miR-146a-5p Promotes Angiogenesis and Confers Trastuzumab Resistance in HER2+ Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture
2.2. miRNA Microarray and miRNA Target Prediction
2.3. miRNA Mimics/Inhibitors and siRNA Transfection
2.4. Cell Viability Assay
2.5. Clinical Samples
2.6. RNA Isolation
2.7. miRNA and Messenger RNA (mRNA) Expression via Real-Time Quantitative PCR (RT-qPCR)
2.8. Exosome Isolation and Transmission Electron Microscopy (TEM) Analysis
2.9. Exosome Internalization
2.10. Western Blot
2.11. Cell Cycle Analysis
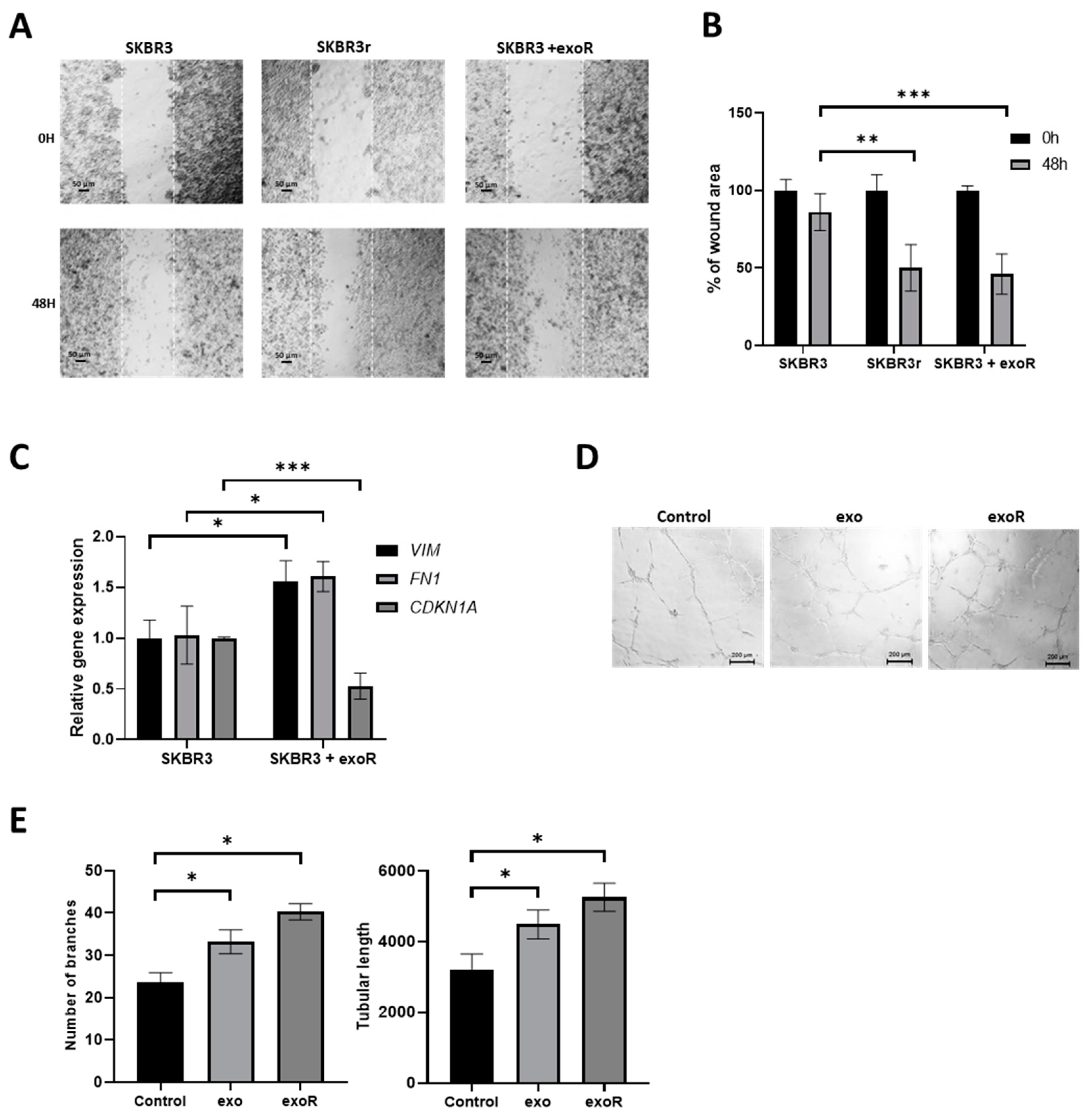
2.12. Wound-Healing Assay
2.13. Angiogenesis Assay
2.14. Survival Analysis
2.15. TCGA Data Analysis
2.16. Statistical Analysis
3. Results
3.1. miR-146a Is Upregulated in Trastuzumab-Resistant Cells
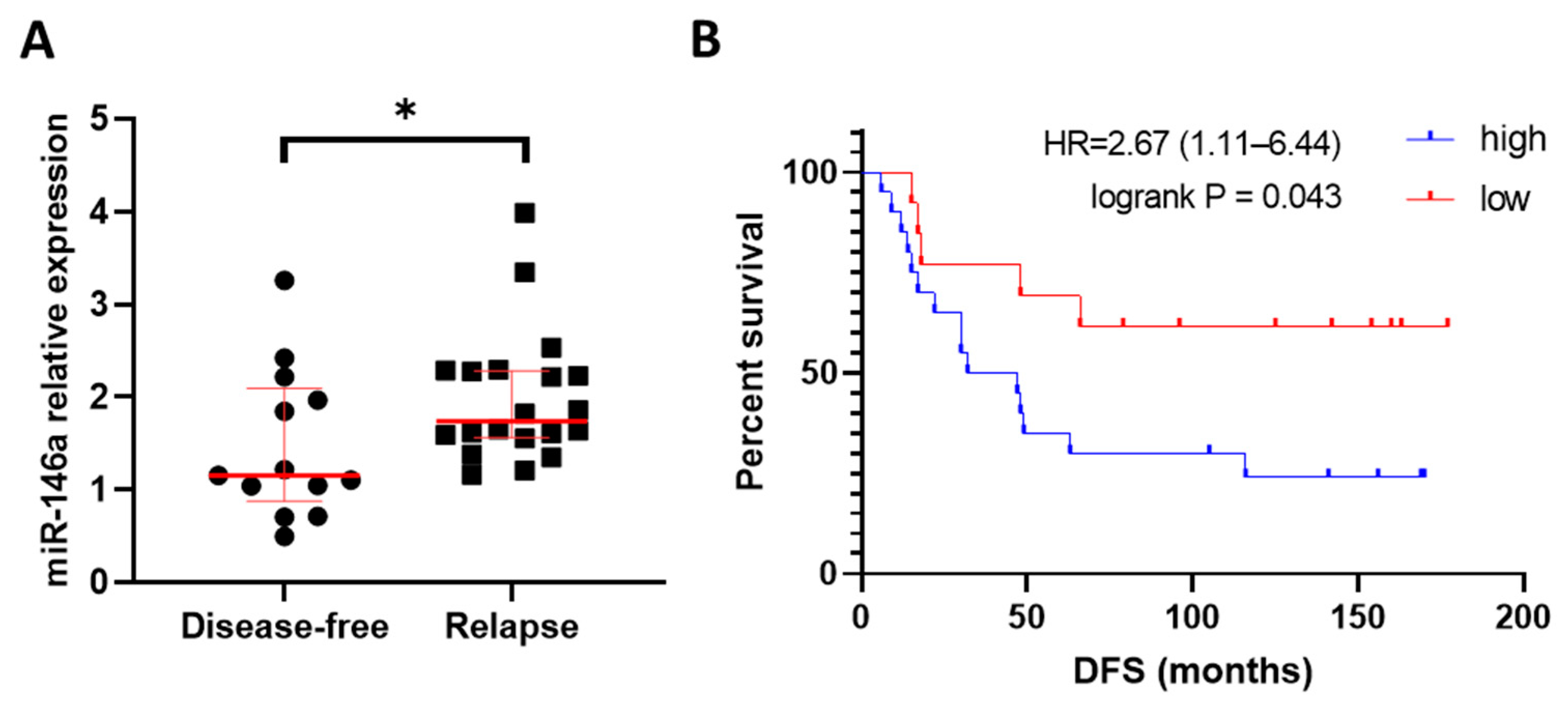
3.2. High Expression of miR-146a-5p Associated with Relapse and Shorter Disease-Free Survival in HER2+ BC Patients
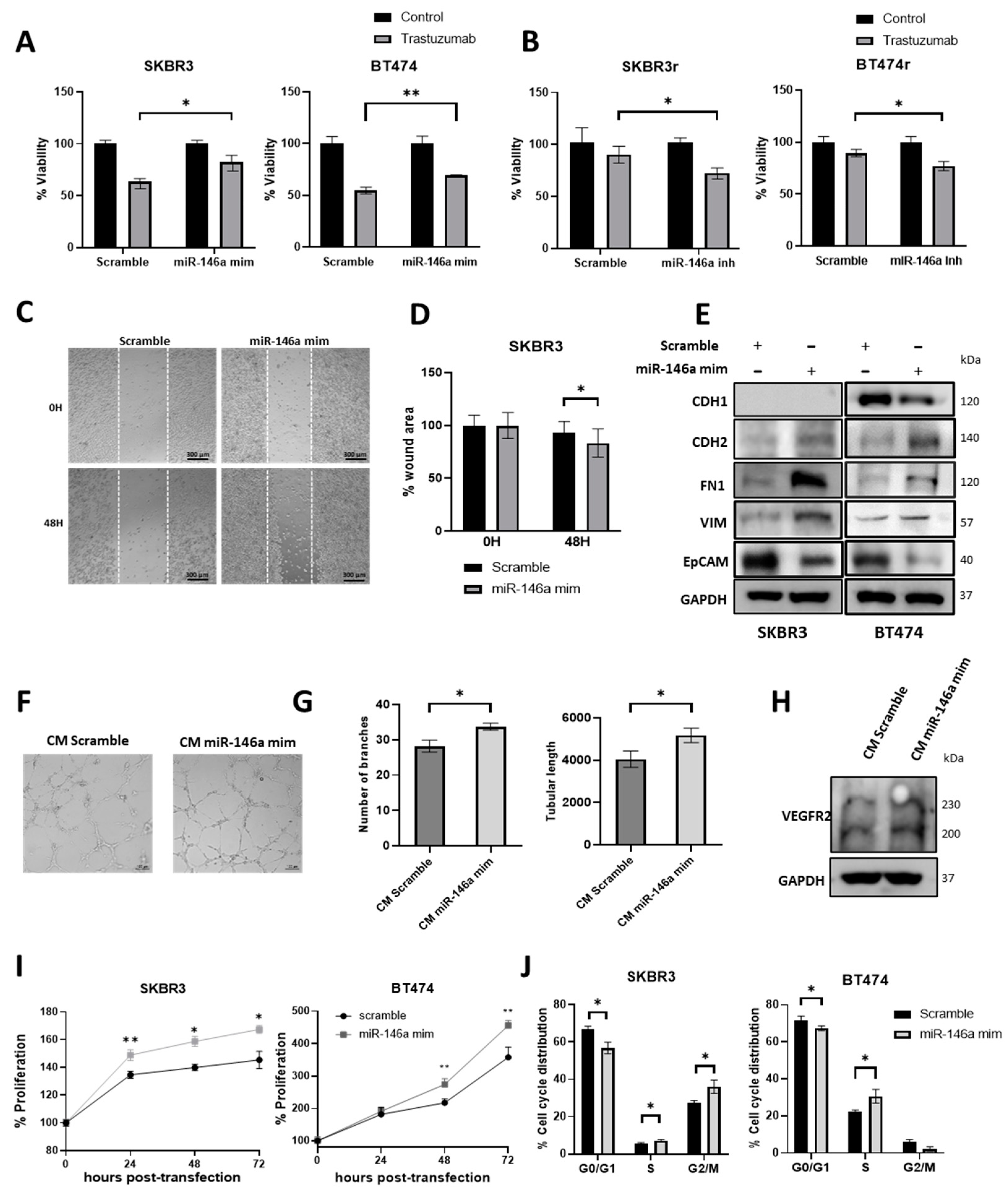
3.3. miR-146a-5p Expression Modulates the Trastuzumab Response in HER2+ BC Cell Lines
3.4. miR-146a-5p Overexpression Increases Migration, Angiogenesis, and Proliferation
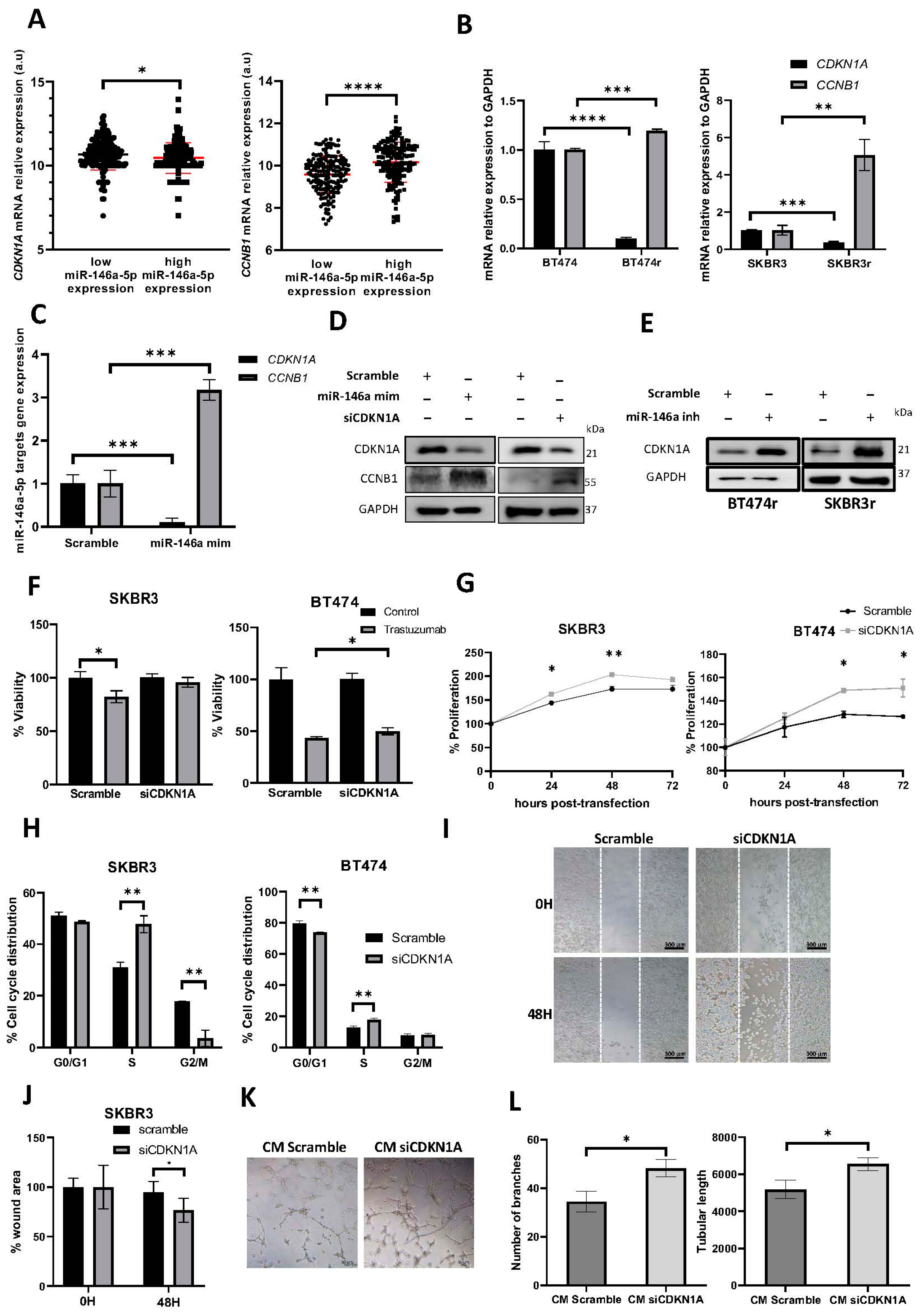
3.5. miR-146a-5p Overexpression Increases the Cell Cycle S and G2/M Phases
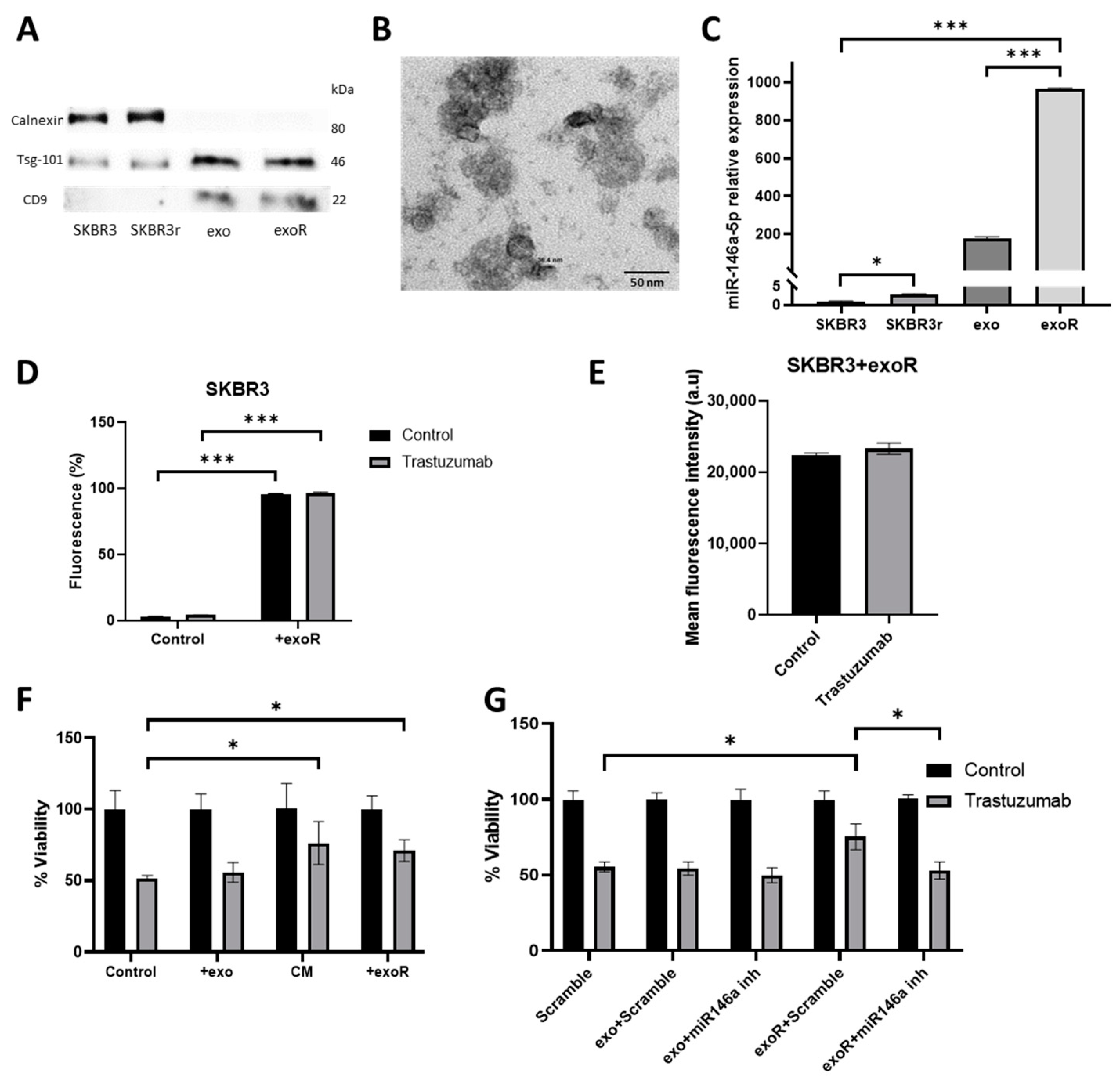
3.6. miR-146a-5p Is Enriched in Exosomes from Trastuzumab-Resistant Cells
3.7. Effects of Exosomes from Resistant Cells in Response to Trastuzumab by Parental Cells
3.8. Effect of Exosomes from Resistant Cells in the Migration Capacities of Parental Cells
3.9. Effect of Exosomes in Angiogenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Baselga, J. Treatment of HER2-overexpressing breast cancer. Ann. Oncol. 2010, 21 (Suppl. S7), vii36–vii40. [Google Scholar] [CrossRef]
- Tai, W.; Mahato, R.; Cheng, K. The role of HER2 in cancer therapy and targeted drug delivery. J. Control. Release 2010, 146, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Saini, K.S.; Azim, H.A., Jr.; Metzger-Filho, O.; Loi, S.; Sotiriou, C.; de Azambuja, E.; Piccart, M. Beyond trastuzumab: New treatment options for HER2-positive breast cancer. Breast 2011, 20 (Suppl. S3), S20–S27. [Google Scholar] [CrossRef]
- de Melo Gagliato, D.; Jardim, D.L.F.; Marchesi, M.S.P.; Hortobagyi, G.N. Mechanisms of resistance and sensitivity to anti-HER2 therapies in HER2+ breast cancer. Oncotarget 2016, 7, 64431–64446. [Google Scholar] [CrossRef]
- Fessler, S.P.; Wotkowicz, M.T.; Mahanta, S.K.; Bamdad, C. MUC1* is a determinant of trastuzumab (Herceptin) resistance in breast cancer cells. Breast Cancer Res. Treat. 2009, 118, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Gajria, D.; Chandarlapaty, S. HER2-amplified breast cancer: Mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev. Anticancer Ther. 2011, 11, 263–275. [Google Scholar] [CrossRef]
- Arribas, J.; Baselga, J.; Pedersen, K. p95HER2 and breast cancer. Cancer Res. 2011, 71, 1515–1519. [Google Scholar] [CrossRef] [PubMed]
- Carpten, J.D.; Faber, A.L.; Horn, C.; Donoho, G.P.; Briggs, S.L.; Robbins, C.M.; Hostetter, G.; Boguslawski, S.; Moses, T.Y.; Savage, S.; et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007, 448, 439–444. [Google Scholar] [CrossRef]
- Chakrabarty, A.; Rexer, B.N.; Wang, S.E.; Cook, R.S.; Engelman, J.A.; Arteaga, C.L. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene 2010, 29, 5193–5203. [Google Scholar] [CrossRef]
- Luque-Cabal, M.; García-Teijido, P.; Fernández-Pérez, Y.; Sánchez-Lorenzo, L.; Palacio-Vázquez, I. Mechanisms Behind the Resistance to Trastuzumab in HER2-Amplified Breast Cancer and Strategies to Overcome It. Clin. Med. Insights Oncol. 2016, 10 (Suppl. S1), 21–30. [Google Scholar] [CrossRef]
- Montero, J.C.; Rodríguez-Barrueco, R.; Ocaña, A.; Díaz-Rodríguez, E.; Esparís-Ogando, A.; Pandiella, A. Neuregulins and cancer. Clin. Cancer Res. 2008, 14, 3237–3241. [Google Scholar] [CrossRef]
- Mukohara, T. Mechanisms of resistance to anti-human epidermal growth factor receptor 2 agents in breast cancer. Cancer Sci. 2011, 102, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pandolfi, P.P. Breast cancer—Loss of PTEN predicts resistance to treatment. N. Engl. J. Med. 2004, 351, 2337–2338. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Nam, J.W. Genomics of microRNA. Trends Genet. 2006, 22, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Kim, H.; Ying, S.Y. Intron-mediated RNA interference and microRNA (miRNA). Front. Biosci. 2008, 13, 2216–2230. [Google Scholar] [CrossRef] [PubMed]
- Caramuta, S.; Egyházi, S.; Rodolfo, M.; Witten, D.; Hansson, J.; Larsson, C.; Lui, W.O. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J. Investig. Dermatol. 2010, 130, 2062–2070. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Zhang, Y.; Ding, J.; Wu, K.; Fan, D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut 2010, 59, 579–585. [Google Scholar] [CrossRef]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef]
- Adam-Artigues, A.; Garrido-Cano, I.; Carbonell-Asins, J.A.; Lameirinhas, A.; Simón, S.; Ortega-Morillo, B.; Martínez, M.T.; Hernando, C.; Constâncio, V.; Burgues, O.; et al. Identification of a Two-MicroRNA Signature in Plasma as a Novel Biomarker for Very Early Diagnosis of Breast Cancer. Cancers 2021, 13, 2848. [Google Scholar] [CrossRef]
- Adam-Artigues, A.; Garrido-Cano, I.; Simón, S.; Ortega, B.; Moragón, S.; Lameirinhas, A.; Constâncio, V.; Salta, S.; Burgués, O.; Bermejo, B.; et al. Circulating miR-30b-5p levels in plasma as a novel potential biomarker for early detection of breast cancer. ESMO Open 2021, 6, 100039. [Google Scholar] [CrossRef]
- Garrido-Cano, I.; Constâncio, V.; Adam-Artigues, A.; Lameirinhas, A.; Simón, S.; Ortega, B.; Martínez, M.T.; Hernando, C.; Bermejo, B.; Lluch, A.; et al. Circulating miR-99a-5p Expression in Plasma: A Potential Biomarker for Early Diagnosis of Breast Cancer. Int. J. Mol. Sci. 2020, 21, 7427. [Google Scholar] [CrossRef]
- Garrido-Cano, I.; Pattanayak, B.; Adam-Artigues, A.; Lameirinhas, A.; Torres-Ruiz, S.; Tormo, E.; Cervera, R.; Eroles, P. MicroRNAs as a clue to overcome breast cancer treatment resistance. Cancer Metastasis Rev. 2022, 41, 77–105. [Google Scholar] [CrossRef]
- Blenkiron, C.; Goldstein, L.D.; Thorne, N.P.; Spiteri, I.; Chin, S.F.; Dunning, M.J.; Barbosa-Morais, N.L.; Teschendorff, A.E.; Green, A.R.; Ellis, I.O.; et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007, 8, R214. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Christensen, M.; Silahtaroglu, A.; Bak, M.; Heath, C.V.; Schwartz, G.; Wells, W.; Kauppinen, S.; Cole, C.N. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007, 67, 11612–11620. [Google Scholar] [CrossRef]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Lowery, A.J.; Miller, N.; Devaney, A.; McNeill, R.E.; Davoren, P.A.; Lemetre, C.; Benes, V.; Schmidt, S.; Blake, J.; Ball, G.; et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009, 11, R27. [Google Scholar] [CrossRef] [PubMed]
- Mattie, M.D.; Benz, C.C.; Bowers, J.; Sensinger, K.; Wong, L.; Scott, G.K.; Fedele, V.; Ginzinger, D.; Getts, R.; Haqq, C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer 2006, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Grange, C.; Tapparo, M.; Collino, F.; Vitillo, L.; Damasco, C.; Deregibus, M.C.; Tetta, C.; Bussolati, B.; Camussi, G. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011, 71, 5346–5356. [Google Scholar] [CrossRef]
- Janowska-Wieczorek, A.; Wysoczynski, M.; Kijowski, J.; Marquez-Curtis, L.; Machalinski, B.; Ratajczak, J.; Ratajczak, M.Z. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int. J. Cancer 2005, 113, 752–760. [Google Scholar] [CrossRef]
- Bobrie, A.; Krumeich, S.; Reyal, F.; Recchi, C.; Moita, L.F.; Seabra, M.C.; Ostrowski, M.; Théry, C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012, 72, 4920–4930. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.X.; Cai, Y.Q.; Lv, M.M.; Chen, L.; Zhong, S.L.; Ma, T.F.; Zhao, J.H.; Tang, J.H. Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumour Biol. 2014, 35, 9649–9659. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Li, J.; Chen, W.X.; Cai, Y.Q.; Yu, D.D.; Zhong, S.L.; Zhao, J.H.; Zhou, J.W.; Tang, J.H. Exosomes decrease sensitivity of breast cancer cells to adriamycin by delivering microRNAs. Tumour Biol. 2016, 37, 5247–5256. [Google Scholar] [CrossRef]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef] [PubMed]
- Rak, J. Extracellular vesicles—Biomarkers and effectors of the cellular interactome in cancer. Front. Pharmacol. 2013, 4, 21. [Google Scholar] [CrossRef]
- Zazo, S.; González-Alonso, P.; Martín-Aparicio, E.; Chamizo, C.; Cristóbal, I.; Arpí, O.; Rovira, A.; Albanell, J.; Eroles, P.; Lluch, A.; et al. Generation, characterization, and maintenance of trastuzumab-resistant HER2+ breast cancer cell lines. Am. J. Cancer Res. 2016, 6, 2661–2678. [Google Scholar]
- Lánczky, A.; Nagy, Á.; Bottai, G.; Munkácsy, G.; Szabó, A.; Santarpia, L.; Győrffy, B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016, 160, 439–446. [Google Scholar] [CrossRef]
- Gooding, A.J.; Schiemann, W.P. Epithelial-Mesenchymal Transition Programs and Cancer Stem Cell Phenotypes: Mediators of Breast Cancer Therapy Resistance. Mol. Cancer Res. 2020, 18, 1257–1270. [Google Scholar] [CrossRef]
- Oliveras-Ferraros, C.; Corominas-Faja, B.; Cufí, S.; Vazquez-Martin, A.; Martin-Castillo, B.; Iglesias, J.M.; López-Bonet, E.; Martin, A.G.; Menendez, J.A. Epithelial-to-mesenchymal transition (EMT) confers primary resistance to trastuzumab (Herceptin). Cell Cycle 2012, 11, 4020–4032. [Google Scholar] [CrossRef]
- Hori, A.; Shimoda, M.; Naoi, Y.; Kagara, N.; Tanei, T.; Miyake, T.; Shimazu, K.; Kim, S.J.; Noguchi, S. Vasculogenic mimicry is associated with trastuzumab resistance of HER2-positive breast cancer. Breast Cancer Res. 2019, 21, 88. [Google Scholar] [CrossRef]
- Kodack, D.P.; Chung, E.; Yamashita, H.; Incio, J.; Duyverman, A.M.; Song, Y.; Farrar, C.T.; Huang, Y.; Ager, E.; Kamoun, W.; et al. Combined targeting of HER2 and VEGFR2 for effective treatment of HER2-amplified breast cancer brain metastases. Proc. Natl. Acad. Sci. USA 2012, 109, E3119–E3127. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Kashima, R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Kandettu, A.; Radhakrishnan, R.; Chakrabarty, S.; Sriharikrishnaa, S.; Kabekkodu, S.P. The emerging role of miRNA clusters in breast cancer progression. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188413. [Google Scholar] [CrossRef]
- Wang, W.T.; Han, C.; Sun, Y.M.; Chen, T.Q.; Chen, Y.Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 55. [Google Scholar] [CrossRef]
- Chen, X.; Lu, P.; Wu, Y.; Wang, D.D.; Zhou, S.; Yang, S.J.; Shen, H.Y.; Zhang, X.H.; Zhao, J.H.; Tang, J.H. MiRNAs-mediated cisplatin resistance in breast cancer. Tumour Biol. 2016, 37, 12905–12913. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Han, Y.; Lu, Q.; Zhang, H.; Zhao, Q.; Wu, K.; Fan, D. Drug resistance-related microRNAs in esophageal cancer. Expert Opin. Biol. Ther. 2012, 12, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Tormo, E.; Adam-Artigues, A.; Ballester, S.; Pineda, B.; Zazo, S.; González-Alonso, P.; Albanell, J.; Rovira, A.; Rojo, F.; Lluch, A.; et al. The role of miR-26a and miR-30b in HER2+ breast cancer trastuzumab resistance and regulation of the CCNE2 gene. Sci. Rep. 2017, 7, 41309. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Kanzaki, H.; Mukhopadhya, N.K.; Cui, X.; Ramanujan, V.K.; Murali, R. Trastuzumab-Resistant Luminal B Breast Cancer Cells Show Basal-Like Cell Growth Features Through NF-kappaB-Activation. Monoclon. Antibodies Immunodiagn. Immunother. 2016, 35, 1–11. [Google Scholar] [CrossRef]
- Pogribny, I.P.; Filkowski, J.N.; Tryndyak, V.P.; Golubov, A.; Shpyleva, S.I.; Kovalchuk, O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer 2010, 127, 1785–1794. [Google Scholar] [CrossRef]
- Tordonato, C.; Marzi, M.J.; Giangreco, G.; Freddi, S.; Bonetti, P.; Tosoni, D.; Di Fiore, P.P.; Nicassio, F. miR-146 connects stem cell identity with metabolism and pharmacological resistance in breast cancer. J. Cell Biol. 2021, 220, e202009053. [Google Scholar] [CrossRef] [PubMed]
- Bhaumik, D.; Scott, G.K.; Schokrpur, S.; Patil, C.K.; Campisi, J.; Benz, C.C. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene 2008, 27, 5643–5647. [Google Scholar] [CrossRef]
- Hurst, D.R.; Edmonds, M.D.; Scott, G.K.; Benz, C.C.; Vaidya, K.S.; Welch, D.R. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009, 69, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Wang, J.; Zhang, C.Y.; Shen, Y.Q.; Wang, H.M.; Ding, L.; Gu, Y.C.; Lou, J.T.; Zhao, X.T.; Ma, Z.L.; et al. MiR-146a-5p inhibits cell proliferation and cell cycle progression in NSCLC cell lines by targeting CCND1 and CCND2. Oncotarget 2016, 7, 59287–59298. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, S.; Yang, J.; Chen, X.; Huang, W. Identification of miR-146a is Associated with the Aggressiveness and Suppresses Proliferation via Targeting CDKN2A in Breast Cancer. Pathol. Oncol. Res. 2020, 26, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Song, J.; Ding, B.; Cui, Y.; Liang, J.; Han, S. miR-146a promotes cervical cancer cell viability via targeting IRAK1 and TRAF6. Oncol. Rep. 2018, 39, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Gang, X.; Xu, H.; Si, L.; Zhu, X.; Yu, T.; Jiang, Z.; Wang, Y. Treatment effect of CDKN1A on rheumatoid arthritis by mediating proliferation and invasion of fibroblast-like synoviocytes cells. Clin. Exp. Immunol. 2018, 194, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W.; Elledge, S.J.; Keyomarsi, K.; Dynlacht, B.; Tsai, L.H.; Zhang, P.; Dobrowolski, S.; Bai, C.; Connell-Crowley, L.; Swindell, E. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell 1995, 6, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, A.; You, Z.; Xu, J.; Zhu, S. Epigenetic silencing of CDKN1A and CDKN2B by SNHG1 promotes the cell cycle, migration and epithelial-mesenchymal transition progression of hepatocellular carcinoma. Cell Death Dis. 2020, 11, 823. [Google Scholar] [CrossRef]
- Namee, N.M.; O’Driscoll, L. Extracellular vesicles and anti-cancer drug resistance. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 123–136. [Google Scholar] [CrossRef]
- Samuel, P.; Fabbri, M.; Carter, D.R.F. Mechanisms of Drug Resistance in Cancer: The Role of Extracellular Vesicles. Proteomics 2017, 17, 1600375. [Google Scholar] [CrossRef]
- Yu, D.D.; Wu, Y.; Shen, H.Y.; Lv, M.M.; Chen, W.X.; Zhang, X.H.; Zhong, S.L.; Tang, J.H.; Zhao, J.H. Exosomes in development, metastasis and drug resistance of breast cancer. Cancer Sci. 2015, 106, 959–964. [Google Scholar] [CrossRef]
- Zhang, H.D.; Jiang, L.H.; Hou, J.C.; Zhong, S.L.; Zhu, L.P.; Wang, D.D.; Zhou, S.Y.; Yang, S.J.; Wang, J.Y.; Zhang, Q.; et al. Exosome: A novel mediator in drug resistance of cancer cells. Epigenomics 2018, 10, 1499–1509. [Google Scholar] [CrossRef]
- Bach, D.H.; Hong, J.Y.; Park, H.J.; Lee, S.K. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int. J. Cancer 2017, 141, 220–230. [Google Scholar] [CrossRef]
- Chen, W.X.; Xu, L.Y.; Qian, Q.; He, X.; Peng, W.T.; Zhu, Y.L.; Cheng, L. Analysis of miRNA signature differentially expressed in exosomes from adriamycin-resistant and parental human breast cancer cells. Biosci. Rep. 2018, 38, BSR20181090. [Google Scholar] [CrossRef] [PubMed]
- Najminejad, H.; Kalantar, S.M.; Abdollahpour-Alitappeh, M.; Karimi, M.H.; Seifalian, A.M.; Gholipourmalekabadi, M.; Sheikhha, M.H. Emerging roles of exosomal miRNAs in breast cancer drug resistance. IUBMB Life 2019, 71, 1672–1684. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Hong, X.; Lai, J.; Cheng, L.; Cheng, Y.; Yao, M.; Wang, R.; Hu, N. Exosomal MicroRNA-221-3p Confers Adriamycin Resistance in Breast Cancer Cells by Targeting PIK3R1. Front. Oncol. 2020, 10, 441. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, Y.; Ye, M.; Wu, J.; Ma, L.; Chen, H. Cisplatin-resistant MDA-MB-231 Cell-derived Exosomes Increase the Resistance of Recipient Cells in an Exosomal miR-423-5p-dependent Manner. Curr. Drug Metab. 2019, 20, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, W.; Xiao, J.; Cao, B. The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett. 2015, 356, 339–346. [Google Scholar] [CrossRef]
- Dong, H.; Wang, W.; Chen, R.; Zhang, Y.; Zou, K.; Ye, M.; He, X.; Zhang, F.; Han, J. Exosome-mediated transfer of lncRNASNHG14 promotes trastuzumab chemoresistance in breast cancer. Int. J. Oncol. 2018, 53, 1013–1026. [Google Scholar]
- Han, M.; Gu, Y.; Lu, P.; Li, J.; Cao, H.; Li, X.; Qian, X.; Yu, C.; Yang, Y.; Yang, X.; et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol. Cancer 2020, 19, 26. [Google Scholar] [CrossRef]
- Han, M.; Hu, J.; Lu, P.; Cao, H.; Yu, C.; Li, X.; Qian, X.; Yang, X.; Yang, Y.; Han, N.; et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020, 11, 43. [Google Scholar] [CrossRef]
- Song, Y.; Dou, H.; Li, X.; Zhao, X.; Li, Y.; Liu, D.; Ji, J.; Liu, F.; Ding, L.; Ni, Y.; et al. Exosomal miR-146a Contributes to the Enhanced Therapeutic Efficacy of Interleukin-1beta-Primed Mesenchymal Stem Cells Against Sepsis. Stem Cells 2017, 35, 1208–1221. [Google Scholar] [CrossRef]
- Yin, W.; Ouyang, S.; Luo, Z.; Zeng, Q.; Hu, B.; Xu, L.; Li, Y.; Xiao, B. Immature Exosomes Derived from MicroRNA-146a Overexpressing Dendritic Cells Act as Antigen-Specific Therapy for Myasthenia Gravis. Inflammation 2017, 40, 1460–1473. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhang, L.I.; Zhang, F.; Tang, T.; Zhou, Q.; Feng, C.; Jin, Y.; Wu, Z. Exosome-mediated miR-146a transfer suppresses type I interferon response and facilitates EV71 infection. PLoS Pathog. 2017, 13, e1006611. [Google Scholar] [CrossRef]
- Dong, C.; Zhou, Q.; Fu, T.; Zhao, R.; Yang, J.; Kong, X.; Zhang, Z.; Sun, C.; Bao, Y.; Ge, X.; et al. Circulating Exosomes Derived-miR-146a from Systemic Lupus Erythematosus Patients Regulates Senescence of Mesenchymal Stem Cells. Biomed. Res. Int. 2019, 2019, 6071308. [Google Scholar] [CrossRef]
- Pan, J.; Alimujiang, M.; Chen, Q.; Shi, H.; Luo, X. Exosomes derived from miR-146a-modified adipose-derived stem cells attenuate acute myocardial infarction-induced myocardial damage via downregulation of early growth response factor 1. J. Cell Biochem. 2019, 120, 4433–4443. [Google Scholar] [CrossRef]
- Perez-Hernandez, J.; Olivares, D.; Forner, M.J.; Ortega, A.; Solaz, E.; Martinez, F.; Chaves, F.J.; Redon, J.; Cortes, R. Urinary exosome miR-146a is a potential marker of albuminuria in essential hypertension. J. Transl. Med. 2018, 16, 228. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Reddy, P.H. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim. Biophys. Acta 2016, 1862, 1617–1627. [Google Scholar] [CrossRef] [PubMed]
- Yuwen, D.; Sheng, B.B.; Liu, J.; Wenyu, W.; Shu, Y.Q. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2650–2658. [Google Scholar]
- Cheng, W.C.; Liao, T.T.; Lin, C.C.; Yuan, L.T.E.; Lan, H.Y.; Lin, H.H.; Teng, H.W.; Chang, H.C.; Lin, C.H.; Yang, C.Y.; et al. RAB27B-activated secretion of stem-like tumor exosomes delivers the biomarker microRNA-146a-5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int. J. Cancer 2019, 145, 2209–2224. [Google Scholar] [CrossRef] [PubMed]
- Zare, N.; Eskandari, N.; Mehrzad, V.; Javanmard, S.H. The expression level of hsa-miR-146a-5p in plasma-derived exosomes of patients with diffuse large B-cell lymphoma. J. Res. Med. Sci. 2019, 2, 10. [Google Scholar]
- Warnecke-Eberz, U.; Chon, S.H.; Hölscher, A.H.; Drebber, U.; Bollschweiler, E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: Comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 2015, 36, 4643–4653. [Google Scholar] [CrossRef]
- Milano, G.; Biemmi, V.; Lazzarini, E.; Balbi, C.; Ciullo, A.; Bolis, S.; Ameri, P.; Di Silvestre, D.; Mauri, P.; Barile, L.; et al. Intravenous administration of cardiac progenitor cell-derived exosomes protects against doxorubicin/trastuzumab-induced cardiac toxicity. Cardiovasc. Res. 2020, 116, 383–392. [Google Scholar] [CrossRef]
- Yang, S.S.; Ma, S.; Dou, H.; Liu, F.; Zhang, S.Y.; Jiang, C.; Xiao, M.; Huang, Y.X. Breast cancer-derived exosomes regulate cell invasion and metastasis in breast cancer via miR-146a to activate cancer associated fibroblasts in tumor microenvironment. Exp. Cell Res. 2020, 391, 111983. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, H.; Wei, X.; Li, H.; Yu, Z.; Zhang, H.; Liu, W. LPS induces HUVEC angiogenesis in vitro through miR-146a-mediated TGF-beta1 inhibition. Am. J. Transl. Res. 2017, 9, 591–600. [Google Scholar]
- Zhu, H.Y.; Bai, W.D.; Liu, J.Q.; Zheng, Z.; Guan, H.; Zhou, Q.; Su, L.L.; Xie, S.T.; Wang, Y.C.; Li, J.; et al. Up-regulation of FGFBP1 signaling contributes to miR-146a-induced angiogenesis in human umbilical vein endothelial cells. Sci. Rep. 2016, 6, 25272. [Google Scholar] [CrossRef]
- Heuslein, J.L.; McDonnell, S.P.; Song, J.; Annex, B.H.; Price, R.J. MicroRNA-146a Regulates Perfusion Recovery in Response to Arterial Occlusion via Arteriogenesis. Front. Bioeng. Biotechnol. 2018, 6, 1. [Google Scholar] [CrossRef]
- Simanovich, E.; Brod, V.; Rahat, M.M.; Rahat, M.A. Function of miR-146a-5p in Tumor Cells as a Regulatory Switch between Cell Death and Angiogenesis: Macrophage Therapy Revisited. Front. Immunol. 2017, 8, 1931. [Google Scholar] [CrossRef]
- Horie, K.; Kawakami, K.; Fujita, Y.; Sugaya, M.; Kameyama, K.; Mizutani, K.; Deguchi, T.; Ito, M. Exosomes expressing carbonic anhydrase 9 promote angiogenesis. Biochem. Biophys. Res. Commun. 2017, 492, 356–361. [Google Scholar] [CrossRef]
- Gong, M.; Yu, B.; Wang, J.; Wang, Y.; Liu, M.; Paul, C.; Millard, R.W.; Xiao, D.S.; Ashraf, M.; Xu, M. Mesenchymal stem cells release exosomes that transfer miRNAs to endothelial cells and promote angiogenesis. Oncotarget 2017, 8, 45200–45212. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, L.; Wang, S.; Han, Q.; Zhao, R.C. Exosomes secreted by mesenchymal stem cells promote endothelial cell angiogenesis by transferring miR-125a. J. Cell Sci. 2016, 129, 2182–2189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef]
- Guo, W.; Gao, Y.; Li, N.; Shao, F.; Wang, C.; Wang, P.; Yang, Z.; Li, R.; He, J. Exosomes: New players in cancer (Review). Oncol. Rep. 2017, 38, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.; Shin, E.; Seong, K.M.; Jin, Y.W.; Youn, H.; Youn, B. The Emerging Roles of Exosomes as EMT Regulators in Cancer. Cells 2020, 9, 861. [Google Scholar] [CrossRef]
- Xiao, D.; Barry, S.; Kmetz, D.; Egger, M.; Pan, J.; Rai, S.N.; Qu, J.; McMasters, K.M.; Hao, H. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016, 376, 318–327. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef]

| Characteristics | HER2+ BC Patients |
|---|---|
| Number | 33 |
| Median age in years (range) | 51.5 (35–70) |
| HR | |
| HR+ | 17 (51.52%) |
| HR− | 16 (48.48%) |
| Stage, N (%) | |
| I | 3 (9.09%) |
| II | 23 (69.70%) |
| III | 6 (18.18%) |
| IV | 1 (3.03%) |
| Grade | |
| II | 11 (33.33%) |
| III | 21 (63.64%) |
| NA | 1 (3.03%) |
| Treatment | |
| Adjuvant | 18 (54.55%) |
| Neoadjuvant | 15 (45.45%) |
| Response | |
| Disease-free | 13 (39.40%) |
| Relapsed | 20 (60.60%) |
| Median follow up (months) | 97 |
| Median DFS | 49 (6–177) |
| Median OS | 96 (7–177) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabello, P.; Torres-Ruiz, S.; Adam-Artigues, A.; Forés-Martos, J.; Martínez, M.T.; Hernando, C.; Zazo, S.; Madoz-Gúrpide, J.; Rovira, A.; Burgués, O.; et al. miR-146a-5p Promotes Angiogenesis and Confers Trastuzumab Resistance in HER2+ Breast Cancer. Cancers 2023, 15, 2138. https://doi.org/10.3390/cancers15072138
Cabello P, Torres-Ruiz S, Adam-Artigues A, Forés-Martos J, Martínez MT, Hernando C, Zazo S, Madoz-Gúrpide J, Rovira A, Burgués O, et al. miR-146a-5p Promotes Angiogenesis and Confers Trastuzumab Resistance in HER2+ Breast Cancer. Cancers. 2023; 15(7):2138. https://doi.org/10.3390/cancers15072138
Chicago/Turabian StyleCabello, Paula, Sandra Torres-Ruiz, Anna Adam-Artigues, Jaume Forés-Martos, María Teresa Martínez, Cristina Hernando, Sandra Zazo, Juan Madoz-Gúrpide, Ana Rovira, Octavio Burgués, and et al. 2023. "miR-146a-5p Promotes Angiogenesis and Confers Trastuzumab Resistance in HER2+ Breast Cancer" Cancers 15, no. 7: 2138. https://doi.org/10.3390/cancers15072138
APA StyleCabello, P., Torres-Ruiz, S., Adam-Artigues, A., Forés-Martos, J., Martínez, M. T., Hernando, C., Zazo, S., Madoz-Gúrpide, J., Rovira, A., Burgués, O., Rojo, F., Albanell, J., Lluch, A., Bermejo, B., Cejalvo, J. M., & Eroles, P. (2023). miR-146a-5p Promotes Angiogenesis and Confers Trastuzumab Resistance in HER2+ Breast Cancer. Cancers, 15(7), 2138. https://doi.org/10.3390/cancers15072138






