The Epigenesis of Salivary Glands Carcinoma: From Field Cancerization to Carcinogenesis
Abstract
Simple Summary
Abstract
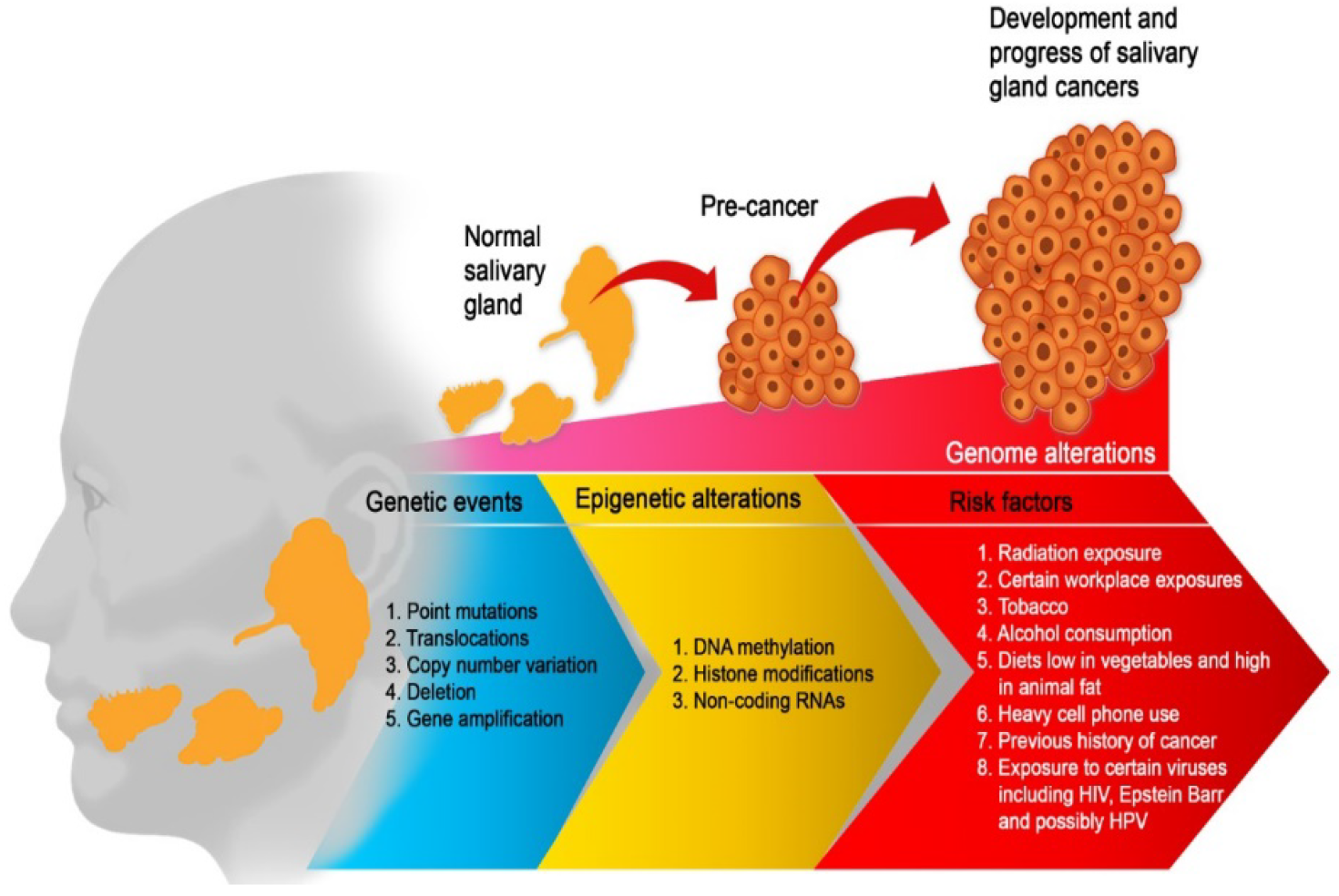
1. Introduction
2. Epigenetics Mechanisms
2.1. DNA Methylation
2.1.1. CpG Islands
2.1.2. DNA Methylases
2.2. Histone Modifications
2.2.1. Histone Acetylation and Deacetylation
2.2.2. Histone Methylation
2.3. Non-Coding RNAs
3. Epigenetic Alterations in Salivary Gland Tumors
3.1. DNA Methylation in Mucoepidermoid Carcinomas (MECs)
3.2. DNA Methylation in Adenoid Cystic Carcinoma (AdCC)
3.3. DNA Methylation in Carcinoma Ex-Pleomorphic Adenoma (Ca Ex-PA)
3.4. DNA Methylation in Acinic Cell Carcinoma
3.5. Non-Coding RNAs Contributing to SGCs
3.6. Tumour to Tumour Interactions
4. Epigenetic Biomarkers for Diagnosis, Prognosis and Treatment Response Prediction of SGCs
5. Epigenetic Drugs in SGC
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iyer, J.; Hariharan, A.; Cao, U.M.N.; Mai, C.T.T.; Wang, A.; Khayambashi, P.; Nguyen, B.H.; Safi, L.; Tran, S.D. An Overview on the Histogenesis and Morphogenesis of Salivary Gland Neoplasms and Evolving Diagnostic Approaches. Cancers 2021, 13, 3910. [Google Scholar] [CrossRef]
- Sreeja, C.; Shahela, T.; Aesha, S.; Satish, M.K. Taxonomy of salivary gland neoplasm. J. Clin. Diagn. Res. 2014, 8, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, E.S.; Ramos, J.C.; Normando, A.G.C.; Mariano, F.V.; Paes Leme, A.F. Epigenetic alterations in salivary gland tumors. Oral Dis. 2020, 26, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, A.K.; Batsakis, J.G. Tumors of the Salivary Glands. In Essentials of Anatomic Pathology; Cheng, L., Bostwick, D.G., Eds.; Humana Press: Totowa, NJ, USA, 2006. [Google Scholar] [CrossRef]
- Da Silva, L.P.; Serpa, M.S.; Viveiros, S.K.; Sena, D.A.C.; de Carvalho Pinho, R.F.; de Abreu Guimarães, L.D.; de Sousa Andrade, E.S.; Dias Pereira, J.R.; Silveira, M.; Sobral, A.P.V.; et al. Salivary gland tumors in a Brazilian population: A 20-year retrospective and multicentric study of 2292 cases. J. Craniomaxillofac. Surg. 2018, 46, 2227–2233. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Hao, Y.; Huang, M.X.; Ma, D.Q.; Chen, Y.; Luo, H.Y.; Gao, Y.; Cao, Z.Q.; Peng, X.; Yu, G.Y. Salivary gland tumours in a northern Chinese population: A 50-year retrospective study of 7190 cases. Int. J. Oral Maxillofac. Surg. 2017, 46, 343–349. [Google Scholar] [CrossRef]
- Wang, X.D.; Meng, L.J.; Hou, T.T.; Huang, S.H. Tumours of the salivary glands in northeastern China: A retrospective study of 2508 patients. Br. J. Oral Maxillofac. Surg. 2015, 53, 132–137. [Google Scholar] [CrossRef]
- Barnes, L.; Eveson, J.W.; Reichart, P.; Sidransky, D. Pathology and Genetics of Head and Neck Tumours. In WHO/IARC Classification of Tumours, 3rd ed.; WHO Press: Geneva, Switzerland, 2005; Volume 9. [Google Scholar]
- Sadetzki, S.; Chetrit, A.; Jarus-Hakak, A.; Cardis, E.; Deutch, Y.; Duvdevani, S.; Zultan, A.; Novikov, I.; Freedman, L.; Wolf, M. Cellular phone use and risk of benign and malignant parotid gland tumors—A nationwide case-control study. Am. J. Epidemiol. 2008, 167, 457–467. [Google Scholar] [CrossRef]
- Dong, C.; Hemminki, K. Second primary neoplasms among 53 159 haematolymphoproliferative malignancy patients in Sweden, 1958–1996: A search for common mechanisms. Br. J. Cancer 2001, 85, 997–1005. [Google Scholar] [CrossRef]
- Isayeva, T.; Said-Al-Naief, N.; Ren, Z.; Li, R.; Gnepp, D.; Brandwein-Gensler, M. Salivary mucoepidermoid carcinoma: Demonstration of transcriptionally active human papillomavirus 16/18. Head Neck Pathol. 2013, 7, 135–148. [Google Scholar] [CrossRef]
- Shebl, F.M.; Bhatia, K.; Engels, E.A. Salivary gland and nasopharyngeal cancers in individuals with acquired immunodeficiency syndrome in United States. Int. J. Cancer 2010, 126, 2503–2508. [Google Scholar] [CrossRef]
- Toper, M.H.; Sarioglu, S. Molecular Pathology of Salivary Gland Neoplasms: Diagnostic, Prognostic, and Predictive Perspective. Adv. Anat. Pathol. 2021, 28, 81–93. [Google Scholar] [CrossRef]
- Costa, F.F. Epigenomics in cancer management. Cancer Manag. Res. 2010, 2, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef]
- Li, Y.; Tollefsbol, T.O. DNA methylation detection: Bisulfite genomic sequencing analysis. Methods Mol. Biol. 2011, 791, 11–21. [Google Scholar] [CrossRef]
- Ramazi, S.; Allahverdi, A.; Zahiri, J. Evaluation of post-translational modifications in histone proteins: A review on histone modification defects in developmental and neurological disorders. J. Biosci. 2020, 45, 135. [Google Scholar] [CrossRef]
- Hanna, C.W.; Demond, H.; Kelsey, G. Epigenetic regulation in development: Is the mouse a good model for the human? Hum. Reprod. Update 2018, 24, 556–576. [Google Scholar] [CrossRef]
- Pal, A. Epigenetics and DNA Methylation. In Protocols in Advanced Genomics and Allied Techniques; Springer: New York, NY, USA, 2022; pp. 245–278. [Google Scholar]
- Fenech, M.; El-Sohemy, A.; Cahill, L.; Ferguson, L.R.; French, T.A.; Tai, E.S.; Milner, J.; Koh, W.P.; Xie, L.; Zucker, M.; et al. Nutrigenetics and nutrigenomics: Viewpoints on the current status and applications in nutrition research and practice. J. Nutr. Nutr. 2011, 4, 69–89. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, N.; Carkic, J.; Ilic Dimitrijevic, I.; Eljabo, N.; Radunovic, M.; Anicic, B.; Tanic, N.; Falk, M.; Milasin, J. P14 methylation: An epigenetic signature of salivary gland mucoepidermoid carcinoma in the Serbian population. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 125, 52–58. [Google Scholar] [CrossRef]
- Wang, Z.; Ling, S.; Rettig, E.; Sobel, R.; Tan, M.; Fertig, E.J.; Considine, M.; El-Naggar, A.K.; Brait, M.; Fakhry, C.; et al. Epigenetic screening of salivary gland mucoepidermoid carcinoma identifies hypomethylation of CLIC3 as a common alteration. Oral Oncol. 2015, 51, 1120–1125. [Google Scholar] [CrossRef]
- Shieh, Y.S.; Shiah, S.G.; Jeng, H.H.; Lee, H.S.; Wu, C.W.; Chang, L.C. DNA methyltransferase 1 expression and promoter methylation of E-cadherin in mucoepidermoid carcinoma. Cancer 2005, 104, 1013–1021. [Google Scholar] [CrossRef]
- Daa, T.; Kashima, K.; Kondo, Y.; Yada, N.; Suzuki, M.; Yokoyama, S. Aberrant methylation in promoter regions of cyclin-dependent kinase inhibitor genes in adenoid cystic carcinoma of the salivary gland. Apmis 2008, 116, 21–26. [Google Scholar] [CrossRef]
- Williams, M.D.; Chakravarti, N.; Kies, M.S.; Maruya, S.; Myers, J.N.; Haviland, J.C.; Weber, R.S.; Lotan, R.; El-Naggar, A.K. Implications of methylation patterns of cancer genes in salivary gland tumors. Clin. Cancer Res. 2006, 12, 7353–7358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Y.; Mao, L.; Li, L.; Tian, Z.; Zhou, X.J.; Zhang, Z.Y.; Li, J. Promoter methylation as a common mechanism for inactivating E-cadherin in human salivary gland adenoid cystic carcinoma. Cancer 2007, 110, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, A.; Sulong, S.; Abdullah, B.; Lazim, N.M. Heterogeneity of Genetic Landscapes in Salivary Gland Tumors and Their Critical Roles in Current Management. Medeni. Med. J. 2022, 37, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.S.; Issa, J.P.; Roberts, D.B.; Williams, M.D.; Weber, R.S.; Kies, M.S.; El-Naggar, A.K. Quantitative promoter hypermethylation analysis of cancer-related genes in salivary gland carcinomas: Comparison with methylation-specific PCR technique and clinical significance. Clin. Cancer Res. 2008, 14, 2664–2672. [Google Scholar] [CrossRef]
- Li, J.; El-Naggar, A.; Mao, L. Promoter methylation of p16INK4a, RASSF1A, and DAPK is frequent in salivary adenoid cystic carcinoma. Cancer 2005, 104, 771–776. [Google Scholar] [CrossRef]
- Durr, M.L.; Mydlarz, W.K.; Shao, C.; Zahurak, M.L.; Chuang, A.Y.; Hoque, M.O.; Westra, W.H.; Liegeois, N.J.; Califano, J.A.; Sidransky, D.; et al. Quantitative Methylation Profiles for Multiple Tumor Suppressor Gene Promoters in Salivary Gland Tumors. PLoS ONE 2010, 5, e10828. [Google Scholar] [CrossRef]
- Uchida, D.; Begum, N.M.; Almofti, A.; Kawamata, H.; Yoshida, H.; Sato, M. Frequent downregulation of 14-3-3 sigma protein and hypermethylation of 14-3-3 sigma gene in salivary gland adenoid cystic carcinoma. Br. J. Cancer 2004, 91, 1131–1138. [Google Scholar] [CrossRef]
- Fan, X.; Chen, B.; Xu, J.; Zhang, H.; Deng, F.; Xiang, X. Methylation status of the PTEN gene in adenoid cystic carcinoma cells. Mol. Med. Rep. 2010, 3, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Maruya, S.; Kurotaki, H.; Wada, R.; Saku, T.; Shinkawa, H.; Yagihashi, S. Promoter methylation and protein expression of the E-cadherin gene in the clinicopathologic assessment of adenoid cystic carcinoma. Mod. Pathol. 2004, 17, 637–645. [Google Scholar] [CrossRef]
- Kanazawa, T.; Misawa, K.; Fukushima, H.; Misawa, Y.; Sato, Y.; Maruta, M.; Imayoshi, S.; Kusaka, G.; Kawabata, K.; Mineta, H.; et al. Epigenetic inactivation of galanin receptors in salivary duct carcinoma of the parotid gland: Potential utility as biomarkers for prognosis. Oncol. Lett. 2018, 15, 9043–9050. [Google Scholar] [CrossRef]
- Tan, M.; Shao, C.; Bishop, J.A.; Feng, Z.; Trock, B.J.; Westra, W.H.; Ha, P.K. Aquaporin-1 promoter hypermethylation is associated with improved prognosis in salivary gland adenoid cystic carcinoma. Otolaryngol. Head Neck Surg. 2014, 150, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Sun, W.; Tan, M.; Glazer, C.A.; Bhan, S.; Zhong, X.; Fakhry, C.; Sharma, R.; Westra, W.H.; Hoque, M.O.; et al. Integrated, genome-wide screening for hypomethylated oncogenes in salivary gland adenoid cystic carcinoma. Clin. Cancer Res. 2011, 17, 4320–4330. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.; Rettig, E.M.; Tan, M.; Chang, X.; Wang, Z.; Brait, M.; Bishop, J.A.; Fertig, E.J.; Considine, M.; Wick, M.J.; et al. Identification of methylated genes in salivary gland adenoid cystic carcinoma xenografts using global demethylation and methylation microarray screening. Int. J. Oncol. 2016, 49, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.H.; Chen, C.; Xu, J.J.; Ling, Z.Q. Critical regions and spreading of runt-related transcription factor-3 C-phosphate-G (CpG) island methylation in human salivary gland adenoid cystic carcinoma. Hum. Pathol. 2011, 42, 1862–1872. [Google Scholar] [CrossRef]
- Hu, Y.H.; Zhang, C.Y.; Tian, Z.; Wang, L.Z.; Li, J. Aberrant protein expression and promoter methylation of p16 gene are correlated with malignant transformation of salivary pleomorphic adenoma. Arch. Pathol. Lab. Med. 2011, 135, 882–889. [Google Scholar] [CrossRef]
- Guo, X.L.; Sun, S.Z.; Wang, W.X.; Wei, F.C.; Yu, H.B.; Ma, B.L. Alterations of p16INK4a tumour suppressor gene in mucoepidermoid carcinoma of the salivary glands. Int. J. Oral Maxillofac. Surg. 2007, 36, 350–353. [Google Scholar] [CrossRef]
- Sasahira, T.; Kurihara, M.; Yamamoto, K.; Bhawal, U.K.; Kirita, T.; Kuniyasu, H. Downregulation of runt-related transcription factor 3 associated with poor prognosis of adenoid cystic and mucoepidermoid carcinomas of the salivary gland. Cancer Sci. 2011, 102, 492–497. [Google Scholar] [CrossRef]
- Wagner, V.P.; Martins, M.D.; Guimaraes, D.M.; Vasconcelos, A.C.; Meurer, L.; Vargas, P.A.; Fonseca, F.P.; Squarize, C.H.; Castilho, R.M. Reduced chromatin acetylation of malignant salivary gland tumors correlates with enhanced proliferation. J. Oral Pathol. Med. 2017, 46, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Zhou, R.; Tian, Z.; Zhang, C.; Wang, L.; Hu, Y.; Han, J.; Li, J. High expression of H3K9me3 is a strong predictor of poor survival in patients with salivary adenoid cystic carcinoma. Arch. Pathol. Lab. Med. 2013, 137, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Binmadi, N.O.; Basile, J.R.; Perez, P.; Gallo, A.; Tandon, M.; Elias, W.; Jang, S.I.; Alevizos, I. miRNA expression profile of mucoepidermoid carcinoma. Oral Dis. 2018, 24, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.L.; Al-Samadi, A.; Sperandio, M.; Soares, A.B.; Teixeira, L.N.; Martinez, E.F.; Demasi, A.P.D.; Araújo, V.C.; Leivo, I.; Salo, T.; et al. MiR-455-3p, miR-150 and miR-375 are aberrantly expressed in salivary gland adenoid cystic carcinoma and polymorphous adenocarcinoma. J. Oral Pathol. Med. 2019, 48, 840–845. [Google Scholar] [CrossRef]
- Mitani, Y.; Roberts, D.B.; Fatani, H.; Weber, R.S.; Kies, M.S.; Lippman, S.M.; El-Naggar, A.K. MicroRNA Profiling of Salivary Adenoid Cystic Carcinoma: Association of miR-17-92 Upregulation with Poor Outcome. PLoS ONE 2013, 8, e66778. [Google Scholar] [CrossRef]
- Persson, M.; Andrén, Y.; Mark, J.; Horlings, H.M.; Persson, F.; Stenman, G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc. Natl. Acad. Sci. USA 2009, 106, 18740–18744. [Google Scholar] [CrossRef]
- Xu, W.; Liu, L.; Lu, H.; Fu, J.; Zhang, C.; Yang, W.; Shen, S. Dysregulated long non-coding RNAs in pleomorphic adenoma tissues of pleomorphic adenoma gene 1 transgenic mice. Mol. Med. Rep. 2019, 19, 4735–4742. [Google Scholar] [CrossRef]
- Flores, B.C.; Lourenço, S.V.; Damascena, A.S.; Kowaslki, L.P.; Soares, F.A.; Coutinho-Camillo, C.M. Altered expression of apoptosis-regulating miRNAs in salivary gland tumors suggests their involvement in salivary gland tumorigenesis. Virchows Arch. 2017, 470, 291–299. [Google Scholar] [CrossRef]
- Lu, H.; Han, N.; Xu, W.; Zhu, Y.; Liu, L.; Liu, S.; Yang, W. Screening and bioinformatics analysis of mRNA, long non-coding RNA and circular RNA expression profiles in mucoepidermoid carcinoma of salivary gland. Biochem. Biophys. Res. Commun. 2019, 508, 66–71. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Kriaucionis, S.; Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009, 324, 929–930. [Google Scholar] [CrossRef] [PubMed]
- Pfaffeneder, T.; Hackner, B.; Truss, M.; Münzel, M.; Müller, M.; Deiml, C.A.; Hagemeier, C.; Carell, T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chem. Int. Ed. Engl. 2011, 50, 7008–7012. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Klose, R. CpG island chromatin: A platform for gene regulation. Epigenetics 2011, 6, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Weber, A.; Langhanki, L.; Schütz, A.; Wittekind, C.; Bootz, F.; Tannapfel, A. Alterations of the INK4a-ARF gene locus in pleomorphic adenoma of the parotid gland. J. Pathol. 2002, 198, 326–334. [Google Scholar] [CrossRef]
- Farthing, C.R.; Ficz, G.; Ng, R.K.; Chan, C.F.; Andrews, S.; Dean, W.; Hemberger, M.; Reik, W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008, 4, e1000116. [Google Scholar] [CrossRef]
- Mohn, F.; Weber, M.; Rebhan, M.; Roloff, T.C.; Richter, J.; Stadler, M.B.; Bibel, M.; Schübeler, D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 2008, 30, 755–766. [Google Scholar] [CrossRef]
- Meissner, A.; Mikkelsen, T.S.; Gu, H.; Wernig, M.; Hanna, J.; Sivachenko, A.; Zhang, X.; Bernstein, B.E.; Nusbaum, C.; Jaffe, D.B.; et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008, 454, 766–770. [Google Scholar] [CrossRef]
- Carey, N.; Marques, C.J.; Reik, W. DNA demethylases: A new epigenetic frontier in drug discovery. Drug. Discov. Today 2011, 16, 683–690. [Google Scholar] [CrossRef]
- Pradhan, S.; Bacolla, A.; Wells, R.D.; Roberts, R.J. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 1999, 274, 33002–33010. [Google Scholar] [CrossRef] [PubMed]
- Sarma, K.; Reinberg, D. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 2005, 6, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Meissner, A.; Lander, E.S. The mammalian epigenome. Cell 2007, 128, 669–681. [Google Scholar] [CrossRef]
- Sun, W.J.; Zhou, X.; Zheng, J.H.; Lu, M.D.; Nie, J.Y.; Yang, X.J.; Zheng, Z.Q. Histone acetyltransferases and deacetylases: Molecular and clinical implications to gastrointestinal carcinogenesis. Acta Biochim. Biophys. Sin. 2012, 44, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.W. Histone-deacetylase inhibitors: Novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 2002, 1, 287–299. [Google Scholar] [CrossRef]
- Janzen, W.P.; Wigle, T.J.; Jin, J.; Frye, S.V. Epigenetics: Tools and Technologies. Drug Discov. Today Technol. 2010, 7, e59–e65. [Google Scholar] [CrossRef]
- Chuang, J.C.; Jones, P.A. Epigenetics and microRNAs. Pediatr. Res. 2007, 61, 24r–29r. [Google Scholar] [CrossRef]
- Koerner, M.V.; Pauler, F.M.; Huang, R.; Barlow, D.P. The function of non-coding RNAs in genomic imprinting. Development 2009, 136, 1771–1783. [Google Scholar] [CrossRef]
- Costa, F.F. Non-coding RNAs, epigenetics and complexity. Gene 2008, 410, 9–17. [Google Scholar] [CrossRef]
- Gutschenritter, T.; Machiorlatti, M.; Vesely, S.; Ahmad, B.; Razaq, W.; Razaq, M. Outcomes and Prognostic Factors of Resected Salivary Gland Malignancies: Examining a Single Institution’s 12-year Experience. Anticancer Res. 2017, 37, 5019–5025. [Google Scholar] [CrossRef]
- Fonseca, F.P.; Sena Filho, M.; Altemani, A.; Speight, P.M.; Vargas, P.A. Molecular signature of salivary gland tumors: Potential use as diagnostic and prognostic marker. J. Oral Pathol. Med. 2016, 45, 101–110. [Google Scholar] [CrossRef]
- Fonseca, F.P.; Carvalho Mde, V.; de Almeida, O.P.; Rangel, A.L.; Takizawa, M.C.; Bueno, A.G.; Vargas, P.A. Clinicopathologic analysis of 493 cases of salivary gland tumors in a Southern Brazilian population. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2012, 114, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Herman, J.G. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J. Pathol. 2002, 196, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, N.; Anicic, B.; Carkic, J.; Simonovic, J.; Toljic, B.; Tanic, N.; Tepavcevic, Z.; Vukadinovic, M.; Konstantinovic, V.S.; Milasin, J. High frequency of p16 and p14 promoter hypermethylation and marked telomere instability in salivary gland tumors. Arch. Oral Biol. 2015, 60, 1662–1666. [Google Scholar] [CrossRef]
- Nishimine, M.; Nakamura, M.; Kishi, M.; Okamoto, M.; Shimada, K.; Ishida, E.; Kirita, T.; Konishi, N. Alterations of p14ARF and p16INK4a genes in salivary gland carcinomas. Oncol. Rep. 2003, 10, 555–560. [Google Scholar]
- Ishida, E.; Nakamura, M.; Ikuta, M.; Shimada, K.; Matsuyoshi, S.; Kirita, T.; Konishi, N. Promotor hypermethylation of p14ARF is a key alteration for progression of oral squamous cell carcinoma. Oral Oncol. 2005, 41, 614–622. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, S.R.; Da Silva, I.C.; Mariz, B.A.; Pereira, A.M.; De Oliveira, N.F. DNA methylation analysis of cancer-related genes in oral epithelial cells of healthy smokers. Arch. Oral Biol. 2015, 60, 825–833. [Google Scholar] [CrossRef]
- Saldaña-Meyer, R.; Recillas-Targa, F. Transcriptional and epigenetic regulation of the p53 tumor suppressor gene. Epigenetics 2011, 6, 1068–1077. [Google Scholar] [CrossRef]
- Daniel, M.; Peek, G.W.; Tollefsbol, T.O. Regulation of the human catalytic subunit of telomerase (hTERT). Gene 2012, 498, 135–146. [Google Scholar] [CrossRef]
- Renaud, S.; Loukinov, D.; Abdullaev, Z.; Guilleret, I.; Bosman, F.T.; Lobanenkov, V.; Benhattar, J. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007, 35, 1245–1256. [Google Scholar] [CrossRef]
- Roh, J.L.; Wang, X.V.; Manola, J.; Sidransky, D.; Forastiere, A.A.; Koch, W.M. Clinical correlates of promoter hypermethylation of four target genes in head and neck cancer: A cooperative group correlative study. Clin. Cancer Res. 2013, 19, 2528–2540. [Google Scholar] [CrossRef] [PubMed]
- Virani, S.; Bellile, E.; Bradford, C.R.; Carey, T.E.; Chepeha, D.B.; Colacino, J.A.; Helman, J.I.; McHugh, J.B.; Peterson, L.A.; Sartor, M.A.; et al. NDN and CD1A are novel prognostic methylation markers in patients with head and neck squamous carcinomas. BMC Cancer 2015, 15, 825. [Google Scholar] [CrossRef]
- Al-Kaabi, A.; van Bockel, L.W.; Pothen, A.J.; Willems, S.M. p16INK4A and p14ARF gene promoter hypermethylation as prognostic biomarker in oral and oropharyngeal squamous cell carcinoma: A review. Dis. Markers 2014, 2014, 260549. [Google Scholar] [CrossRef]
- Toyota, M.; Ahuja, N.; Ohe-Toyota, M.; Herman, J.G.; Baylin, S.B.; Issa, J.P. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA 1999, 96, 8681–8686. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.G.; Baylin, S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003, 349, 2042–2054. [Google Scholar] [CrossRef]
- Bell, A.; Bell, D.; Weber, R.S.; El-Naggar, A.K. CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer 2011, 117, 2898–2909. [Google Scholar] [CrossRef]
- Phuchareon, J.; Ohta, Y.; Woo, J.M.; Eisele, D.W.; Tetsu, O. Genetic profiling reveals cross-contamination and misidentification of 6 adenoid cystic carcinoma cell lines: ACC2, ACC3, ACCM, ACCNS, ACCS and CAC2. PLoS ONE 2009, 4, e6040. [Google Scholar] [CrossRef]
- Clapham, D.E. Not so funny anymore: Pacing channels are cloned. Neuron 1998, 21, 5–7. [Google Scholar] [CrossRef]
- Pape, H.C. Queer current and pacemaker: The hyperpolarization-activated cation current in neurons. Annu. Rev. Physiol. 1996, 58, 299–327. [Google Scholar] [CrossRef] [PubMed]
- Michels, G.; Brandt, M.C.; Zagidullin, N.; Khan, I.F.; Larbig, R.; van Aaken, S.; Wippermann, J.; Hoppe, U.C. Direct evidence for calcium conductance of hyperpolarization-activated cyclic nucleotide-gated channels and human native If at physiological calcium concentrations. Cardiovasc. Res. 2008, 78, 466–475. [Google Scholar] [CrossRef]
- Maruya, S.; Kurotaki, H.; Shimoyama, N.; Kaimori, M.; Shinkawa, H.; Yagihashi, S. Expression of p16 protein and hypermethylation status of its promoter gene in adenoid cystic carcinoma of the head and neck. ORL J. Otorhinolaryngol. Relat. Spec. 2003, 65, 26–32. [Google Scholar] [CrossRef]
- Takata, T.; Kudo, Y.; Zhao, M.; Ogawa, I.; Miyauchi, M.; Sato, S.; Cheng, J.; Nikai, H. Reduced expression of p27(Kip1) protein in relation to salivary adenoid cystic carcinoma metastasis. Cancer 1999, 86, 928–935. [Google Scholar] [CrossRef]
- Sherr, C.J.; Roberts, J.M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995, 9, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Song, S.H.; Jong, H.S.; Choi, H.H.; Kang, S.H.; Ryu, M.H.; Kim, N.K.; Kim, W.H.; Bang, Y.J. Methylation of specific CpG sites in the promoter region could significantly down-regulate p16(INK4a) expression in gastric adenocarcinoma. Int. J. Cancer 2000, 87, 236–240. [Google Scholar] [CrossRef]
- Benassi, M.S.; Molendini, L.; Gamberi, G.; Magagnoli, G.; Ragazzini, P.; Gobbi, G.A.; Sangiorgi, L.; Pazzaglia, L.; Asp, J.; Brantsing, C.; et al. Involvement of INK4A gene products in the pathogenesis and development of human osteosarcoma. Cancer 2001, 92, 3062–3067. [Google Scholar] [CrossRef]
- Yakushiji, T.; Uzawa, K.; Shibahara, T.; Noma, H.; Tanzawa, H. Over-expression of DNA methyltransferases and CDKN2A gene methylation status in squamous cell carcinoma of the oral cavity. Int. J. Oncol. 2003, 22, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Kim, M.J.; Hong, S.P.; Hong, S.D. Inactivation patterns of p16/INK4A in oral squamous cell carcinomas. Exp. Mol. Med. 2004, 36, 165–171. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetic lesions causing genetic lesions in human cancer: Promoter hypermethylation of DNA repair genes. Eur. J. Cancer 2000, 36, 2294–2300. [Google Scholar] [CrossRef] [PubMed]
- Mokarram, P.; Kumar, K.; Brim, H.; Naghibalhossaini, F.; Saberi-firoozi, M.; Nouraie, M.; Green, R.; Lee, E.; Smoot, D.T.; Ashktorab, H. Distinct high-profile methylated genes in colorectal cancer. PLoS ONE 2009, 4, e7012. [Google Scholar] [CrossRef]
- Devaney, J.M.; Wang, S.; Furbert-Harris, P.; Apprey, V.; Ittmann, M.; Wang, B.D.; Olender, J.; Lee, N.H.; Kwabi-Addo, B. Genome-wide differentially methylated genes in prostate cancer tissues from African-American and Caucasian men. Epigenetics 2015, 10, 319–328. [Google Scholar] [CrossRef]
- Ambrosone, C.B.; Young, A.C.; Sucheston, L.E.; Wang, D.; Yan, L.; Liu, S.; Tang, L.; Hu, Q.; Freudenheim, J.L.; Shields, P.G.; et al. Genome-wide methylation patterns provide insight into differences in breast tumor biology between American women of African and European ancestry. Oncotarget 2014, 5, 237–248. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Yuan, Y.; Jiang, S.; Jiang, H. Promoter methylation and expression of CDH1 and susceptibility and prognosis of eyelid squamous cell carcinoma. Tumour Biol. 2016, 37, 9521–9526. [Google Scholar] [CrossRef]
- Shargh, S.A.; Sakizli, M.; Khalaj, V.; Movafagh, A.; Yazdi, H.; Hagigatjou, E.; Sayad, A.; Mansouri, N.; Mortazavi-Tabatabaei, S.A.; Khorram Khorshid, H.R. Downregulation of E-cadherin expression in breast cancer by promoter hypermethylation and its relation with progression and prognosis of tumor. Med. Oncol. 2014, 31, 250. [Google Scholar] [CrossRef]
- Li, G.; Liu, Y.; Yin, H.; Zhang, X.; Mo, X.; Tang, J.; Chen, W. E-cadherin gene promoter hypermethylation may contribute to the risk of bladder cancer among Asian populations. Gene 2014, 534, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Lu, Y.; Li, C.Y.; Yuan, P.; Lin, S.S. Role of CDH1 promoter methylation in colorectal carcinogenesis: A meta-analysis. DNA Cell Biol. 2014, 33, 455–462. [Google Scholar] [CrossRef]
- Lombaerts, M.; van Wezel, T.; Philippo, K.; Dierssen, J.W.; Zimmerman, R.M.; Oosting, J.; van Eijk, R.; Eilers, P.H.; van de Water, B.; Cornelisse, C.J.; et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br. J. Cancer 2006, 94, 661–671. [Google Scholar] [CrossRef]
- Corso, G.; Figueiredo, J.; Biffi, R.; Trentin, C.; Bonanni, B.; Feroce, I.; Serrano, D.; Cassano, E.; Annibale, B.; Melo, S.; et al. E-cadherin germline mutation carriers: Clinical management and genetic implications. Cancer Metastasis Rev. 2014, 33, 1081–1094. [Google Scholar] [CrossRef]
- Liu, J.; Sun, X.; Qin, S.; Wang, H.; Du, N.; Li, Y.; Pang, Y.; Wang, C.; Xu, C.; Ren, H. CDH1 promoter methylation correlates with decreased gene expression and poor prognosis in patients with breast cancer. Oncol. Lett. 2016, 11, 2635–2643. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Shen, N.; Pang, J.; Molina, J.R.; Yang, P.; Liu, S. The DNA Methyltransferase DNMT1 and Tyrosine-Protein Kinase KIT Cooperatively Promote Resistance to 5-Aza-2’-deoxycytidine (Decitabine) and Midostaurin (PKC412) in Lung Cancer Cells. J. Biol. Chem. 2015, 290, 18480–18494. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Tanigami, A.; Yamakawa, K.; Akiyama, F.; Kasumi, F.; Sakamoto, G.; Nakamura, Y. Allelotype of breast cancer: Cumulative allele losses promote tumor progression in primary breast cancer. Cancer Res. 1990, 50, 7184–7189. [Google Scholar] [PubMed]
- Cui, H.; Wang, L.; Gong, P.; Zhao, C.; Zhang, S.; Zhang, K.; Zhou, R.; Zhao, Z.; Fan, H. Deregulation between miR-29b/c and DNMT3A is associated with epigenetic silencing of the CDH1 gene, affecting cell migration and invasion in gastric cancer. PLoS ONE 2015, 10, e0123926. [Google Scholar] [CrossRef]
- Dong, C.; Wu, Y.; Wang, Y.; Wang, C.; Kang, T.; Rychahou, P.G.; Chi, Y.I.; Evers, B.M.; Zhou, B.P. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene 2013, 32, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Kashibuchi, K.; Tomita, K.; Schalken, J.A.; Kume, H.; Takeuchi, T.; Kitamura, T. The prognostic value of E-cadherin, alpha-, beta- and gamma-catenin in bladder cancer patients who underwent radical cystectomy. Int. J. Urol. 2007, 14, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, S.; Sasaki, A.; Mese, H.; Alcalde, R.E.; Tsuji, T.; Matsumura, T. The E-cadherin gene is silenced by CpG methylation in human oral squamous cell carcinomas. Int. J. Cancer 2001, 93, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Augello, C.; Gregorio, V.; Bazan, V.; Cammareri, P.; Agnese, V.; Cascio, S.; Corsale, S.; Calò, V.; Gullo, A.; Passantino, R.; et al. TP53 and p16INK4A, but not H-KI-Ras, are involved in tumorigenesis and progression of pleomorphic adenomas. J. Cell Physiol. 2006, 207, 654–659. [Google Scholar] [CrossRef]
- Stephens, P.J.; Davies, H.R.; Mitani, Y.; Van Loo, P.; Shlien, A.; Tarpey, P.S.; Papaemmanuil, E.; Cheverton, A.; Bignell, G.R.; Butler, A.P.; et al. Whole exome sequencing of adenoid cystic carcinoma. J. Clin. Investig. 2013, 123, 2965–2968. [Google Scholar] [CrossRef]
- Ho, A.S.; Kannan, K.; Roy, D.M.; Morris, L.G.; Ganly, I.; Katabi, N.; Ramaswami, D.; Walsh, L.A.; Eng, S.; Huse, J.T.; et al. The mutational landscape of adenoid cystic carcinoma. Nat. Genet. 2013, 45, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Pouloudi, D.; Manou, M.; Sarantis, P.; Tsoukalas, N.; Tsourouflis, G.; Dana, E.; Karamouzis, M.V.; Klijanienko, J.; Theocharis, S. Clinical Significance of Histone Deacetylase (HDAC)-1, -2, -4 and -6 Expression in Salivary Gland Tumors. Diagnostics 2021, 11, 517. [Google Scholar] [CrossRef]
- Giaginis, C.; Alexandrou, P.; Delladetsima, I.; Giannopoulou, I.; Patsouris, E.; Theocharis, S. Clinical significance of histone deacetylase (HDAC)-1, HDAC-2, HDAC-4, and HDAC-6 expression in human malignant and benign thyroid lesions. Tumour Biol. 2014, 35, 61–71. [Google Scholar] [CrossRef]
- Mutze, K.; Langer, R.; Becker, K.; Ott, K.; Novotny, A.; Luber, B.; Hapfelmeier, A.; Göttlicher, M.; Höfler, H.; Keller, G. Histone deacetylase (HDAC) 1 and 2 expression and chemotherapy in gastric cancer. Ann. Surg. Oncol. 2010, 17, 3336–3343. [Google Scholar] [CrossRef]
- Seo, J.; Min, S.K.; Park, H.R.; Kim, D.H.; Kwon, M.J.; Kim, L.S.; Ju, Y.S. Expression of Histone Deacetylases HDAC1, HDAC2, HDAC3, and HDAC6 in Invasive Ductal Carcinomas of the Breast. J. Breast Cancer 2014, 17, 323–331. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef]
- Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007, 8, 286–298. [Google Scholar] [CrossRef]
- Chen, Z.; Lin, S.; Li, J.L.; Ni, W.; Guo, R.; Lu, J.; Kaye, F.J.; Wu, L. CRTC1-MAML2 fusion-induced lncRNA LINC00473 expression maintains the growth and survival of human mucoepidermoid carcinoma cells. Oncogene 2018, 37, 1885–1895. [Google Scholar] [CrossRef] [PubMed]
- Hiss, S.; Eckstein, M.; Segschneider, P.; Mantsopoulos, K.; Iro, H.; Hartmann, A.; Agaimy, A.; Haller, F.; Mueller, S.K. Tumour-Infiltrating Lymphocytes (TILs) and PD-L1 Expression Correlate with Lymph Node Metastasis, High-Grade Transformation and Shorter Metastasis-Free Survival in Patients with Acinic Cell Carcinoma (AciCC) of the Salivary Glands. Cancers 2021, 13, 965. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wen, Z.; Kou, N.; Liu, J.; Jin, D.; Wang, L.; Wang, F.; Gao, L. LIS1 interacts with CLIP170 to promote tumor growth and metastasis via the Cdc42 signaling pathway in salivary gland adenoid cystic carcinoma. Int. J. Oncol. 2022, 61, 129. [Google Scholar] [CrossRef]
- Zhao, D.; Yang, K.; Tang, X.F.; Lin, N.N.; Liu, J.Y. Expression of integrin-linked kinase in adenoid cystic carcinoma of salivary glands correlates with epithelial-mesenchymal transition markers and tumor progression. Med. Oncol. 2013, 30, 619. [Google Scholar] [CrossRef]
- Dodd, R.L.; Slevin, N.J. Salivary gland adenoid cystic carcinoma: A review of chemotherapy and molecular therapies. Oral Oncol. 2006, 42, 759–769. [Google Scholar] [CrossRef]
- Kaira, K.; Toyoda, M.; Shino, M.; Sakakura, K.; Takahashi, K.; Tominaga, H.; Oriuchi, N.; Kanai, Y.; Oyama, T.; Chikamatsu, K. Clinicopathological significance of L-type amino acid transporter 1 (LAT1) expression in patients with adenoid cystic carcinoma. Pathol. Oncol. Res. 2013, 19, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shao, C.; Tan, M.L.; Mu, D.; Ferris, R.L.; Ha, P.K. Molecular biology of adenoid cystic carcinoma. Head Neck 2012, 34, 1665–1677. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Zhao, Y.X.; Xia, R.H.; Han, J.; Wang, B.S.; Tian, Z.; Wang, L.Z.; Hu, Y.H.; Li, J. RASSF1A promoter hypermethylation is a strong biomarker of poor survival in patients with salivary adenoid cystic carcinoma in a Chinese population. PLoS ONE 2014, 9, e110159. [Google Scholar] [CrossRef]
- Zhou, X.Q.; Huang, S.Y.; Zhang, D.S.; Zhang, S.Z.; Li, W.G.; Chen, Z.W.; Wu, H.W. Effects of 5-aza-2’deoxycytidine on RECK gene expression and tumor invasion in salivary adenoid cystic carcinoma. Braz. J. Med. Biol. Res. 2015, 48, 254–260. [Google Scholar] [CrossRef]
- Lam-Ubol, A.; Phattarataratip, E. Distinct histone H3 modification profiles correlate with aggressive characteristics of salivary gland neoplasms. Sci. Rep. 2022, 12, 15063. [Google Scholar] [CrossRef]
- Takahashi, H.; Tada, Y.; Saotome, T.; Akazawa, K.; Ojiri, H.; Fushimi, C.; Masubuchi, T.; Matsuki, T.; Tani, K.; Osamura, R.Y.; et al. Phase II Trial of Trastuzumab and Docetaxel in Patients With Human Epidermal Growth Factor Receptor 2-Positive Salivary Duct Carcinoma. J. Clin. Oncol. 2019, 37, 125–134. [Google Scholar] [CrossRef]
- Majchrzak-Celińska, A.; Warych, A.; Szoszkiewicz, M. Novel Approaches to Epigenetic Therapies: From Drug Combinations to Epigenetic Editing. Genes 2021, 12, 208. [Google Scholar] [CrossRef]
- Burkitt, K.; Saloura, V. Epigenetic Modifiers as Novel Therapeutic Targets and a Systematic Review of Clinical Studies Investigating Epigenetic Inhibitors in Head and Neck Cancer. Cancers 2021, 13, 5241. [Google Scholar] [CrossRef] [PubMed]
- Di Villeneuve, L.; Souza, I.L.; Tolentino, F.D.S.; Ferrarotto, R.; Schvartsman, G. Salivary Gland Carcinoma: Novel Targets to Overcome Treatment Resistance in Advanced Disease. Front. Oncol. 2020, 10, 580141. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H.; Kusaba, T.; Kawamura, K.; Oyama, Y.; Daa, T. Histopathological Aspects of the Prognostic Factors for Salivary Gland Cancers. Cancers 2023, 15, 1236. [Google Scholar] [CrossRef]
- Gargano, S.M.; Senarathne, W.; Feldman, R.; Florento, E.; Stafford, P.; Swensen, J.; Vranic, S.; Gatalica, Z. Novel therapeutic targets in salivary duct carcinoma uncovered by comprehensive molecular profiling. Cancer Med. 2019, 8, 7322–7329. [Google Scholar] [CrossRef] [PubMed]
- Kurzrock, R.; Bowles, D.W.; Kang, H.; Meric-Bernstam, F.; Hainsworth, J.; Spigel, D.R.; Bose, R.; Burris, H.; Sweeney, C.J.; Beattie, M.S.; et al. Targeted therapy for advanced salivary gland carcinoma based on molecular profiling: Results from MyPathway, a phase IIa multiple basket study. Ann. Oncol. 2019, 31, 412–421. [Google Scholar] [CrossRef]
- Park, J.C.; Ma, T.M.; Rooper, L.; Hembrough, T.; Foss, R.D.; Schmitt, N.C.; Sawhney, R.; Flanders, A.; Kang, H. Exceptional responses to pertuzumab, trastuzumab, and docetaxel in human epidermal growth factor receptor-2 high expressing salivary duct carcinomas. Head Neck 2018, 40, E100–E106. [Google Scholar] [CrossRef] [PubMed]
- Uijen, M.J.M.; Lassche, G.; van Engen-van Grunsven, A.C.H.; Driessen, C.M.L.; van Herpen, C.M.L. Case series of docetaxel, trastuzumab, and pertuzumab (DTP) as first line anti-HER2 therapy and ado-trastuzumab emtansine (T-DM1) as second line for recurrent or metastatic HER2-positive salivary duct carcinoma. Oral Oncol. 2022, 125, 105703. [Google Scholar] [CrossRef] [PubMed]

| First Author/Year | Country | Gene/Genome Elements | Genome Region | SGC Tumor (Malignant and/or Benign) | Sample Type | Molecular Alteration | Biological Function Associated with Molecular Alteration | References |
|---|---|---|---|---|---|---|---|---|
| DNA Methylation | ||||||||
| Nikolic et al., 2018 | Serbia | p14ARF/p16INK4a | 9p21.3 | Pleomorphic Adenoma/Carcinoma Ex Pleomorphic Adenoma | formalin-fixed, paraffin-embedded (FFPE) samples | hypermethylation | Transcriptional silencing of the p14/ARF gene | [23] |
| Wang et al., 2015 | USA | CLIC3 | 9q34.3 | Mucoepidermoid carcinomas (MECs) | FFPE MEC tumor samples | hypermethylation | Oncogenic ole | [24] |
| Shieh et al., 2005 | Taiwan | CDH1 | 16q22.1 | Mucoepidermoid carcinomas (MECs) | FFPE MEC tumor samples | hypermethylation | Loss of E-cadherin expression | [25] |
| Daa et al., 2008 | Japan | p15, p18, p19, p21, & p27 | 9p21.3, 1p32.3, 19p13.2, 6p21.2, 12p13.1 | Adenoid cystic carcinoma (AdCC) | FFPE AdCC tumor samples | hypermethylation | Cell cycle disruption | [26] |
| Williams et al., 2006 | Texas | DAPK, MGMT, RARβ2, anRASSF1 (Ras association domain family protein1 isoform A) | 9q21.33, | Adenoid Cystic Carcinoma, Mucoepidermoid Carcinoma, Acinic cell Carcinoma, Salivary Duct Carcinomas | FFPE tissues from Adenoid cystic carcinoma (AdCC), Mucoepidermoid carcinoma (MECs), Salivary duct carcinoma (SDCs), and acinic cell carcinoma samples | RASSF1 (Ras association domain family protein1 isoform A) and RARβ2 were highly methylated in malignant tumors and MGMT and DAPK in benign neoplasm. | Low or absent protein expression | [27] |
| Zhang et al., 2007 | China | E-cadherin | Cadherin 1: 16q22.1 | Adenoid cystic carcinoma (AdCC) | Tissue samples | Hypermethylation | E-cadherin plays a critical role in transducing signals to influence several important biologic processes. Reduced expression of E-cad, caused by genetic and epigenetic events, has been observed in aggressive carcinoma types. promoter methylation of E-cadherin is a more common mechanism for its inactivation. | [28] |
| Xia et al., 2017 | China | CDH1 | CDH1 gene located on 16q 22.1 | Salivary carcinoma ex pleomorphic adenoma (CXPA) | Formalin-fixed and paraffin-embedded tissues | Promoter hypermethylation | CDH1 silencing is directly related to advanced tumor stage and an aggressive phenotype. The association of CDH1 methylation with cervical lymph node metastasis, histological grade and advanced tumor stage suggests that the CDH1 gene may be particularly important in salivary CXPA tumor progression. | [29] |
| Lee et al., 2008 | Republic of Korea | RARβ2 and RASSF1 | chromosomal region 3p24 and chromosome 3p21.3 | AdCC, adenoid cystic carcinoma, and salivary duct carcinoma. | Tissue samples | Promoter hypermethylation | These genes are tumor suppressor genes and known for their ability to suppress vital cellular processes, including cell-cycle regulation, apoptosis, DNA repair, differentiation, and metastasis. The hypermethylation of the two genes may synergistically involve in the carcinogenesis of these two entities. | [30] |
| Li et al., 2005 | Texas | p16INK4a, RASSF1A, and DAPK | chromosome 9p21, chromosome 3p21. 3 and chromosome 9q21.33 | Adenoid cystic carcinoma (AdCC) | Formalin-fixed and paraffin-embedded tissues | Promoter hypermethylation | Promoter methylation of these gene often results in silencing of its expression and is acommon mechanism to inactivate tumor suppressor genes in tumorigenesis. | [31] |
| Durr et al., 2010 | United States of America | APC, Mint 1, PGP9.5, RAR-b, andTimp3 | Chromosome 5q22. 2, SPEN gene, chromosome 4p14, chromosome 17q21.2 and Chromosome 22 | Malignant SGTs | Paraffin embedded tissues | Promoter hypermethylation | Promoter methylation of these gene may contribute to salivary gland carcinogenesis | [32] |
| Uchida et al., 2004 | Japan | 14-3-3 σ | chromosome 8q22.3 | Adenoid cystic carcinoma (AdCC) | Tissue sample | Hypermethylation | The downregulation of 14-3-3 σ by hypermethylation of the CpG island may contribute to salivary gland carcinogenesis | [33] |
| Fan et al., 2010 | China | PTEN | Chromosome 10q23. 31 | Adenoid cystic carcinoma (AdCC) | ACC-2 cell lines | Promoter hypermethylation | The hypermethylation of the PTEN promoter region is one of the major mechanisms leading to reduced expression of PTen in adenoid cystic carcinomas. This indicates that PTen is an important candidate gene involved in the pathogenesis of adenoid cystic carcinomas | [34] |
| Maruya et al., 2004 | Japan | E-cadherin | Cadherin 1: 16q22.1 | Adenoid cystic carcinoma (AdCC) | Paraffin-embedded tumor tissues | Hypermethylation | E-cadherin plays a critical role in transducing signals to influence several important biologic processes. Reduced expression of E-cad, caused by genetic and epigenetic events, has been observed in high grade and aggressive tumors. promoter methylation of E-cadherin is a more common mechanism for its inactivation. | [35] |
| Kanazawa et al., 2018 | USA | GALR1 and GALR2 | G-protein coupled receptors family | Salivary duct carcinoma (SDC) | Tissue samples | Promoter hypermethylation | The galanin receptors, GALR1 and GALR2, are members of the GPCR superfamily, and serve as important tumor suppressor genes. The silencing of the GALR1 and GALR2 genes by methylation may constitute a critical event in SDC. | [36] |
| Tan et al., 2014; Shao et al., 2011 | USA | AQP1 | Aquaporins: located on chromosome 6 in a region with homology of synteny with human 7p14. | Adenoid cystic carcinoma (AdCC) | Tumor tissue sample | Hypomethylation | AQP1 is a small transmembrane protein that selectively transports water across cell membranes. It is highly expressed in several tumor types and has been implicated in tumor cell proliferation, extravasation, migration, and metastasis. AQP1 was significantly hypomethylated in ACC tumors compared with controls | [37,38] |
| Ling et al., 2016 | USA | HCN2 | Chromosome 19 | Adenoid cystic carcinoma (AdCC) | Formalin-fixed, paraffin-embedded tissue sample | Promoter hypomethylation | HCN2 normally conducts K+ and Na+, it has been reported that HCN2 was permeable to Ca2+, and it was suggested that they may participate in pathological Ca2+ signaling when HCN2 is overexpressed. Hypomethylation of HCN2 as a potential biomarker for ACC that may be associated with more aggressive disease | [39] |
| Ge et al., 2011 | China | RUNX3 gene | Chromosomal region 1p36 | Adenoid cystic carcinoma (AdCC) | Tissue samples | Hypermethylation | The methylation of the promoter 5′-CpG island in the RUNX3 gene is a major gene-silencing mechanism. RUNX3 protein expression is significantly related to metastasis and T stage. | [40] |
| HU et al., 2011 | China | P16 | Chromosome 9p21 | Carcinoma ex Pleomorphic adenoma (Ca-ex-PA) | Tissue samples | Promoter hypermethylation | Overexpression of p16 protein in the cytoplasm and decreased expression of p16 protein in the nucleus may play important roles in the evolution of pleomorphic adenoma to Ca-ex-PA | [41] |
| Guo et al., 2007 | China | p16INK4a | Chromosome 9p21 | Mucoepidermoid carcinoma (MEC) | Tissue samples | promoter hypermethylation | Alterations of the p16INK4a tumour suppressor gene are often observed in a variety of human cancers and are considered to play a critical role in the transition to malignant growth. the main mechanisms of inactivation of the p16INK4a gene in MEC of the salivary glands are promoter hypermethylation. | [42] |
| Sasahira et al., 2011 | Japan | RUNX3 | Chromosomal region 1p36 | Pleomorphic adenoma (PA). Adenoid cystic carcinoma (AdCC) andMucoepidermoid carcinoma (MEC) | Formalin-fixed, paraffin embedded samples | Hypermethylation | RUNX3 inactivation is observed more frequently in salivary gland tumors than in normal salivary gland tissues and RUNX3 downregulation is significantly correlated with tumor progression and poor prognosis in AdCC and MEC | [43] |
| Histone modifications | ||||||||
| Wanger et al., 2017 | Brazil | H3lys9 | histone H3 at Lys 9 | Adenoid cystic carcinoma, mucoepidermoid carcinomas and acinic cell carcinoma | FFPE tissue blocks of SGTs | Histones are hypoacetylated | histone deposition, chromatin assembly and gene activation | [44] |
| Xia et al., 2013 | China | H3K9me3 and H3K9Ac | Trimethylation of histone 3 lysine 9 1q42.13 | Adenoid cystic carcinoma (AdCC) | AdCC tumor samples | Histones were acetylated and methylated | Rapid cell proliferation and distant metastasis in ACC | [45] |
| Non-coding RNAs | ||||||||
| Binmadi e al., 2018 | Maryland | miR-302a | 4q25 | Mucoepidermoid carcinomas (MECs) | MECs tumor samples | Non-coding RNA was upregulated | Cancer invasions | [46] |
| Brown et al., 2019 | Finland | miRNA-150, miRNA-375, and miRNA-455-3p | Chromosome 19q13, chromosome 2 and chromosome 9 at locus 9q32 | Mucoepidermoid carcinomas (MEC) and Adenoid cystic carcinoma (AdCC) | Formalin-fixed paraffin embedded | miRNA-150 and miRNA-375 expression was significantly decreased in AdCC and PAC, whilst miRNA-455-3p showed significantly increased expression in AdCC when compared to PAC. | The post-transcriptional protein expression has been shown to play important roles in neoplastic and non-neoplastic processes. These non-coding RNAs presented alternated expression in SGC. | [47] |
| Mitani et al., 2013 | USA | miR-17 and miR-20a | Chromosome 13 | Adenoid cystic carcinoma (AdCC) | Tissue samples | Overexpression of the miR-17 and miR-20a | miRNAs play a role in the regulation of cellular pathways in the ACC tumorigenesis, and this may be influenced by the fusion gene status. Overexpression of the miR-17 and miR-20a were significantly associated with poor outcome in the screening and validation sets | [48] |
| Persson et al., 2009 | The Netherlands | miR-15a/16 and miR-150 | Chromosome 13q14 and chromosome 19 | Adenoid cystic carcinoma (AdCC) | Tissue samples | miR-15a/16 was overexpressed in ACC as compared with normal glandular tissues, whereas the expression of miR-150 was lower in ACC than in normal glandular tissues | The deregulation of the expression of MYB and its target genes is a key oncogenic event in the pathogenesis of ACC. miR-15a/16 and miR-150 recently were shown to regulate MYB expression negatively. The MYB-NFIB fusion is a hallmark of ACC and that deregulation of the expression of MYB and its target genes is a key oncogenic event in the pathogenesis of ACC | [49] |
| Xu et al., 2019 | China | Different lncRNA and mRNA in PLAG1 gene | Chromosome 8q12 | Carcinoma Ex Pleomorphic Adenoma | Mouse tumors glands | lncRNAs and mRNAs were differentially expressed in PA tissues obtained from PLAG1 transgenic mice as compared with those from control mice. | The differentially expressed mRNAs and lncRNA revealed that these mRNAs were closely associated with a number of processes involved in the development of PA. | [50] |
| Flores et al., 2017 | Brazil | miR-15a, miR16, miR-17-5p, miR-21, miR-29, miR-34a and miR-20a | Chromosome 13q14, Chromosome 13q14, chromosome 13, chromosome 17q23.2, chromosome 7q32.3, chromosome 1p36.22 and chromosome 19 | Mucoepidermoid carcinoma (MEC) and Carcinoma Ex Pleomorphic adenoma (PA) | Tissue samples | The expression of miR-21 and miR-34a was upregulated in MEC, respectively. Downregulation of miR-20a was observed in PA and in MEC. The upregulation of miR-15a, miR16, miR-17-5p, miR-21, miR-29, and miR-34a was observed in PA | The expression of apoptosis-regulating miRNAs in salivary gland tumors, suggesting possible involvement of these microRNAs in salivary gland tumorigenesis. | [51] |
| Lu et al., 2019 | China | hsa_circ_00123, NON- HSAT154433.1 and circ012342 | circRNA | Mucoepidermoid carcinoma (MEC) | Tissue samples | The circR- NAs showed the highest fold change in MEC group compared with normal control group. The elevated expression of NON- HSAT154433.1 and decreased expression of circ012342 were observed and closely related to the pathogenesis of MEC | An increasing number of circRNAs have been discovered in various diseases and exhibit cell-type or tissue-specific patterns. | [52] |
| Lam-Ubol et al., 2022 | Thailand | H3K9Me3 and H3K9Ac | Trimethylation of histone 3 lysine 9 1q42.13 | Mucoepidermoid carcinoma (MEC) and Adenoid cystic carcinoma (AdCC) | Tissue samples | Hyperacetylation and trimethylation of histone H3 | Increased H3 trimethylation at lysine residue 9, as well as H3 acetylation at lysine residue 9 and 18, could be involved in the progression of malignancies. | [53] |
| Carcinoma Type | Methylated Genes | References |
|---|---|---|
| Mucoepidermoid Carcinoma | P14, CLIC3, CDH1, APC, Mint1, PGP9.5, Timp3, p16(INK4A), RUNX3, DAPK, MGMT, RARβ2 and RASSF1 | [3,11,23] |
| Adenoid cystic carcinoma | P15, p18, p19, p21, APC, Mint1, PGP9.5, Timp3 Cyclin-dependent kinase inhibitors (p27), HCN2, AQP1, SBSN, RUNX3, DAPK, MGMT, RARβ2 and RASSF1 | [3,26,28,31] |
| CA-Ex-PA | RASSF1, p53, p16(INK4A), promoter methylation in CDH1, P14ARF | [29,32,41] |
| Acinic cell carcinoma | RASSF1 (Ras association domain family protein1 isoform A) and RARβ2, DAPK, and MGMT | [3] |
| Potential Diagnostic Biomarkers | |||||
|---|---|---|---|---|---|
| Gene/Genome Elements | Genome Region | Sample Type | Molecular Alteration | Tumor Types of SGC (Malignant and/or Benign) | References |
| RARβ2 and RASSF1A | Chromosome 3p21. 3 and Chromosome 3p24 | Fresh-frozen tissue specimens | Hypermethylation | Salivary Gland Carcinomas (AdCC, adenoid cystic carcinoma, and salivary duct carcinoma) | [30] |
| p16INK4a | Chromosome 9p21 | Tissue samples | Promoter hypermethylation | Mucoepidermoid carcinoma (MEC) and adenoid cystic carcinoma (AdCC) | [31,42] |
| p15, p18, p19, p21, and p27 | Chromosome 9p21.3, 1p32.3, 19p13.2, 6p21.2, 12p13.1 | Tissue samples | Promoter hypermethylation | Adenoid cystic carcinoma (AdCC) | [26,94] |
| APC, Mint 1, PGP9.5, RAR-b, andTimp3 | Chromosome 5q22. 2, SPEN gene, chromosome 4p14, chromosome 17q21.2 and Chromosome 22 | Tissue samples | Hypermethylation | Malignant SGTs (Pleomorphic adenoma (PA), Mucoepidermoid carcinoma (MEC), adenoid cystic carcinoma AdCC and Salivary duct carcinoma (SDC) | [32] |
| 14-3-3 σ | chromosome 8q22.3 | Tissue samples | Hypermethylation | Adenoid cystic carcinoma (AdCC) | [33] |
| AQP1 | Aquaporins: located on chromosome 6 in a region with homology of synteny with human 7p14. | Tumor tissues | hypomethylation | Adenoid cystic carcinoma (AdCC) | [38] |
| SBSN | Suprabasin: 19q13.12 | The saliva of AdCC patients | hypomethylation | Adenoid cystic carcinoma (AdCC) | [38] |
| acetyl-H3 (lys9) | histone 3 (H3) acetylation at Lys9 | paraffin-embedded tissue | hypoacetylated | Adenoid cystic carcinoma (AdCC), Mucoepidermoid carcinoma (MEC) and Adenoid cystic carcinoma (AdCC) | [44] |
| PTEN | Chromosome 10q23. 31 | ACC-2 cell lines | Promoter hypermethylation | Adenoid cystic carcinoma (AdCC) | [34] |
| RASSF1 and RARβ2 | Chromosome 3p24 and chromosome 3p21. 3 | Tissue samples | hypermethylation | Adenoid cystic carcinoma (AdCC) and Salivary duct carcinoma (SDC) | [27] |
| E-cadherin | Chromosome 16q22. 1 | Tissue samples | Promoter hypermethylation | Adenoid cystic carcinoma (AdCC) | [35] |
| p16INK4a, RASSF1A, and DAPK | chromosome 9p21, chromosome 3p21. 3 and chromosome 9q21.33 | Formalin-fixed and paraffin-embedded tissues | Promoter hypermethylation | Adenoid cystic carcinoma (AdCC) | [31] |
| P14, p16, hTERT and TP53 | chromosome 9p21, chromosome 9p21, chromosome 5p15. 33 | formalin-fixed, paraffin-embedded sample | Hypermethylation | Mucoepidermoid carcinoma (MEC) | [23] |
| CLIC3 | Chromosome 9 | Tissue samples | Promoter hypomethylation | Mucoepidermoid carcinoma (MEC) | [24] |
| HCN2 | Chromosome 19 | Formalin-fixed, paraffin-embedded tissue sample | Promoter hypomethylation | Adenoid cystic carcinoma (AdCC) | [39] |
| RUNX3 gene | Chromosomal region 1p36 | Tissue samples | Hypermethylation | Adenoid cystic carcinoma (AdCC) | [40] |
| P16 | Chromosome 9p21 | Tissue samples | Promoter Hypermethylation | Carcinoma ex Pleomorphic adenoma(Ca-ex-PA) | [41] |
| MiR-455-3p | Chromosome 9 at locus 9q32 | Formalin-fixed paraffin embedded | Significantly increased expression in AdCC | Adenoid cystic carcinoma (AdCC) | [47] |
| Different lncRNA and mRNA in PLAG1 gene | Chromosome 8q12 | Mouse tumors glands | lncRNAs and mRNAs were differentially expressed in PA tissues obtained from PLAG1 transgenic mice as compared with those from control mice. | Pleomorphic adenoma (PA) | [50] |
| Potential biomarkers of treatment response and prognostic | |||||
| Gene/Genome elements | Genome region | Sample type | Molecular alteration | Tumor types of SGC (malignant and/or benign) | References |
| EN1 gene | Engrailed Homeobox 1: 2q14.2 | The saliva of AdCC patients | hypermethylation | Adenoid cystic carcinoma (AdCC) | [133] |
| SBSN | Suprabasin: 19q13.12 | paraffin-embedded samples | hypomethylation | Adenoid cystic carcinoma (AdCC) | [38] |
| AQP1 | Aquaporins: located on chromosome 6 in a region with homology of synteny with human 7p14. | Tumor tissues | hypomethylation | Adenoid cystic carcinoma (AdCC) | [38] |
| inactivation of E-cadherin, encoded by CDH1 | Cadherin 1: 16q22.1 | Tissue samples | Promoter hypermethylation | Adenoid cystic carcinoma (AdCC) | [28] |
| RASSF1A | Ras Association Domain Family Member 1: 3p21.31 | The saliva of AdCC patients | hypermethylation | Adenoid cystic carcinoma (AdCC) | [134] |
| H3k9me3 | 9th lysine residue of the histone H3 protein and is often associated with heterochromatin. | The saliva of AdCC patients | trimethylation of histone 3 lysine 9 | Adenoid cystic carcinoma (AdCC) | [45] |
| galanin receptors (GALRs); GALR1 and GALR2 | G-protein coupled receptors family | Tumor specimens | Hypermethylation | Salivary duct carcinoma (SDS) | [36] |
| RARβ2 and RASSF1A | Chromosome 3p24 and chromosome 3p21. 3 | Fresh-frozen tissue specimens | Promoter hypermethylation | Malignant Salivary Gland Carcinomas (ACC, adenoid cystic carcinoma, and salivary duct carcinoma) | [30] |
| E-cadherin | Cadherin 1: 16q22.1 | Tissue samples | Promoter hypermethylation | Adenoid cystic carcinoma (AdCC) | [35] |
| CDH1 | Chromosome 16q22.1 | Formalin-fixed and paraffin-embedded tissues | Promoter hyperrmethylation | Salivary carcinoma ex pleomorphic adenoma (CXPA) | [135] |
| RASSF1A | Chromosome 3p21. 3 | Formalin-fixed and paraffin-embedded tissues | Promoter hypermethylation | Adenoid cystic carcinoma (AdCC) | [31] |
| 14-3-3 σ | Chromosome 8q22.3 | Tissue samples | Hypermethylation | Adenoid cystic carcinoma (AdCC) | [33] |
| H3K9Ac, H3K9Me3 and H3K18Ac | Trimethylation of histone 3 lysine 9 1q42.13 and histone H3 lysine 18 | Tissue samples | Hyperacetylation and trimethylation of histone H3 | mucoepidermoid carcinoma (MEC) and adenoid cystic carcinoma (AdCC) | [136] |
| RUNX3 gene | Chromosomal region 1p36 | Tissue samples | Hypermethylation | Adenoid cystic carcinoma (AdCC) and Mucoepidermoid carcinoma (MEC) | [40,43] |
| miR-17 and miR-20a | Chromosome 13 | Tissue samples | Overexpression of the miR-17 and miR-20a | Adenoid cystic carcinoma (AdCC) | [48] |
| Different lncRNA and mRNAin PLAG1 gene | Chromosome 8q12 | Mouse tumors glands | lncRNAs and mRNAs were differentially expressed in PA tissues obtained from PLAG1 transgenic mice as compared with those from control mice. | Pleomorphic adenoma (PA) | [50] |
| hsa_circ_00123 and NON-HSAT154433.1 | circRNA | Tissue samples | The circR-NAs showed the highest fold change in MEC group compared with normal control group. The elevated expression of NON-HSAT154433.1 and decreased expression of circ012342 were observed and closely related to the pathogenesis of MEC | Mucoepidermoid carcinoma (MEC) | [52] |
| Agent(s) | Cancer Type(s) | Target | FDA-Approved Date | Trial Details | Trial Identifer/Status | Reference |
|---|---|---|---|---|---|---|
| Azacytidine (DNMT inhibitor) | HPV-positive HNSCC (resectable disease) | pending | 2014-ongoing (window study) | Recruiting | [137] | |
| Decitabine (DNMT unhibitor) | HPV-positive Anogenital and HNSCC (R/M) | Pending | 2019-ongoing (phase 1b) | Recruiting | [138,139] | |
| Cabozantinib | All histologies | c-MET | 17 September 2021 | Phase II | Active, not recruiting | [140] |
| Nivolumab | All histologies | PD-1 | 4 March 2022 | Phase II | Active, not recruiting | [140,141] |
| ivolumab + ipilimumab | All histologies | PD-1 CTLA-4 | 26 May 2020 | Phase II | Active, not recruiting | [142] |
| Pembrolizumab | All histologies | PD-1 | 26 July 2021 | Phase II | Recruiting | [142,143] |
| Nivolumab + ipilimumab | All histologies | PD-1 CTLA-4 | 26 May 2020 | Phase II | Recruiting | [142] |
| Pembrolizumab + lenvatinib | All histologies | PD-1 VEGFR | 10 August 2021 | Phase II | Not yet Recruiting | [142] |
| Lutetium-177 PSMA | All histologies | PSMA | 23 March 2022 | Phase II | Not yet Recruiting | [142] |
| Axitinib + Avelumab | Adenoid cystic only | VEGFR PD-L1, | 14 March 2019 | Phase II | Recruiting | [142] |
| CB-103 | Adenoid cystic carcinoma + other tumors | NOTCH | - | Phase I/II | Recruiting | [142] |
| BB1503 (amcarsetinib) | All histologies + other tumors | NOTCH | 5 April 2022 | Phase Ib/II | Active, not recruiting | [142] |
| AL101 | Adenoid cystic only | NOTCH | 16 September 2021 | Phase II | Recruiting | [142] |
| TeTMYB + BGBA 17 | Solid Tumors | MYB | - | Phase I | Not Yet Recruiting | [142] |
| Transtuzumab | Salivary duct carcinoma + other solid tumours | HER2 positive | 2002 | Phase II | Active | [144,145] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mat Lazim, N.; Yousaf, A.; Abusalah, M.A.H.; Sulong, S.; Mohd Ismail, Z.I.; Mohamud, R.; Abu-Harirah, H.A.; AlRamadneh, T.N.; Hassan, R.; Abdullah, B. The Epigenesis of Salivary Glands Carcinoma: From Field Cancerization to Carcinogenesis. Cancers 2023, 15, 2111. https://doi.org/10.3390/cancers15072111
Mat Lazim N, Yousaf A, Abusalah MAH, Sulong S, Mohd Ismail ZI, Mohamud R, Abu-Harirah HA, AlRamadneh TN, Hassan R, Abdullah B. The Epigenesis of Salivary Glands Carcinoma: From Field Cancerization to Carcinogenesis. Cancers. 2023; 15(7):2111. https://doi.org/10.3390/cancers15072111
Chicago/Turabian StyleMat Lazim, Norhafiza, Anam Yousaf, Mai Abdel Haleem Abusalah, Sarina Sulong, Zul Izhar Mohd Ismail, Rohimah Mohamud, Hashem A. Abu-Harirah, Tareq Nayef AlRamadneh, Rosline Hassan, and Baharudin Abdullah. 2023. "The Epigenesis of Salivary Glands Carcinoma: From Field Cancerization to Carcinogenesis" Cancers 15, no. 7: 2111. https://doi.org/10.3390/cancers15072111
APA StyleMat Lazim, N., Yousaf, A., Abusalah, M. A. H., Sulong, S., Mohd Ismail, Z. I., Mohamud, R., Abu-Harirah, H. A., AlRamadneh, T. N., Hassan, R., & Abdullah, B. (2023). The Epigenesis of Salivary Glands Carcinoma: From Field Cancerization to Carcinogenesis. Cancers, 15(7), 2111. https://doi.org/10.3390/cancers15072111







