Imaging the WHO 2021 Brain Tumor Classification: Fully Automated Analysis of Imaging Features of Newly Diagnosed Gliomas
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Neuropathology and Methylation Analysis
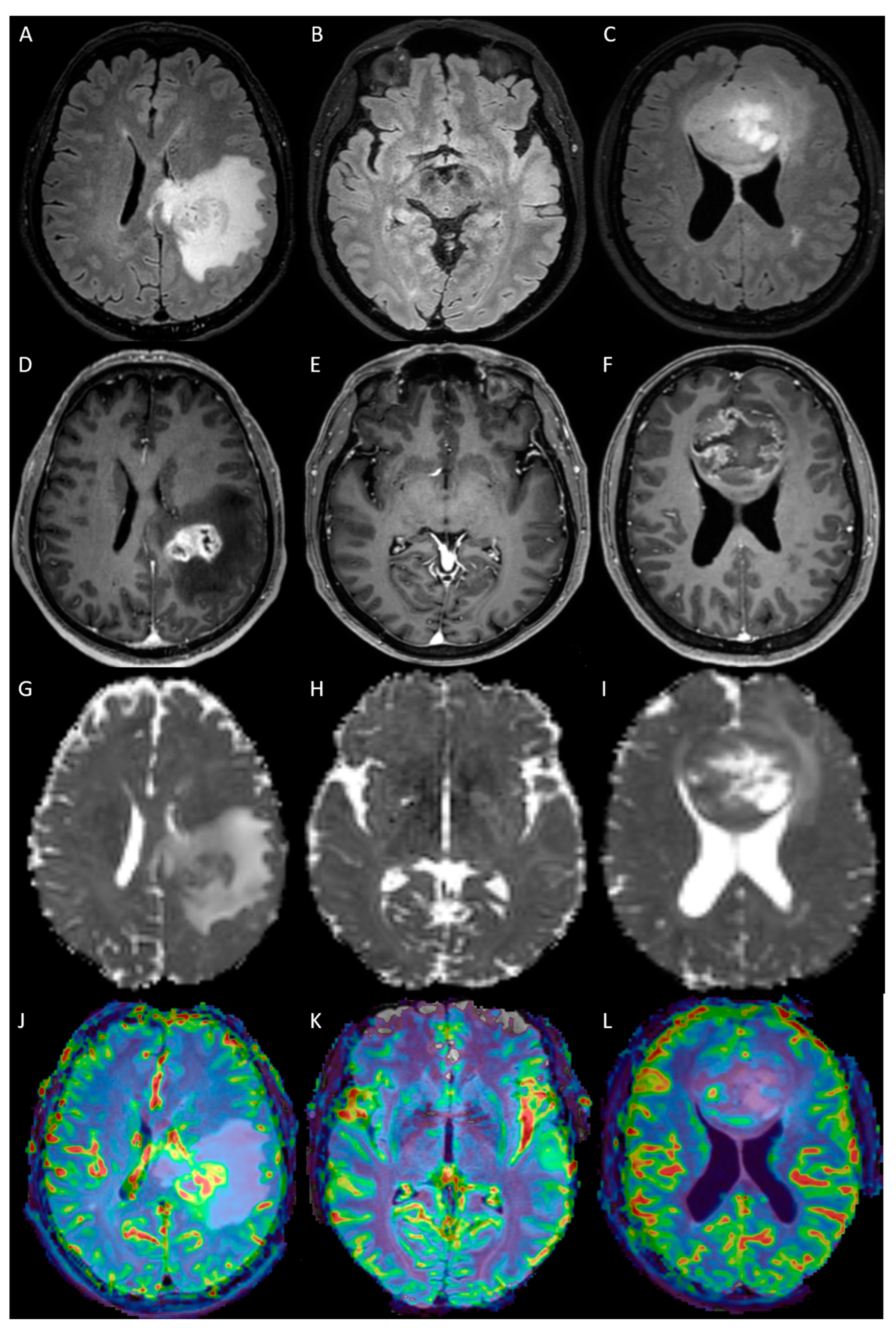
2.3. MRI Analysis
2.4. Statistical Analysis
3. Results
3.1. Characteristics of the Study Population
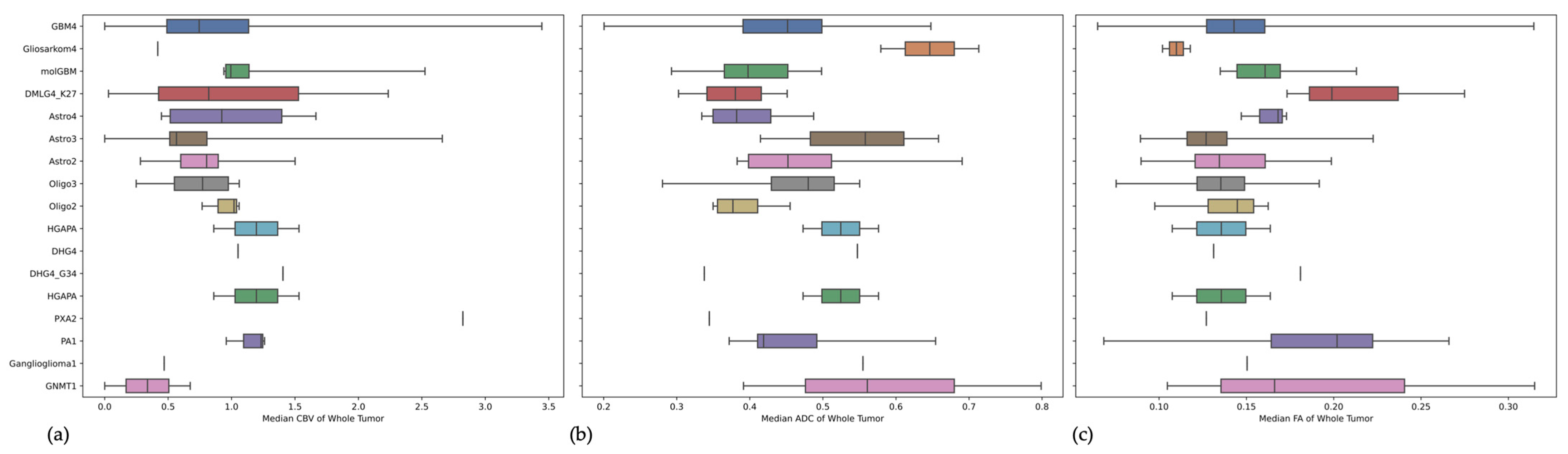
3.2. Comparison of Imaging Metrics between the Three Types of Adult-Type Diffuse Gliomas
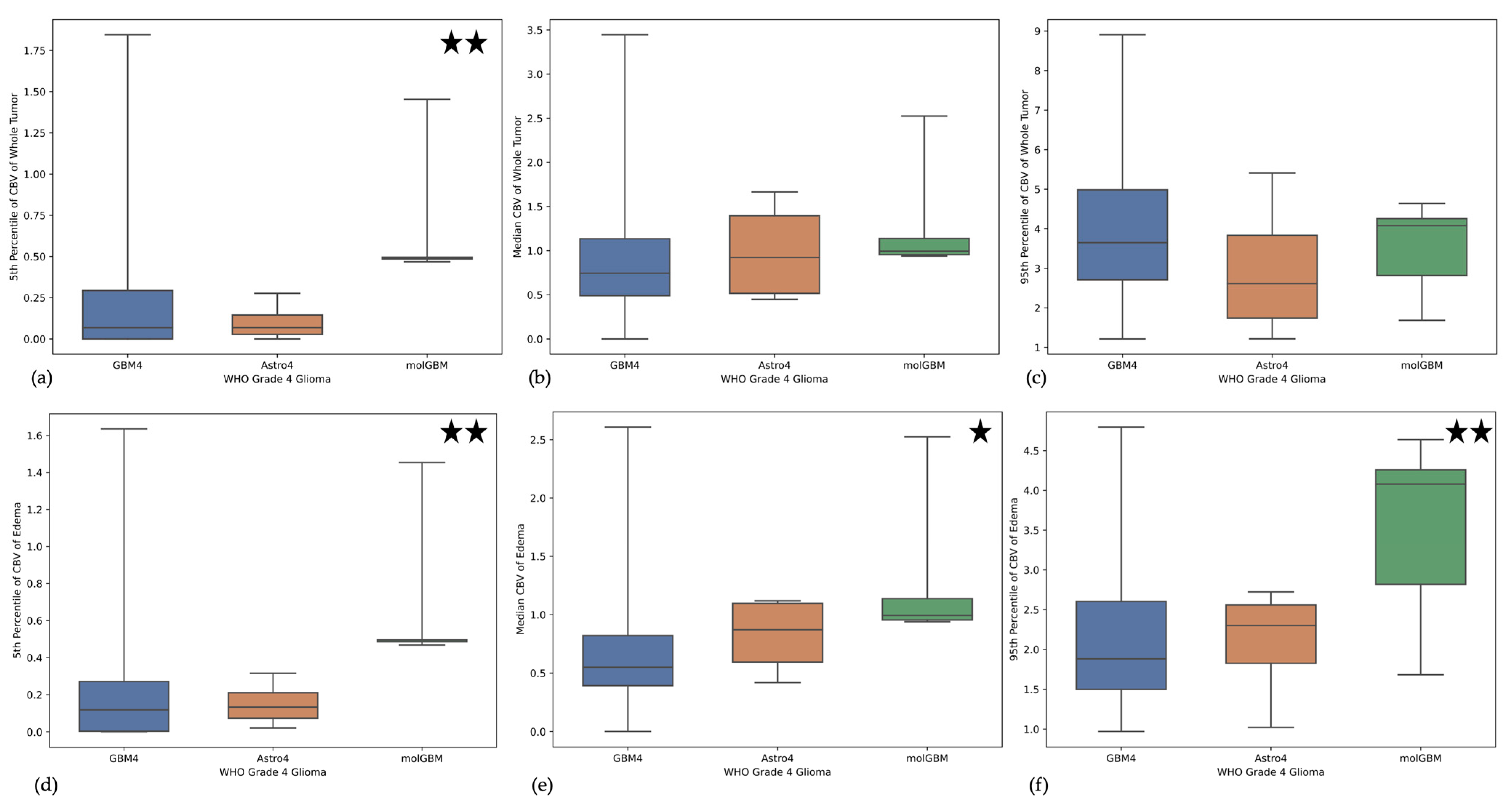
3.3. Imaging the Novel Three Different Types of WHO Grade 4 Gliomas
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.R.; Giannini, C.; Vaubel, R.A.; Morris, J.M.; Eckel, L.J.; Kaufmann, T.J.; Guerin, J.B. A Radiologist’s Guide to the 2021 WHO Central Nervous System Tumor Classification: Part I-Key Concepts and the Spectrum of Diffuse Gliomas. Radiology 2022, 304, 494–508. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xing, Z.; She, D.; Lin, Y.; Zhang, H.; Su, Y.; Cao, D. Grading of IDH-Mutant Astrocytoma Using Diffusion, Susceptibility and Perfusion-Weighted Imaging. BMC Med. Imaging 2022, 22, 105. [Google Scholar] [CrossRef]
- Lasocki, A.; Anjari, M.; Ӧrs Kokurcan, S.; Thust, S.C. Conventional MRI Features of Adult Diffuse Glioma Molecular Subtypes: A Systematic Review. Neuroradiology 2021, 63, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Delfanti, R.L.; Piccioni, D.E.; Handwerker, J.; Bahrami, N.; Krishnan, A.; Karunamuni, R.; Hattangadi-Gluth, J.A.; Seibert, T.M.; Srikant, A.; Jones, K.A.; et al. Imaging Correlates for the 2016 Update on WHO Classification of Grade II/III Gliomas: Implications for IDH, 1p/19q and ATRX Status. J. Neurooncol. 2017, 135, 601–609. [Google Scholar] [CrossRef]
- Joyner, D.A.; Garrett, J.; Batchala, P.P.; Rama, B.; Ravicz, J.R.; Patrie, J.T.; Lopes, M.-B.; Fadul, C.E.; Schiff, D.; Jain, R.; et al. MRI Features Predict Tumor Grade in Isocitrate Dehydrogenase (IDH)-Mutant Astrocytoma and Oligodendroglioma. Neuroradiology 2023, 65, 121–129. [Google Scholar] [CrossRef]
- Hilario, A.; Ramos, A.; Perez-Nuñez, A.; Salvador, E.; Millan, J.M.; Lagares, A.; Sepulveda, J.M.; Gonzalez-Leon, P.; Hernandez-Lain, A.; Ricoy, J.R. The Added Value of Apparent Diffusion Coefficient to Cerebral Blood Volume in the Preoperative Grading of Diffuse Gliomas. AJNR Am. J. Neuroradiol. 2012, 33, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Jain, R.; Radmanesh, A.; Poisson, L.M.; Guo, W.-Y.; Zagzag, D.; Snuderl, M.; Placantonakis, D.G.; Golfinos, J.; Chi, A.S. Predicting Genotype and Survival in Glioma Using Standard Clinical MR Imaging Apparent Diffusion Coefficient Images: A Pilot Study from The Cancer Genome Atlas. AJNR Am. J. Neuroradiol. 2018, 39, 1814–1820. [Google Scholar] [CrossRef]
- Ene, C.I.; Fine, H.A. Many Tumors in One: A Daunting Therapeutic Prospect. Cancer Cell 2011, 20, 695–697. [Google Scholar] [CrossRef]
- 10 Hu, L.S.; Ning, S.; Eschbacher, J.M.; Baxter, L.C.; Gaw, N.; Ranjbar, S.; Plasencia, J.; Dueck, A.C.; Peng, S.; Smith, K.A.; et al. Radiogenomics to Characterize Regional Genetic Heterogeneity in Glioblastoma. Neuro Oncol. 2017, 19, 128–137. [Google Scholar] [CrossRef]
- Barajas, R.F.; Hodgson, J.G.; Chang, J.S.; Vandenberg, S.R.; Yeh, R.-F.; Parsa, A.T.; McDermott, M.W.; Berger, M.S.; Dillon, W.P.; Cha, S. Glioblastoma Multiforme Regional Genetic and Cellular Expression Patterns: Influence on Anatomic and Physiologic MR Imaging. Radiology 2010, 254, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA Methylation-Based Classification of Central Nervous System Tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Capper, D.; Stichel, D.; Sahm, F.; Jones, D.T.W.; Schrimpf, D.; Sill, M.; Schmid, S.; Hovestadt, V.; Reuss, D.E.; Koelsche, C.; et al. Practical Implementation of DNA Methylation and Copy-Number-Based CNS Tumor Diagnostics: The Heidelberg Experience. Acta Neuropathol. 2018, 136, 181–210. [Google Scholar] [CrossRef] [PubMed]
- Rohlfing, T.; Zahr, N.M.; Sullivan, E.V.; Pfefferbaum, A. The SRI24 Multichannel Atlas of Normal Adult Human Brain Structure. Hum. Brain Mapp. 2010, 31, 798–819. [Google Scholar] [CrossRef]
- Modat, M.; Cash, D.M.; Daga, P.; Winston, G.P.; Duncan, J.S.; Ourselin, S. Global Image Registration Using a Symmetric Block-Matching Approach. J. Med. Imaging 2014, 1, 024003. [Google Scholar] [CrossRef]
- Isensee, F.; Schell, M.; Pflueger, I.; Brugnara, G.; Bonekamp, D.; Neuberger, U.; Wick, A.; Schlemmer, H.-P.; Heiland, S.; Wick, W.; et al. Automated Brain Extraction of Multisequence MRI Using Artificial Neural Networks. Hum. Brain Mapp. 2019, 40, 4952–4964. [Google Scholar] [CrossRef] [PubMed]
- Kofler, F.; Berger, C.; Waldmannstetter, D.; Lipkova, J.; Ezhov, I.; Tetteh, G.; Kirschke, J.; Zimmer, C.; Wiestler, B.; Menze, B.H. BraTS Toolkit: Translating BraTS Brain Tumor Segmentation Algorithms Into Clinical and Scientific Practice. Front. Neurosci. 2020, 14, 125. [Google Scholar] [CrossRef]
- Thomas, M.F.; Kofler, F.; Grundl, L.; Finck, T.; Li, H.; Zimmer, C.; Menze, B.; Wiestler, B. Improving Automated Glioma Segmentation in Routine Clinical Use Through Artificial Intelligence-Based Replacement of Missing Sequences With Synthetic Magnetic Resonance Imaging Scans. Investig. Radiol. 2022, 57, 187–193. [Google Scholar] [CrossRef]
- Garyfallidis, E.; Brett, M.; Amirbekian, B.; Rokem, A.; Van Der Walt, S.; Descoteaux, M.; Nimmo-Smith, I. Dipy, a Library for the Analysis of Diffusion MRI Data. Front. Neuroinf. 2014, 8, 8. [Google Scholar] [CrossRef]
- Arzanforoosh, F.; Croal, P.L.; van Garderen, K.A.; Smits, M.; Chappell, M.A.; Warnert, E.A.H. Effect of Applying Leakage Correction on RCBV Measurement Derived From DSC-MRI in Enhancing and Nonenhancing Glioma. Front. Oncol. 2021, 11, 648528. [Google Scholar] [CrossRef]
- Avants, B.B.; Tustison, N.J.; Wu, J.; Cook, P.A.; Gee, J.C. An Open Source Multivariate Framework for N-Tissue Segmentation with Evaluation on Public Data. Neuroinformatics 2011, 9, 381–400. [Google Scholar] [CrossRef]
- Qin, J.-B.; Zhang, H.; Wang, X.-C.; Tan, Y.; Wu, X.-F. Combination Value of Diffusion-Weighted Imaging and Dynamic Susceptibility Contrast-Enhanced MRI in Astrocytoma Grading and Correlation with GFAP, Topoisomerase IIα and MGMT. Oncol. Lett. 2019, 18, 2763–2770. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Tan, Y.; Qin, J.; Wu, X.; Wang, L.; Zhang, L. Combined Value of Susceptibility-Weighted and Perfusion-Weighted Imaging in Assessing Who Grade for Brain Astrocytomas. J. Magn. Reson. Imaging 2014, 39, 1569–1574. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Ma, X.; Cheng, K.; Cheng, G.; Li, C.; Lyu, J.; Lan, Y.; Duan, C.; Bian, X.; Zhang, J.; Lou, X. Apparent Diffusion Coefficient as Imaging Biomarker for Identifying IDH Mutation, 1p19q Codeletion, and MGMT Promoter Methylation Status in Patients With Glioma. J. Magn. Reson. Imaging 2023. [Google Scholar] [CrossRef] [PubMed]
- White, M.L.; Zhang, Y.; Yu, F.; Jaffar Kazmi, S.A. Diffusion Tensor MR Imaging of Cerebral Gliomas: Evaluating Fractional Anisotropy Characteristics. AJNR Am. J. Neuroradiol. 2011, 32, 374–381. [Google Scholar] [CrossRef]
- Metz, M.-C.; Molina-Romero, M.; Lipkova, J.; Gempt, J.; Liesche-Starnecker, F.; Eichinger, P.; Grundl, L.; Menze, B.; Combs, S.E.; Zimmer, C.; et al. Predicting Glioblastoma Recurrence from Preoperative MR Scans Using Fractional-Anisotropy Maps with Free-Water Suppression. Cancers 2020, 12, 728. [Google Scholar] [CrossRef]

| Entity | Male | Female | Total |
|---|---|---|---|
| Glioblastomas (IDH wild type) WHO CNS grade 4 | 91 | 56 | 147 |
| Oligodenrogliomas (IDH mutant, 1p/19q codeleted) WHO CNS grade 3 | 10 | 6 | 16 |
| Oligodendrogliomas (IDH mutant, 1p/19q codeleted) WHO CNS grade 2 | 1 | 4 | 5 |
| Astrocytomas (IDH mutant) WHO CNS grade 4 | 1 | 3 | 4 |
| Astrocytomas (IDH mutant) WHO CNS grade 3 (Astro3) | 7 | 4 | 11 |
| Astrocytomas (IDH mutant) WHO CNS grade 2 (Astro2) | 4 | 5 | 9 |
| Pilocytic astrocytomas WHO CNS grade 1 | 6 | 3 | 9 |
| Diffuse gliomas (IDH wild type) with molecular characteristics of glioblastoma WHO CNS grade 4 | 6 | 0 | 6 |
| Gliosarcomas (IDH wild type) WHO CNS grade 4 | 1 | 2 | 3 |
| Glioneural mixed tumors WHO CNS grade 1 | 1 | 2 | 3 |
| Diffuse midline gliomas (H3 K27M mutant) WHO CNS grade 4 | 3 | 0 | 3 |
| High grade astrocytomas with piloid features | 0 | 2 | 2 |
| IDH mutant gliomas WHO CNS grade 2 | 1 | 2 | 3 |
| Diffuse high grade pediatric type glioma WHO CNS grade 4 | 1 | 0 | 1 |
| Diffuse hemispheric glioma; H3 G34- mutant WHO CNS grade 4 | 1 | 0 | 1 |
| Ganglioglioma WHO CNS grade 1 | 0 | 1 | 1 |
| Higher grade glioma; IDH wild type | 1 | 1 | 1 |
| Pleomorphic xanthoastrocytoma WHO CNS grade 2 | 1 | 0 | 1 |
| All gliomas | 136 | 90 | 226 |
| CBV | ADC | FA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5th Percentile | Median | 95th Percentile | 5th Percentile | Median | 95th Percentile | 5th Percentile | Median | 95th Percentile | ||
| Whole Tumor | p-value ANOVA | 0.70068 | 0.37781 | <0.01 | 0.15987 | 0.05027 | 0.41801 | 0.64256 | 0.28855 | 0.38134 |
| p-value Levine | 0.70946 | 0.15586 | 0.05249 | 0.57551 | 0.31109 | 0.80807 | 0.11212 | 0.83200 | 0.59977 | |
| Edema/non-enhancing tumor | p-value ANOVA | 0.98535 | 0.56095 | 0.38532 | 0.70645 | 0.51986 | 0.45611 | 0.01222 | 0.12756 | 0.82143 |
| p-value Levine | 0.98536 | 0.51305 | 0.53941 | 0.86890 | 0.18209 | 0.90134 | 0.37560 | 0.69175 | 0.72108 | |
| CET | p-value ANOVA | 0.03270 | 0.00003 | 0.00026 | 0.15085 | 0.41333 | 0.39038 | 0.81616 | 0.55772 | 0.24551 |
| p-value Levine | 0.01596 | 0.12627 | 0.43610 | 0.04164 | 0.87101 | 0.01402 | 0.43759 | 0.65382 | 0.06869 | |
| CBV | ADC | FA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5th Percentile | Median | 95th Percentile | 5th Percentile | Median | 95th Percentile | 5th Percentile | Median | 95th Percentile | ||
| Whole Tumor | p-value ANOVA | 0.00220 | 0.58685 | 0.40175 | 0.34625 | 0.21793 | 0.02810 | 0.00994 | 0.61366 | 0.57205 |
| p-value Levine | 0.76759 | 0.87670 | 0.68798 | 0.85383 | 0.76440 | 0.46239 | 0.39268 | 0.55937 | 0.52161 | |
| Edema/non-enhancing tumor | p-value ANOVA | 0.00111 | 0.01309 | 0.00295 | 0.28981 | 0.079690 | 0.03295 | 0.81053 | 0.86418 | 0.27737 |
| p-value Levine | 0.77104 | 0.95884 | 0.64370 | 0.94306 | 0.65422 | 0.28295 | 0.36378 | 0.36073 | 0.56071 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Griessmair, M.; Delbridge, C.; Ziegenfeuter, J.; Bernhardt, D.; Gempt, J.; Schmidt-Graf, F.; Kertels, O.; Thomas, M.; Meyer, H.S.; Zimmer, C.; et al. Imaging the WHO 2021 Brain Tumor Classification: Fully Automated Analysis of Imaging Features of Newly Diagnosed Gliomas. Cancers 2023, 15, 2355. https://doi.org/10.3390/cancers15082355
Griessmair M, Delbridge C, Ziegenfeuter J, Bernhardt D, Gempt J, Schmidt-Graf F, Kertels O, Thomas M, Meyer HS, Zimmer C, et al. Imaging the WHO 2021 Brain Tumor Classification: Fully Automated Analysis of Imaging Features of Newly Diagnosed Gliomas. Cancers. 2023; 15(8):2355. https://doi.org/10.3390/cancers15082355
Chicago/Turabian StyleGriessmair, Michael, Claire Delbridge, Julian Ziegenfeuter, Denise Bernhardt, Jens Gempt, Friederike Schmidt-Graf, Olivia Kertels, Marie Thomas, Hanno S. Meyer, Claus Zimmer, and et al. 2023. "Imaging the WHO 2021 Brain Tumor Classification: Fully Automated Analysis of Imaging Features of Newly Diagnosed Gliomas" Cancers 15, no. 8: 2355. https://doi.org/10.3390/cancers15082355
APA StyleGriessmair, M., Delbridge, C., Ziegenfeuter, J., Bernhardt, D., Gempt, J., Schmidt-Graf, F., Kertels, O., Thomas, M., Meyer, H. S., Zimmer, C., Meyer, B., Combs, S. E., Yakushev, I., Wiestler, B., & Metz, M.-C. (2023). Imaging the WHO 2021 Brain Tumor Classification: Fully Automated Analysis of Imaging Features of Newly Diagnosed Gliomas. Cancers, 15(8), 2355. https://doi.org/10.3390/cancers15082355








