Classifying Malignancy in Prostate Glandular Structures from Biopsy Scans with Deep Learning
Abstract
Simple Summary
Abstract
1. Introduction
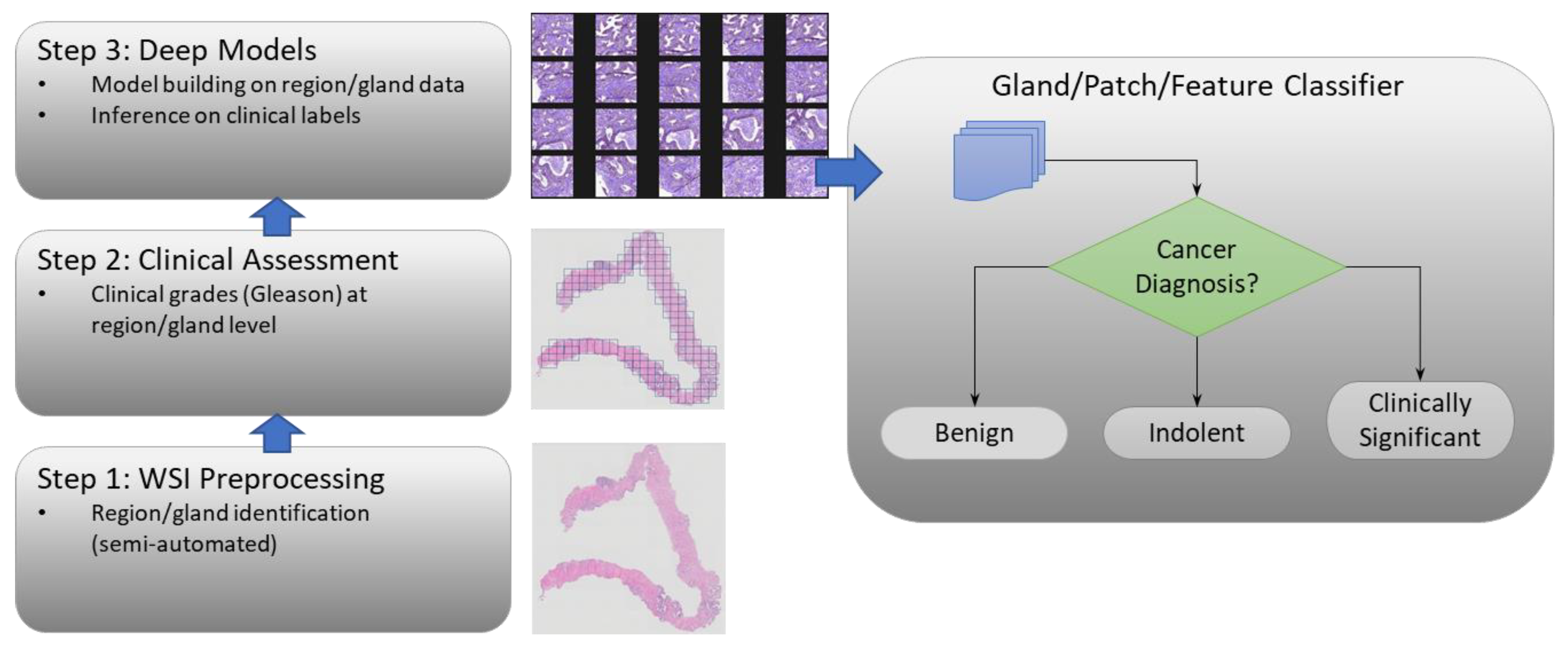
2. Materials and Methods
2.1. Data Cohorts
2.1.1. Gland Level Patient Data Cohort
2.1.2. PANDA Radboud Data Cohort
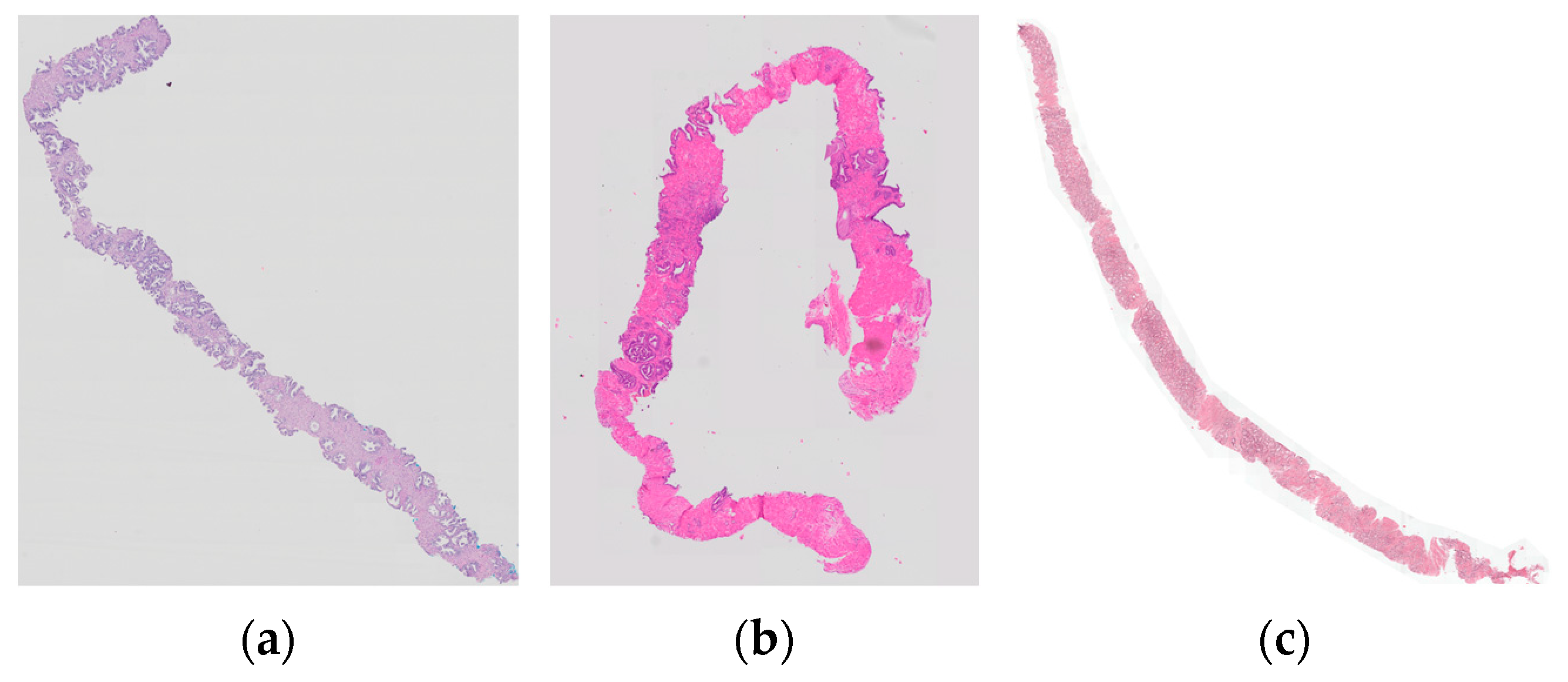
2.2. Image Preprocessing
2.2.1. Sample-Mix
2.2.2. Standardization
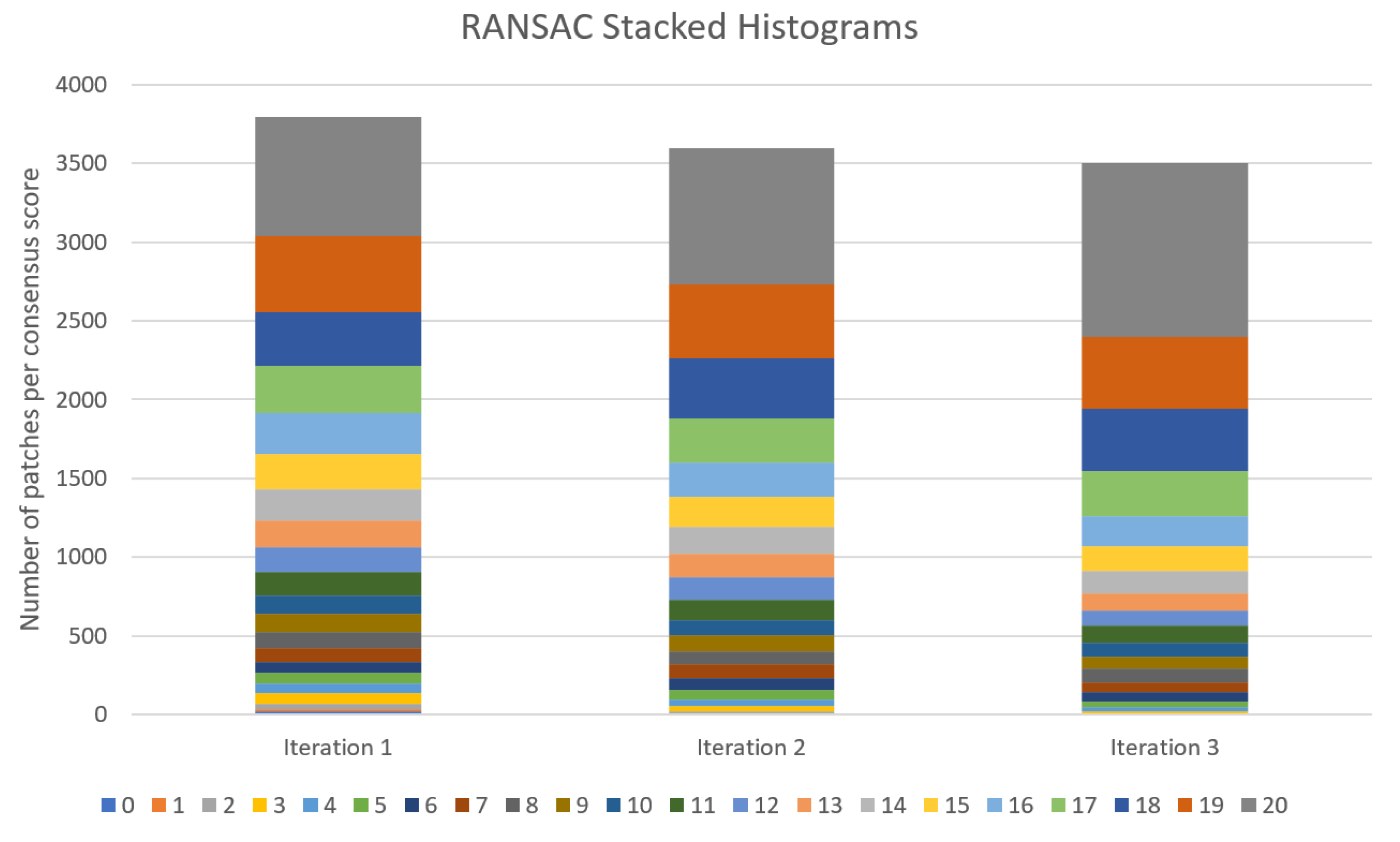
2.2.3. Eliminating Outliers
2.2.4. Balancing Data
2.3. Deep Learning
2.3.1. Optimization Technique
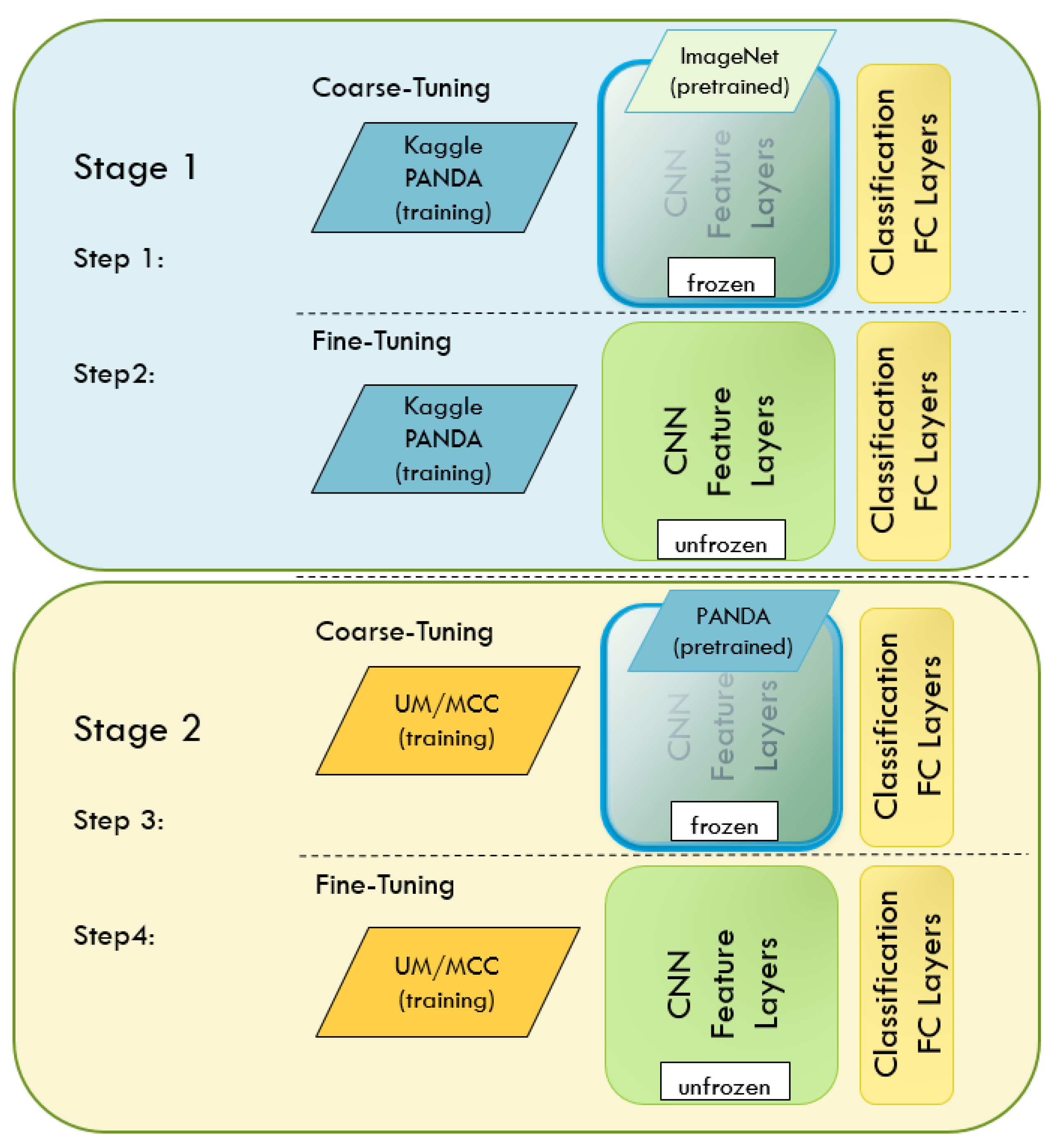
2.3.2. Transfer Learning
2.4. Measuring Performance
2.5. Implementation Challenges
3. Results
Deep Network Performance
4. Discussion
Limitations and Future Improvements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | Area Under (ROC) Curve |
| CNN | Convolutional Neural Network |
| CV | Cross Validation |
| DL | Deep Learning |
| DSS | Decision Support System |
| FC | Fully Connected |
| GS | Gleason Score |
| ISUP | International Society of Urological Pathology |
| MCCV | Monte Carlo Cross-Validation |
| MCC | Moffitt Cancer Center |
| MIL | Multi-Instance Learning |
| ML | Machine Learning |
| NN | Neural Network |
| OD | Outlier Detection |
| PANDA | Prostate cANcer graDe Assessment |
| RANSAC | Random Sampling and Consensus |
| RNN | Recurrent Neural Network |
| ROC | Receiver Operating Characteristic |
| SGD | Stochastic Gradient Descent |
| UM | University of Miami |
| WSI | Whole-Slide Image |
References
- Humphrey, P.A. Histopathology of Prostate Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a030411. [Google Scholar] [CrossRef]
- Marini, N.; Otalora, S.; Wodzinski, M.; Tomassini, S.; Dragoni, A.F.; Marchand-Maillet, S.; Morales, J.P.D.; Duran-Lopez, L.; Vatrano, S.; Müller, H.; et al. Data-driven color augmentation for H&E stained images in computational pathology. J. Pathol. Inform. 2023, 14, 100183. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, P.A.; Moch, H.; Cubilla, A.L.; Ulbright, T.M.; Reuter, V.E. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part B: Prostate and Bladder Tumours. Eur. Urol. 2016, 70, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J.I.; Zelefsky, M.J.; Sjoberg, D.D.; Nelson, J.B.; Egevad, L.; Magi-Galluzzi, C.; Vickers, A.J.; Parwani, A.V.; Reuter, V.E.; Fine, S.W.; et al. A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. Eur. Urol. 2016, 69, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Sehn, J.K. Prostate Cancer Pathology: Recent Updates and Controversies. MO Med. 2018, 115, 151–155. [Google Scholar] [PubMed]
- Allsbrook, W.C.; Mangold, K.A.; Johnson, M.H.; Lane, R.B.; Lane, C.G.; Amin, M.B.; Bostwick, D.G.; Humphrey, P.A.; Jones, E.C.; Reuter, V.E.; et al. Interobserver reproducibility of Gleason grading of prostatic carcinoma: Urologic pathologists. Hum. Pathol. 2001, 32, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Allsbrook, W.C.; Mangold, K.A.; Johnson, M.H.; Lane, R.B.; Lane, C.G.; Epstein, J.I. Interobserver reproducibility of Gleason grading of prostatic carcinoma: General pathologist. Hum. Pathol. 2001, 32, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Egevad, L.; Ahmad, A.S.; Algaba, F.; Berney, D.M.; Boccon-Gibod, L.; Compérat, E.; Evans, A.J.; Griffiths, D.; Grobholz, R.; Kristiansen, G.; et al. Standardization of Gleason grading among 337 European pathologists: Gleason grading in Europe. Histopathology 2013, 62, 247–256. [Google Scholar] [CrossRef]
- Ozkan, T.A.; Eruyar, A.T.; Cebeci, O.O.; Memik, O.; Ozcan, L.; Kuskonmaz, I. Interobserver variability in Gleason histological grading of prostate cancer. Scand. J. Urol. 2016, 50, 420–424. [Google Scholar] [CrossRef]
- Oyama, T.; Allsbrook, W.C., Jr.; Kurokawa, K.; Matsuda, H.; Segawa, A.; Sano, T.; Suzuki, K.; Epstein, J.I. A comparison of interobserver reproducibility of Gleason grading of prostatic carcinoma in Japan and the United States. Arch. Pathol. Lab. Med. 2005, 129, 1004–1010. [Google Scholar] [CrossRef]
- Zarella, M.D.; Bowman, D.; Aeffner, F.; Farahani, N.; Xthona, A.; Absar, S.F.; Parwani, A.; Bui, M.; Hartman, D.J. A Practical Guide to Whole Slide Imaging: A White Paper From the Digital Pathology Association. Arch. Pathol. Lab. Med. 2019, 143, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.; Elder, J.H. Deep learning models fail to capture the configural nature of human shape perception. iScience 2022, 25, 104913. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.P.; Samala, R.K.; Hadjiiski, L.M.; Zhou, C. Deep Learning in Medical Image Analysis. Adv. Exp. Med. Biol. 2020, 1213, 3–21. [Google Scholar] [CrossRef]
- De Haan, K.; Zhang, Y.; Zuckerman, J.E.; Liu, T.; Sisk, A.E.; Diaz, M.F.P.; Jen, K.-Y.; Nobori, A.; Liou, S.; Zhang, S.; et al. Deep learning-based transformation of H&E stained tissues into special stains. Nat. Commun. 2021, 12, 4884. [Google Scholar] [CrossRef]
- Deng, S.; Zhang, X.; Yan, W.; Chang, E.I.; Fan, Y.; Lai, M.; Xu, Y. Deep learning in digital pathology image analysis: A survey. Front. Med. 2020, 14, 470–487. [Google Scholar] [CrossRef] [PubMed]
- Bulten, W.; Kartasalo, K.; Chen, P.-H.C.; Ström, P.; Pinckaers, H.; Nagpal, K.; Cai, Y.; Steiner, D.F.; van Boven, H.; Vink, R.; et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: The PANDA challenge. Nat. Med. 2022, 28, 154–163. [Google Scholar] [CrossRef]
- Paul, R.; Hawkins, S.; Balagurunathan, Y.; Schabath, M.; Gillies, R.; Hall, L.; Goldgof, D. Deep Feature Transfer Learning in Combination with Traditional Features Predicts Survival among Patients with Lung Adenocarcinoma. Tomography 2016, 2, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.; Kariev, S.; Cherezov, D.; Schabath, M.; Gillies, R.; Hall, L.; Goldgof, D.; Drukker, K.; Mazurowski, M.A. Deep radiomics: Deep learning on radiomics texture images. In Medical Imaging 2021: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2021. [Google Scholar] [CrossRef]
- Bulten, W.; Pinckaers, H.; van Boven, H.; Vink, R.; de Bel, T.; van Ginneken, B.; van der Laak, J.; Hulsbergen-van de Kaa, C.; Litjens, G. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: A diagnostic study. Lancet Oncol. 2020, 21, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Bulten, W.; Balkenhol, M.; Belinga, J.-J.A.; Brilhante, A.; Çakır, A.; Egevad, L.; Eklund, M.; Farré, X.; Geronatsiou, K.; Molinié, V.; et al. Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists. Mod. Pathol. 2021, 34, 660–671. [Google Scholar] [CrossRef]
- Pinckaers, H.; Bulten, W.; van der Laak, J.; Litjens, G. Detection of Prostate Cancer in Whole-Slide Images Through End-to-End Training With Image-Level Labels. IEEE Trans. Med. Imaging 2021, 40, 1817–1826. [Google Scholar] [CrossRef]
- Ström, P.; Kartasalo, K.; Olsson, H.; Solorzano, L.; Delahunt, B.; Berney, D.M.; Bostwick, D.G.; Evans, A.J.; Grignon, D.J.; Humphrey, P.A.; et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: A population-based, diagnostic study. Lancet Oncol. 2020, 21, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, K.; Foote, D.; Liu, Y.; Chen, P.-H.C.; Wulczyn, E.; Tan, F.; Olson, N.; Smith, J.L.; Mohtashamian, A.; Wren, J.H.; et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit. Med. 2019, 2, 48. [Google Scholar] [CrossRef] [PubMed]
- Rana, A.; Lowe, A.; Lithgow, M.; Horback, K.; Janovitz, T.; Da Silva, A.; Tsai, H.; Shanmugam, V.; Bayat, A.; Shah, P. Use of Deep Learning to Develop and Analyze Computational Hematoxylin and Eosin Staining of Prostate Core Biopsy Images for Tumor Diagnosis. JAMA Netw. Open 2020, 3, e205111. [Google Scholar] [CrossRef] [PubMed]
- Varoquaux, G.; Cheplygina, V. Machine learning for medical imaging: Methodological failures and recommendations for the future. Npj Digit. Med. 2022, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [PubMed]
- Krizhevshy, A.; Sutskever, I.; Hilton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Thrall, J.H.; Li, X.; Li, Q.; Cruz, C.; Do, S.; Dreyer, K.; Brink, J. Artificial Intelligence and Machine Learning in Radiology: Opportunities, Challenges, Pitfalls, and Criteria for Success. J. Am. Coll. Radiol. 2018, 15, 504–508. [Google Scholar] [CrossRef]
- Zhu, Y.; Wei, R.; Gao, G.; Ding, L.; Zhang, X.; Wang, X.; Zhang, J. Fully automatic segmentation on prostate MR images based on cascaded fully convolution network. J. Magn. Reson. Imaging 2019, 49, 1149–1156. [Google Scholar] [CrossRef]
- Morid, M.A.; Borjali, A.; Del Fiol, G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput. Biol. Med. 2021, 128, 104115. [Google Scholar] [CrossRef]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding Transfer Learning for Medical Imaging. In Proceedings of the NeurIPS, Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Alzubaidi, L.; Duan, Y.; Al-Dujaili, A.; Ibraheem, I.K.; Alkenani, A.H.; Santamaria, J.; Fadhel, M.A.; Al-Shamma, O.; Zhang, J. Deepening into the suitability of using pre-trained models of ImageNet against a lightweight convolutional neural network in medical imaging: An experimental study. PeerJ Comput. Sci. 2021, 7, e715. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; Volume 97, pp. 6105–6114. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Las Vegas, NV, USA, 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Linkon, A.H.M.; Labib, M.M.; Hasan, T.; Hossain, M.; Jannat, M.-E. Deep learning in prostate cancer diagnosis and Gleason grading in histopathology images: An extensive study. Inform. Med. Unlocked 2021, 24, 100582. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Xu, Q.-S.; Liang, Y.-Z. Monte Carlo cross validation. Chemom. Intell. Lab. Syst. 2001, 56, 1–11. [Google Scholar] [CrossRef]
- Talebi, H.; Milanfar, P. Learning to Resize Images for Computer Vision Tasks. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021; IEEE: Montreal, QC, Canada, 2021; pp. 487–496. [Google Scholar]
- Hashemi, M. Enlarging smaller images before inputting into convolutional neural network: Zero-padding vs. interpolation. J. Big Data 2019, 6, 98. [Google Scholar] [CrossRef]
- Kim, J.; Jang, J.; Seo, S.; Jeong, J.; Na, J.; Kwak, N. MUM: Mix Image Tiles and UnMix Feature Tiles for Semi-Supervised Object Detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 14512–14521. [Google Scholar]
- Yun, S.; Han, D.; Oh, S.J.; Chun, S.; Choe, J.; Yoo, Y. CutMix: Regularization Strategy to Train Strong Classifiers with Localizable Features. arXiv 2019, arXiv:1905.04899. [Google Scholar]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. Mixup: Beyond Empirical Risk Minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Kleczek, P.; Jaworek-Korjakowska, J.; Gorgon, M. A novel method for tissue segmentation in high-resolution H&E-stained histopathological whole-slide images. Comput. Med. Imaging Graph. 2020, 79, 101686. [Google Scholar] [CrossRef]
- Chan, J.K. The wonderful colors of the hematoxylin-eosin stain in diagnostic surgical pathology. Int. J. Surg. Pathol. 2014, 22, 12–32. [Google Scholar] [CrossRef]
- Cooper, L.A.; Demicco, E.G.; Saltz, J.H.; Powell, R.T.; Rao, A.; Lazar, A.J. PanCancer insights from The Cancer Genome Atlas: The pathologist’s perspective. J. Pathol. 2018, 244, 512–524. [Google Scholar] [CrossRef]
- Hutchison, D.; Kanade, T.; Kittler, J.; Kleinberg, J.M.; Mattern, F.; Mitchell, J.C.; Naor, M.; Nierstrasz, O.; Pandu Rangan, C.; Steffen, B.; et al. Image Segmentation with Implicit Color Standardization Using Spatially Constrained Expectation Maximization: Detection of Nuclei. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2012; Springer: Berlin/Heidelberg, Germany, 2012; Volume 7510, pp. 365–372. [Google Scholar]
- Hoffman, R.A.; Kothari, S.; Wang, M.D. Comparison of normalization algorithms for cross-batch color segmentation of histopathological images. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; IEEE: Chicago, IL, USA, 2014; pp. 194–197. [Google Scholar] [CrossRef]
- Magee, D.; Treanor, D.; Crellin, D.; Shires, M.; Mohee, K.; Quirke, P. Colour Normalisation in Digital Histopathology Images. In Optical Tissue Image Analysis in Microscopy, Histopathology and Endoscopy; Daniel Elson: London, UK, 2009; Volume 100, pp. 100–111. [Google Scholar]
- Fernández, Á.; Bella, J.; Dorronsoro, J.R. Supervised outlier detection for classification and regression. Neurocomputing 2022, 486, 77–92. [Google Scholar] [CrossRef]
- Fischler, M.A.; Bolles, R.C. Random sample consensus: A paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM 1981, 24, 381–395. [Google Scholar] [CrossRef]
- Ur Rehman, A.; Belhaouari, S.B. Unsupervised outlier detection in multidimensional data. J. Big Data 2021, 8, 80. [Google Scholar] [CrossRef]
- Li, Y.; Pei, W.; He, Z. SSORN: Self-Supervised Outlier Removal Network for Robust Homography Estimation. arXiv 2022, arXiv:2208.14093. [Google Scholar]
- Davison, A.C.; Hinkley, D.V.; Schechtman, E. Efficient Bootstrap Simulation. Biometrika 1986, 73, 555–566. [Google Scholar] [CrossRef]
- D’Ascoli, S.; Touvron, H.; Leavitt, M.; Morcos, A.; Biroli, G.; Sagun, L. ConViT: Improving Vision Transformers with Soft Convolutional Inductive Biases. arXiv 2021, arXiv:2103.10697. [Google Scholar] [CrossRef]
- Abnar, S.; Dehghani, M.; Zuidema, W. Transferring Inductive Biases through Knowledge Distillation. arXiv 2020, arXiv:2006.00555. [Google Scholar]
- Feng, H.; Yang, B.; Wang, J.; Liu, M.; Yin, L.; Zheng, W.; Yin, Z.; Liu, C. Identifying Malignant Breast Ultrasound Images Using ViT-Patch. Appl. Sci. 2023, 13, 3489. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Zhou, Y.T.; Chellappa, R. Computation of optical flow using a neural network. In Proceedings of the IEEE International Conference on Neural Networks, Anchorage, AK, USA, 4–9 May 1998; IEEE: San Diego, CA, USA, 1988; Volume 2, pp. 71–78. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. PMLR 2015, 37, 448–456. [Google Scholar]
- Loshchilov, I.; Hutter, F. SGDR: Stochastic Gradient Descent with Warm Restarts. arXiv 2017, arXiv:1608.03983. [Google Scholar]
- Huang, G.; Li, Y.; Pleiss, G.; Liu, Z.; Hopcroft, J.E.; Weinberger, K.Q. Snapshot Ensembles: Train 1, get M for free. arXiv 2017, arXiv:1704.00109. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Region-Based Convolutional Networks for Accurate Object Detection and Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 38, 17. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Hiruta, N.; Terai, K.; Nosato, H.; Murakawa, M.; Sakanashi, H. Gastric Pathology Image Classification Using Stepwise Fine-Tuning for Deep Neural Networks. J. Healthc. Eng. 2018, 2018, 8961781. [Google Scholar] [CrossRef]
- Trevethan, R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in Research and Practice. Front. Public Health 2017, 5, 307. [Google Scholar] [CrossRef] [PubMed]
- Forman, G.; Scholz, M. Apples-to-Apples in Cross-Validation Studies: Pitfalls in Classifier Performance Measurement. Assoc. Comput. Mach. 2010, 12, 49–57. [Google Scholar] [CrossRef]
- DiCiccio, T.J.; Efron, B. Bootstrap Confidence Intervals. Stat. Sci. 1996, 11, 40. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef]
- Gao, J.; Jiang, Q.; Zhou, B.; Chen, D. Convolutional neural networks for computer-aided detection or diagnosis in medical image analysis: An overview. Math. Biosci. Eng. 2019, 16, 6536–6561. [Google Scholar] [CrossRef]
- Pujar, A.; Pereira, T.; Tamgadge, A.; Bhalerao, S.; Tamgadge, S. Comparing The Efficacy of Hematoxylin and Eosin, Periodic Acid Schiff and Fluorescent Periodic Acid Schiff-Acriflavine Techniques for Demonstration of Basement Membrane in Oral Lichen Planus: A Histochemical Study. Indian J. Dermatol. 2015, 60, 450–456. [Google Scholar] [CrossRef]
- Azevedo Tosta, T.A.; de Faria, P.R.; Neves, L.A.; do Nascimento, M.Z. Computational normalization of H&E-stained histological images: Progress, challenges and future potential. Artif. Intell. Med. 2019, 95, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Geirhos, R.; Rubisch, P.; Michaelis, C.; Bethge, M.; Wichmann, F.A.; Brendel, W. ImageNet-trained CNNs are biased towards texture; Increasing Shape Bias Improves Accuracy and Robustness. arXiv 2019, arXiv:1811.12231. [Google Scholar]
- Ciga, O.; Xu, T.; Martel, A.L. Self supervised contrastive learning for digital histopathology. Mach. Learn. Appl. 2022, 7, 100198. [Google Scholar] [CrossRef]
- Yang, P.; Yin, X.; Lu, H.; Hu, Z.; Zhang, X.; Jiang, R.; Lv, H. CS-CO: A Hybrid Self-Supervised Visual Representation Learning Method for H&E-stained Histopathological Images. Med. Image Anal. 2022, 81, 102539. [Google Scholar] [CrossRef]
- Wang, X.; Yang, S.; Zhang, J.; Wang, M.; Zhang, J.; Yang, W.; Huang, J.; Han, X. Transformer-based unsupervised contrastive learning for histopathological image classification. Med. Image Anal. 2022, 81, 102559. [Google Scholar] [CrossRef]
- Chen, H.; Li, C.; Wang, G.; Li, X.; Mamunur Rahaman, M.; Sun, H.; Hu, W.; Li, Y.; Liu, W.; Sun, C.; et al. GasHis-Transformer: A multi-scale visual transformer approach for gastric histopathological image detection. Pattern Recognit. 2022, 130, 108827. [Google Scholar] [CrossRef]
- Egevad, L.; Swanberg, D.; Delahunt, B.; Ström, P.; Kartasalo, K.; Olsson, H.; Berney, D.M.; Bostwick, D.G.; Evans, A.J.; Humphrey, P.A.; et al. Identification of areas of grading difficulties in prostate cancer and comparison with artificial intelligence assisted grading. Virchows Arch. 2020, 477, 777–786. [Google Scholar] [CrossRef]
- Salvi, M.; Bosco, M.; Molinaro, L.; Gambella, A.; Papotti, M.; Acharya, U.R.; Molinari, F. A hybrid deep learning approach for gland segmentation in prostate histopathological images. Artif. Intell. Med. 2021, 115, 102076. [Google Scholar] [CrossRef]
- Bulten, W.; Bándi, P.; Hoven, J.; Loo, R.v.d.; Lotz, J.; Weiss, N.; Laak, J.V.d.; Ginneken, B.V.; Hulsbergen-van de Kaa, C.; Litjens, G. Epithelium segmentation using deep learning in H&E-stained prostate specimens with immunohistochemistry as reference standard. Sci. Rep. 2019, 9, 864. [Google Scholar] [CrossRef]
- Singhal, N.; Soni, S.; Bonthu, S.; Chattopadhyay, N.; Samanta, P.; Joshi, U.; Jojera, A.; Chharchhodawala, T.; Agarwal, A.; Desai, M.; et al. A deep learning system for prostate cancer diagnosis and grading in whole slide images of core needle biopsies. Sci. Rep. 2022, 12, 3383. [Google Scholar] [CrossRef]
- Paul, R.; Schabath, M.; Gillies, R.; Hall, L.; Goldgof, D. Convolutional Neural Network ensembles for accurate lung nodule malignancy prediction 2 years in the future. Comput. Biol. Med. 2020, 122, 103882. [Google Scholar] [CrossRef] [PubMed]
- McCloskey, M.; Cohen, N.J. Catastrophic Interference in Connectionist Networks: The Sequential Learning Problem. Psychol. Learn. Motiv. 1989, 24, 57. [Google Scholar] [CrossRef]
- Li, Z.; Hoiem, D. Learning without Forgetting. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 2935–2947. [Google Scholar] [CrossRef]
- Rusu, A.A.; Rabinowitz, N.C.; Desjardins, G.; Soyer, H.; Kirkpatrick, J.; Kavukcuoglu, K.; Pascanu, R.; Hadsell, R. Progressive Neural Networks. arXiv 2022, arXiv:1606.04671. [Google Scholar]
- Alahmari, S.S.; Goldgof, D.B.; Mouton, P.R.; Hall, L.O. Challenges for the Repeatability of Deep Learning Models. IEEE Access 2020, 8, 211860–211868. [Google Scholar] [CrossRef]
- Langford, J. Tutorial On Practical Prediction Theory For Classification. J. Mach. Learn. Res. 2005, 6, 273–306. [Google Scholar]
- Hosen, M.A.; Khosravi, A.; Nahavandi, S.; Creighton, D. Improving the Quality of Prediction Intervals Through Optimal Aggregation. IEEE Trans. Ind. Electron. 2015, 62, 4420–4429. [Google Scholar] [CrossRef]
- Khan, A.M.; Rajpoot, N.; Treanor, D.; Magee, D. A Nonlinear Mapping Approach to Stain Normalization in Digital Histopathology Images Using Image-Specific Color Deconvolution. IEEE Trans. Biomed. Eng. 2014, 61, 1729–1738. [Google Scholar] [CrossRef]
- Dave, P.; Alahmari, S.; Goldgof, D.; Hall, L.O.; Morera, H.; Mouton, P.R. An adaptive digital stain separation method for deep learning-based automatic cell profile counts. J. Neurosci. Methods 2021, 354, 109102. [Google Scholar] [CrossRef]
- Ruifrok, A.C.; Johnston, D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001, 23, 291–299. [Google Scholar]
- Danon, D.; Arar, M.; Cohen-Or, D.; Shamir, A. Image resizing by reconstruction from deep features. Comput. Vis. Media 2021, 7, 453–466. [Google Scholar] [CrossRef]
- Ben Ahmed, K.; Hall, L.O.; Goldgof, D.B.; Fogarty, R. Achieving Multisite Generalization for CNN-Based Disease Diagnosis Models by Mitigating Shortcut Learning. IEEE Access 2022, 10, 78726–78738. [Google Scholar] [CrossRef]
- Cauni, V.; Stanescu, D.; Tanase, F.; Mihai, B.; Persu, C. Magnetic Resonance/Ultrasound Fusion Targeted Biopsy of the Prostate Can Be Improved By Adding Systematic Biopsy. Med. Ultrason. 2021, 23, 277–282. [Google Scholar] [CrossRef] [PubMed]




| Total | Benign | GS3 | GS4 | |
|---|---|---|---|---|
| Subjects | 52 | 23 | 38 | 32 |
| Whole-Slide Images | 150 | 72 | 72 | 60 |
| Labeled Glands | 14509 | 6882 | 5143 | 2484 |
| Total | Benign | GS3 | GS4 | GS5 | |
|---|---|---|---|---|---|
| Biopsy scans | 1240 | 310 | 310 | 310 | 310 |
| Patches | 24,800 | 6200 | 6200 | 6200 | 6200 |
| Trained on PANDA Radboud | ||
|---|---|---|
| PANDA Radboud Benign vs. GS3/4/5 | PANDA Radboud GS3 vs. GS4 | |
| Accuracy | 0.941 (0.88, 0.98) | 0.979 (0.95, 1.0) |
| Sensitivity | 0.964 (0.92, 0.99) | 0.980 (0.94, 1.0) |
| Specificity | 0.920 (0.80, 0.98) | 0.979(0.93, 1.0) |
| Precision | 0.927 (0.83, 0.98) | 0.979 (0.94, 1.0) |
| NPV | 0.959 (0.90, 0.99) | 0.980 (0.94, 1.0) |
| F1-score | 0.944 (0.88, 0.98) | 0.980 (0.95, 1.0) |
| AUC | 0.981 (0.93, 1.0) | 0.997 (0.99, 1.0) |
| Trained on UM/MCC | ||||
|---|---|---|---|---|
| Benign vs. GS3/4 (1-Stage ImageNet Transfer-Learning) | Benign vs. GS3/4 (2-Stage ImageNet Plus PANDA Transfer-Learn) | GS3 vs. GS4 (1-Stage ImageNet Transfer-Learning) | GS3 vs. GS4 (2-Stage ImageNet Plus PANDA Transfer-Learn) | |
| Accuracy | 0.901 (0.79, 0.98) | 0.911 (0.81, 0.97) | 0.669 (0.53, 0.84) | 0.680 (0.54, 0.84) |
| Sensitivity | 0.898 (0.75, 0.97) | 0.897 (0.71, 0.97) | 0.732 (0.37, 0.93) | 0.753 (0.47, 0.90) |
| Specificity | 0.898 (0.75, 0.97) | 0.897 (0.71, 0.97) | 0.606 (0.26, 0.87) | 0.606 (0.19, 0.92) |
| Precision | 0.912 (0.75, 1.0) | 0.923 (0.76, 0.99) | 0.660 (0.52, 0.86) | 0.670 (0.52, 0.90) |
| NPV | 0.912 (0.75, 1.0) | 0.923 (0.76, 0.99) | 0.699 (0.54, 0.86) | 0.712 (0.55, 0.83) |
| F1-score | 0.903 (0.78, 0.98) | 0.908 (0.77, 0.98) | 0.686 (0.47, 0.83) | 0.702 (0.51, 0.84) |
| AUC | 0.955 (0.87, 0.99) | 0.955 (0.87, 0.99) | 0.706 (0.43, 0.90) | 0.714 (0.44, 0.90) |
| Trained on Combined PANDA Radboud + UM/MCC | ||||
|---|---|---|---|---|
| PANDA Radboud Benign vs. GS3/4 | UM/MCC Benign vs. GS3/4 | PANDA Radboud GS3 vs. GS4 | UM/MCC GS3 vs. GS4 | |
| Accuracy | 0.961 (0.93, 0.99) | 0.915 (0.80, 0.97) | 0.970 (0.94, 1.0) | 0.668 (0.53, 0.84) |
| Sensitivity | 0.944 (0.89, 0.99) | 0.902 (0.75, 0.99) | 0.971 (0.92, 1.0) | 0.647 (0.36, 0.84) |
| Specificity | 0.978 (0.95, 1.0) | 0.928 (0.82, 0.98) | 0.968 (0.88, 1.0) | 0.689 (0.24, 0.87) |
| Precision | 0.977 (0.95, 1.0) | 0.927 (0.83, 0.98) | 0.970 (0.89, 1.0) | 0.687 (0.52, 0.85) |
| NPV | 0.946 (0.90, 0.99) | 0.908 (0.78, 0.99) | 0.972 (0.92, 1.0) | 0.665 (0.55, 0.83) |
| F1-score | 0.960 (0.92, 0.99) | 0.913 (0.79, 0.97) | 0.970 (0.94, 1.0) | 0.656 (0.47, 0.84) |
| AUC | 0.988 (0.96, 1.0) | 0.963 (0.86, 0.99) | 0.996 (0.99, 1.0) | 0.710 (0.52, 0.90) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fogarty, R.; Goldgof, D.; Hall, L.; Lopez, A.; Johnson, J.; Gadara, M.; Stoyanova, R.; Punnen, S.; Pollack, A.; Pow-Sang, J.; et al. Classifying Malignancy in Prostate Glandular Structures from Biopsy Scans with Deep Learning. Cancers 2023, 15, 2335. https://doi.org/10.3390/cancers15082335
Fogarty R, Goldgof D, Hall L, Lopez A, Johnson J, Gadara M, Stoyanova R, Punnen S, Pollack A, Pow-Sang J, et al. Classifying Malignancy in Prostate Glandular Structures from Biopsy Scans with Deep Learning. Cancers. 2023; 15(8):2335. https://doi.org/10.3390/cancers15082335
Chicago/Turabian StyleFogarty, Ryan, Dmitry Goldgof, Lawrence Hall, Alex Lopez, Joseph Johnson, Manoj Gadara, Radka Stoyanova, Sanoj Punnen, Alan Pollack, Julio Pow-Sang, and et al. 2023. "Classifying Malignancy in Prostate Glandular Structures from Biopsy Scans with Deep Learning" Cancers 15, no. 8: 2335. https://doi.org/10.3390/cancers15082335
APA StyleFogarty, R., Goldgof, D., Hall, L., Lopez, A., Johnson, J., Gadara, M., Stoyanova, R., Punnen, S., Pollack, A., Pow-Sang, J., & Balagurunathan, Y. (2023). Classifying Malignancy in Prostate Glandular Structures from Biopsy Scans with Deep Learning. Cancers, 15(8), 2335. https://doi.org/10.3390/cancers15082335








