynecological Malignancies. Front. Oncol. 2020, 10, 591181. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Luo, X.; Panda, H.; Chegini, N. miR-93/106b and their host gene, MCM7, are differentially expressed in leiomyomas and functionally target F3 and IL-8. Mol. Endocrinol. 2012, 26, 1028–1042. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Panda, H.; Luo, X.; Chegini, N. miR-200c is aberrantly expressed in leiomyomas in an ethnic-dependent manner and targets ZEBs, VEGFA, TIMP2, and FBLN5. Endocr. Relat. Cancer 2012, 19, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Ho, M.; Khorram, O. The regulatory function of miR-200c on inflammatory and cell-cycle associated genes in SK-LMS-1, a leiomyosarcoma cell line. Reprod. Sci. 2015, 22, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Perle, M.A.; Mittal, K.; Chen, H.; Zou, X.; Narita, M.; Hernando, E.; Lee, P.; Wei, J.J. Let-7 repression leads to HMGA2 overexpression in uterine leiomyosarcoma. J. Cell. Mol. Med. 2009, 13, 3898–3905. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Bariani, M.V.; Falahati, A.; Khosh, A.; Lastra, R.R.; Siblini, H.; Boyer, T.G.; Al-Hendy, A. The Functional Role and Regulatory Mechanism of Bromodomain-Containing Protein 9 in Human Uterine Leiomyosarcoma. Cells 2022, 11, 2160. [Google Scholar] [CrossRef] [PubMed]
- Benna, C.; Rajendran, S.; Rastrelli, M.; Mocellin, S. miRNA deregulation targets specific pathways in leiomyosarcoma development: An in silico analysis. J. Transl. Med. 2019, 17, 153. [Google Scholar] [CrossRef]
- Danielson, L.S.; Menendez, S.; Attolini, C.S.; Guijarro, M.V.; Bisogna, M.; Wei, J.; Socci, N.D.; Levine, D.A.; Michor, F.; Hernando, E. A differentiation-based microRNA signature identifies leiomyosarcoma as a mesenchymal stem cell-related malignancy. Am. J. Pathol. 2010, 177, 908–917. [Google Scholar] [CrossRef]
- Kowalewska, M.; Bakula-Zalewska, E.; Chechlinska, M.; Goryca, K.; Nasierowska-Guttmejer, A.; Danska-Bidzinska, A.; Bidzinski, M. microRNAs in uterine sarcomas and mixed epithelial-mesenchymal uterine tumors: A preliminary report. Tumour. Biol. 2013, 34, 2153–2160. [Google Scholar] [CrossRef]
- Nuovo, G.J.; Schmittgen, T.D. Benign metastasizing leiomyoma of the lung: Clinicopathologic, immunohistochemical, and micro-RNA analyses. Diagn. Mol. Pathol. 2008, 17, 145–150. [Google Scholar] [CrossRef]
- Renner, M.; Czwan, E.; Hartmann, W.; Penzel, R.; Brors, B.; Eils, R.; Wardelmann, E.; Buttner, R.; Lichter, P.; Schirmacher, P.; et al. MicroRNA profiling of primary high-grade soft tissue sarcomas. Genes Chromosomes Cancer 2012, 51, 982–996. [Google Scholar] [CrossRef]
- Stope, M.B.; Cernat, V.; Kaul, A.; Diesing, K.; Koensgen, D.; Burchardt, M.; Mustea, A. Functionality of the Tumor Suppressor microRNA-1 in Malignant Tissue and Cell Line Cells of Uterine Leiomyosarcoma. Anticancer Res. 2018, 38, 1547–1550. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, A.; Matsuzaki, J.; Yamamoto, Y.; Tate, K.; Yoneoka, Y.; Shimizu, H.; Uehara, T.; Ishikawa, M.; Takizawa, S.; Aoki, Y.; et al. Serum microRNA profile enables preoperative diagnosis of uterine leiomyosarcoma. Cancer Sci. 2019, 110, 3718–3726. [Google Scholar] [CrossRef] [PubMed]
- Ventura, L.; Gnetti, L.; Rossi, M.; Tiseo, M.; Giordano, G.; Corradi, M.; Silva, M.; Milanese, G.; Minari, R.; Leonetti, A.; et al. The role of miRNA-221 and miRNA-126 in patients with benign metastasizing leiomyoma of the lung: An overview with new interesting scenarios. Mol. Biol. Rep. 2021, 48, 3485–3494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ubago, J.; Li, L.; Guo, H.; Liu, Y.; Qiang, W.; Kim, J.J.; Kong, B.; Wei, J.J. Molecular analyses of 6 different types of uterine smooth muscle tumors: Emphasis in atypical leiomyoma. Cancer 2014, 120, 3165–3177. [Google Scholar] [CrossRef] [PubMed]
- Ravid, Y.; Formanski, M.; Smith, Y.; Reich, R.; Davidson, B. Uterine leiomyosarcoma and endometrial stromal sarcoma have unique miRNA signatures. Gynecol. Oncol. 2016, 140, 512–517. [Google Scholar] [CrossRef]
- Guled, M.; Pazzaglia, L.; Borze, I.; Mosakhani, N.; Novello, C.; Benassi, M.S.; Knuutila, S. Differentiating soft tissue leiomyosarcoma and undifferentiated pleomorphic sarcoma: A miRNA analysis. Genes Chromosomes Cancer 2014, 53, 693–702. [Google Scholar] [CrossRef]
- de Almeida, B.C.; Garcia, N.; Maffazioli, G.; dos Anjos, L.G.; Baracat, E.C.; Carvalho, K.C. Oncomirs Expression Profiling in Uterine Leiomyosarcoma Cells. Int. J. Mol. Sci. 2017, 19, 52. [Google Scholar] [CrossRef]
- de Almeida, B.C.; Dos Anjos, L.G.; Uno, M.; Cunha, I.W.D.; Soares, F.A.; Baiocchi, G.; Baracat, E.C.; Carvalho, K.C. Let-7 miRNA’s Expression Profile and Its Potential Prognostic Role in Uterine Leiomyosarcoma. Cells 2019, 8, 1452. [Google Scholar] [CrossRef]
- Gonzalez Dos Anjos, L.; de Almeida, B.C.; Gomes de Almeida, T.; Mourao Lavorato Rocha, A.; De Nardo Maffazioli, G.; Soares, F.A.; Werneck da Cunha, I.; Baracat, E.C.; Carvalho, K.C. Could miRNA Signatures be Useful for Predicting Uterine Sarcoma and Carcinosarcoma Prognosis and Treatment? Cancers 2018, 10, 315. [Google Scholar] [CrossRef]
- Schiavon, B.N.; Carvalho, K.C.; Coutinho-Camillo, C.M.; Baiocchi, G.; Valieris, R.; Drummond, R.; da Silva, I.T.; De Brot, L.; Soares, F.A.; Werneck da Cunha, I. miRNAs 144-3p, 34a-5p, and 206 are a useful signature for distinguishing uterine leiomyosarcoma from other smooth muscle tumors. Surg. Exp. Pathol. 2019, 2, 5. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Electronic address, e.d.s.c.; Cancer Genome Atlas Research, N. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 2017, 171, 950–965.e928. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Wang, X.; Wang, C.; Li, L. Elevated levels of serum MiR-152 and miR-24 in uterine sarcoma: Potential for inducing autophagy via SIRT1 and deacetylated LC3. Br. J. Biomed. Sci. 2018, 75, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Wiemer, E.A.C.; Wozniak, A.; Burger, H.; Smid, M.; Floris, G.; Nzokirantevye, A.; Sciot, R.; Sleijfer, S.; Schoffski, P. Identification of microRNA biomarkers for response of advanced soft tissue sarcomas to eribulin: Translational results of the EORTC 62052 trial. Eur. J. Cancer 2017, 75, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Schoffski, P.; Chawla, S.; Maki, R.G.; Italiano, A.; Gelderblom, H.; Choy, E.; Grignani, G.; Camargo, V.; Bauer, S.; Rha, S.Y.; et al. Eribulin versus dacarbazine in previously treated patients with advanced liposarcoma or leiomyosarcoma: A randomised, open-label, multicentre, phase 3 trial. Lancet 2016, 387, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Blay, J.Y.; Schoffski, P.; Bauer, S.; Krarup-Hansen, A.; Benson, C.; D’Adamo, D.R.; Jia, Y.; Maki, R.G. Eribulin versus dacarbazine in patients with leiomyosarcoma: Subgroup analysis from a phase 3, open-label, randomised study. Br. J. Cancer 2019, 120, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Survival Rates for Uterine Sarcoma; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Desar, I.M.E.; Ottevanger, P.B.; Benson, C.; van der Graaf, W.T.A. Systemic treatment in adult uterine sarcomas. Crit. Rev. Oncol. Hematol. 2018, 122, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Loganantharaj, R.; Randall, T.A. The Limitations of Existing Approaches in Improving MicroRNA Target Prediction Accuracy. Methods Mol. Biol. 2017, 1617, 133–158. [Google Scholar] [CrossRef]
- Gillespie, P.; Ladame, S.; O’Hare, D. Molecular methods in electrochemical microRNA detection. Analyst 2018, 144, 114–129. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef] [PubMed]
| miRNA | Expression Level | Associated Gene | Reference |
|---|---|---|---|
| miRNA-93/106b | Gain of function | F3 | [37] |
| IL8 | |||
| CTGF | |||
| PAI1 | |||
| miRNA-200c | Gain of function | ZEB1/2 | [38,39] |
| VEGFA | |||
| FBLN5 | |||
| TIMP2 | |||
| IKBKB | |||
| IL8 | |||
| CDK2 | |||
| CCNE2 | |||
| let-7 | Upregulation | HMGA2 | [40] |
| miRNA-4776-5p | Upregulation or downregulation | Genes linked to BRD9 inhibition | [41] |
| miRNA-671-3p | |||
| miRNA-3619-3p | |||
| miRNA-621 | |||
| miRNA-553 | |||
| miRNA-542-5p | |||
| miRNA-4734 | |||
| miRNA-3682-3p | |||
| miRNA-4727-3p |
| miRNA | Differential Expression | Reference |
|---|---|---|
| miRNA-23b | Downregulation in endometrial sarcomas No statistically significant changes between leiomyosarcomas and normal uteri | [44] |
| miRNA-1 | ||
| let-7f | ||
| let-7c | ||
| miRNA-221 | Upregulation in leiomyosarcoma Absence in leiomyoma and benign metastasizing leiomyoma | [45] |
| miRNA-133a | Upregulation in leiomyosarcoma | [46] |
| miRNA-133b | ||
| miRNA-1 | ||
| miRNA-1 | Strong suppression in leiomyosarcoma | [47] |
| miRNA-4430 | Downregulation in leiomyosarcoma | [48] |
| miRNA-6511b-5p | ||
| miRNA-451a | ||
| miRNA-4485-5p | ||
| miRNA-4635 | ||
| miRNA-1246 | ||
| miRNA-191-5p | ||
| miRNA-126 | Downregulation in leiomyosarcoma Hypo-expression between uterine leiomyoma and benign metastasizing leiomyoma | [49] |
| miRNA-221 | Upregulation in leiomyosarcoma | [49] |
| miRNA | Clinical Parameter | Reference |
|---|---|---|
| let-7b | OS DFS Distant metastasis rate | [54] |
| let-7d | ||
| let-7e | ||
| let-7f | ||
| miRNA-196a-5p | CCS DFS Metastasis Tumor relapse | [55] |
| miRNA-34c-5p | ||
| miRNA-125a-5p | ||
| miRNA-10a-5p | ||
| miRNA-373-3p | ||
| miRNA-372-3p | ||
| let-7b-5p | ||
| let-7f-5p | ||
| miRNA-23-3p | ||
| let-7b-5p | ||
| miRNA-138-5p | ||
| miRNA-335-5p | ||
| miRNA-301a-3p | ||
| miRNA-210-3p | ||
| miRNA-146b-5p | ||
| miRNA-218-5p | ||
| miRNA-148a-3p | Tumor relapse Metastasis Patient death DFS | [56] |
| miRNA-27b-3p | ||
| miRNA-124-3p | ||
| miRNA-183-5p | ||
| miRNA135b-5p | ||
| miRNA-181b-5p | RFS | [57] |
| miRNA-152 | Tumor stage Patient survival | [58] |
| miRNA-24 | ||
| miRNA-205 | ||
| miRNA-222 | ||
| miRNA-150 | ||
| miRNA-1271 | Response to eribulin | [59] |
| miRNA-146a | ||
| let-7g | ||
| miRNA-574-3p | ||
| miRNA-362-3p | ||
| miRNA-181a-2 | ||
| miRNA-29b-2 | ||
| miRNA-590-3p |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Psilopatis, I.; Vrettou, K.; Kokkali, S.; Theocharis, S. The Role of MicroRNAs in Uterine Leiomyosarcoma Diagnosis and Treatment. Cancers 2023, 15, 2420. https://doi.org/10.3390/cancers15092420
Psilopatis I, Vrettou K, Kokkali S, Theocharis S. The Role of MicroRNAs in Uterine Leiomyosarcoma Diagnosis and Treatment. Cancers. 2023; 15(9):2420. https://doi.org/10.3390/cancers15092420
Chicago/Turabian StylePsilopatis, Iason, Kleio Vrettou, Stefania Kokkali, and Stamatios Theocharis. 2023. "The Role of MicroRNAs in Uterine Leiomyosarcoma Diagnosis and Treatment" Cancers 15, no. 9: 2420. https://doi.org/10.3390/cancers15092420
APA StylePsilopatis, I., Vrettou, K., Kokkali, S., & Theocharis, S. (2023). The Role of MicroRNAs in Uterine Leiomyosarcoma Diagnosis and Treatment. Cancers, 15(9), 2420. https://doi.org/10.3390/cancers15092420








