Simple Summary
There remains a lack of identification of genes responsible for the most aggressive of prostate cancers. A unique resource consisting of sampled, related men who died from their confirmed prostate cancer and who were members of pedigrees shown to have a significant excess of prostate cancer cases has allowed the identification of multiple rare candidate predisposition variants for prostate cancer. Strong candidate variants are presented, including a rare, validated variant in the gene LRBA, all of which can be examined further in additional studies.
Abstract
There is evidence for contribution of inherited factors to prostate cancer, and more specifically to lethal prostate cancer, but few responsible genes/variants have been identified. We examined genetic sequence data for 51 affected cousin pairs who each died from prostate cancer and who were members of high-risk prostate cancer pedigrees in order to identify rare variants shared by the cousins as candidate predisposition variants. Candidate variants were tested for association with prostate cancer risk in UK Biobank data. Candidate variants were also assayed in 1195 additional sampled Utah prostate cancer cases. We used 3D protein structure prediction methods to analyze structural changes and provide insights into mechanisms of pathogenicity. Almost 4000 rare (<0.005) variants were identified as shared in the 51 affected cousin pairs. One candidate variant was also significantly associated with prostate cancer risk among the 840 variants with data in UK Biobank, in the gene LRBA (p = 3.2 × 10−5; OR = 2.09). The rare risk variant in LRBA was observed to segregate in five pedigrees. The overall predicted structures of the mutant protein do not show any significant overall changes upon mutation, but the mutated structure loses a helical structure for the two residues after the mutation. This unique analysis of closely related individuals with lethal prostate cancer, who were members of high-risk prostate cancer pedigrees, has identified a strong set of candidate predisposition variants which should be pursued in independent studies. Validation data for a subset of the candidates identified are presented, with strong evidence for a rare variant in LRBA.
1. Introduction
There is evidence for an inherited contribution to predisposition to prostate cancer, and stronger evidence for lethal prostate cancer [1,2,3,4,5,6]. While hundreds of common variants in low-risk genes have been associated with risk for prostate cancer (e.g., [7]), few rare variants in moderate- to high-penetrance genes have been identified, and they explain little of familial prostate cancer [8,9]. The genes most consistently recognized to affect prostate cancer risk include ATM, BRCA1, BRCA2, CHEK2, HOXB13, MLH1, MSH2, MSH6, PMS2, and PALB2 [10], not all of which display equivalent penetrance.
We sequenced germline DNA from 51 pairs of cousins who both died from prostate cancer who were also members of pedigrees with a significant excess of prostate cancer cases. We identified the set of rare coding and noncoding variants shared in these high-risk lethal prostate cancer-affected cousin pairs as strong candidate variants for predisposition to prostate cancer. Confirmation of significant association with prostate cancer risk in an independent population and observation of segregation of variants with prostate cancer in multiple high-risk pedigrees provided additional validation for a subset of the candidates considered. We used 3D protein structure prediction methods to analyze structural changes in one outstanding candidate variant that may provide insights on mechanisms of pathogenicity [11].
2. Data/Methods
2.1. Utah Population Database (UPDB)
The UPDB consists of the genealogy of the majority of the Utah population, from its founders in the mid-1800s to their modern-day descendants. The genealogy has been linked to the Utah Cancer Registry (UCR) and to Utah death certificates coded with International Classification of Disease causes of death from 1904. The statewide UCR was created in 1966 and has been an NCI Surveillance, Epidemiology, and End-Results (SEER) registry since 1973. It records and tracks all independent primary cancers diagnosed or treated in Utah, including pathologic confirmation. Of the over 3 million individuals with genealogy of at least 3 generations linking to Utah founders, there were 7727 individuals whose linked Utah death certificate indicated prostate cancer as a cause of death; 6328 of these individuals also had a UCR record confirming a prostate cancer diagnosis.
Using the combined UPDB genealogy and UCR data, the Genetic Epidemiology Program at the University of Utah has previously identified and sampled ~2500 prostate cancer cases and ~7500 relatives who belonged to approximately 500 Utah pedigrees that each had an excess of prostate cancer cases among the descendants (high-risk pedigrees). In total, 422 of these sampled prostate cancer cases were since identified to have died from their prostate cancer based on the presence of prostate cancer as a cause of death on their linked Utah death certificate. These men were termed lethal prostate cancer (LPrCa) cases. All genetic relationships among these cases were identified in the UPDB genealogy to identify descending pedigrees.
2.2. Affected LPrCa Cousin Pairs in High-Risk Pedigrees
All independent descending clusters (pedigrees) including 2 or more of the 422 sampled men who died of their prostate cancer (LPrCa cases) were identified in the UPDB. No pedigrees were completely overlapping, but an LPrCa case could belong to more than one pedigree through different ancestors. Each pedigree that included at least two sampled LPrCa cases was tested for a significant excess of prostate cancer cases as follows. All individuals with biological sex of male and at least three generations of genealogy linking to Utah founders were assigned to a cohort based on five-year birthyear and birth state (Utah or not). Prostate cancer cohort-specific rates were estimated as the number of prostate cancer cases in each cohort divided by the number of males with genealogy data in the cohort. Each pedigree was tested for an excess of prostate cancer cases by comparing the observed number of prostate cancer cases among the descendants to the expected number of prostate cancer cases among the descendants. The expected number of cases among the descendants was estimated by summing the cohort-specific rate of prostate cancer for all male descendants. A pedigree was termed high-risk for prostate cancer in the presence of a significant excess of observed cases (p < 0.05). For each of these high-risk prostate cancer pedigrees identified, which also contained at least 2 sampled LPrCa cases, we selected those LPrCa cases who were related as first or second cousins for sequencing. Each of the 102 LPrCa cases in the 51 cousin pairs identified had a stored sample of germline DNA extracted from whole blood available for whole-genome sequencing.
2.3. Whole-Genome Sequencing/Identification of Candidate Predisposition Variants/Assay Development
The 102 samples of high-molecular-weight DNA (>30 Kb in length) were whole-genome sequenced at MedGenome, Foster City, CA, USA. (http://www.medgenome.com) utilizing 10× Genomics (https://www.10xgenomics.com/) long, linked read sequence technology. A high efficiency microfluidic device mixes functionalized gel beads containing unique barcodes with enzymes and a limiting amount of genomic DNA. These components are encapsulated in oil to produce a GEM, gel bead in emulsion. With up to 4 million barcodes available, GEM particles produce uniquely identifiable high-molecular-weight DNA fragments. These fragments are then sequenced utilizing Illumina sequencers to produce 30× whole-genome phased coverage. Genomes were aligned to GRCh37 with 10× Genomics LongRanger software version 2.2.2. VCFs were merged with BCFTOOLS. The merged VCF was annotated with ANNOVAR ([12]; https://academic.oup.com/nar/article/38/16/e164/1749458).
A total of 114,513 rare (MAF < 0.005 in GnomAD 2.1) coding (nonsynonymous, frameshift, startloss, startgain, stoploss) variants were identified in the 102 LPrCa cases. Of these, 17,859 variants had allele counts >1, and 3251 of these rare coding variants (in 1762 genes) were shared within at least 1 LPrCa cousin pair. Noncoding and UTR variants were prioritized after RegulomeDB scoring. The RegulomeDB score represents evidence that each variant functions in a regulatory role (ranging from 1, indicating strong evidence, to 6, indicating weak evidence). A total of 546 rare noncoding variants with RegulomeDB scores ranging between 2a and 4, and shared in at least 1 cousin pair, were identified.
These 3797 (3251 coding + 546 noncoding) rare, shared candidate predisposition variants were submitted for assay design using the Illumina iSelectHD Custom Genotyping BeadChips (https://www.illumina.com/products/by-type/informatics-products/designstudio.html). A total of 200 base pairs of genomic sequence surrounding each variant was used for the design process. Assays were manufactured for 3559 of these candidate variants (238 failed to design). Illumina iSelectHD Custom Beadchips were processed at the University of Utah HSC Genomics Core.
2.4. Segregation of Candidate Predisposition Variants in Pedigrees
In total, 1298 additional previously sampled Utah prostate cancer cases were identified for assay with the candidate predisposition variants to determine segregation of the candidate variants. These assayed prostate cancer cases included the original affected cousin pairs (n = 102); all other sampled prostate cancer cases whose death certificate indicated prostate cancer as a cause of death (n = 320 LPRCA cases); all sampled prostate cancer cases who were first-, second-, or third-degree relatives of the affected cousin pair cases (n = 168) and all sampled prostate cancer cases who were first-, second-, and third-degree relatives (n = 307) of these 168 cases; and all metastatic prostate cancer cases recruited by author N.A. in the Huntsman Cancer Institute Urology Clinic (n = 401). DNA for 1195 of these 1298 sampled prostate cancer cases passed quality control and was assayed for the candidate variants to test for segregation and to identify additional carriers.
2.5. Case/Control Risk Association in UK Biobank
These rare, shared candidate predisposition variants that were identified in the sequencing experiment and had data in UK Biobank were analyzed for association with prostate cancer risk in a set of 7764 Caucasian prostate cancer cases and 1:1 ancestrally matched controls from among the UK Biobank’s 488,377 total subjects genotyped on the Illumina OmniExpress SNP array [13]. UK Biobank case and control subjects were matched via principal components (PC) using ~27K independent markers that excluded several genomic regions known to adversely affect PC analysis [14]. FLASHPCA2 software was used to generate eigenvectors for control selection [14]. Controls were selected from among 64,284 Caucasian UK Biobank subjects who were male, over 70 years of age, and had no cancer diagnosis. One control, representing the nearest neighbor based on the Euclidean distance of the first two PCs, was selected for each case. Then, 129 outlier cases and controls were removed, leaving 7635 cases and controls.
2.6. UK Biobank Imputation
The selected UK Biobank case and control subjects were imputed to ~40M SNP markers using the Haplotype Reference Consortium’s (HRC) 67 K background genomes [15]. Beginning with 784,256 observed SNP genotypes, pre-imputation quality control using PLINK software [16] required sample genotyping > 98% (no subjects removed); a total of 353,578 markers were removed by filtering for genotyping call rate < 98%, HWE p < 1 × 10−5, MAF < 0.005, duplicated position in the HRC’s reference genome, or site not included in the HRC’s reference genome. The remaining 430,678 SNPs were converted to human genome B37 forward strand orientation using GenotypeHarmonizer software [17] and served as the basis for imputation. Imputation was performed with EAGLE v2.3 software for phasing [18] and MINIMAC3 software for imputation [19]. Post-imputation quality control included removing markers with imputation information score (INFO-r2) < 0.7 [7,20,21].
2.7. Protein Structure Prediction
LRBA (lipopolysaccharide-responsive beige-like anchor protein) is a very large protein with 2863 amino acids that is not tractable using existing 3D structure prediction methods. The only experimental structure available for LRBA is an X-ray structure for positions 2076–2489 [22,23]. There are two isoforms; here, we consider the isoform 1 as the canonical sequence. For this sequence, the single point mutation considered here is T2533P. The protein has multiple domains, and the mutation in question, T2533P, is localized between the second BEACH (2200–2489) domain and the second WD2 (2591–2633) domain. To perform 3D structure prediction with I-TASSER structure prediction software using full homology modeling [24,25,26,27], we constructed a model including the PH domain (2073–2181), the BEACH domain (2200–2489), and the WD2–WD6 domains (2591–2858). For this model, the amino acid substitution under consideration is T461P. The I-TASSER predictions resulted in a set of three candidate structures with C-scores of −3.86, −3.85, and −3.93 for the wild-type sequence, and two with C-scores of −3.74 and −3.80 for the mutant sequence. All the structures obtained were visualized and analyzed using Chimera [28].
3. Results
3.1. Case/Control Risk Association
Only 840 of the 3797 candidate predisposition variants had some data present in UK Biobank, likely due to the very low frequency (<0.005). Only 1 of the 840 variants was significantly associated with prostate cancer risk after correction for multiple testing (p < 0.05/840 or p < 5.95 × 10−5), in the gene LRBA (p = 3.2 × 10−5; OR = 2.09).
3.2. Assay of Candidate Variants in 1195 Prostate Cancer Cases
In total, 102 of the 1195 prostate cancer cases assayed for the candidate variants were the original target pair LPrCa cases; these cases were assayed to validate performance of the assays. Of the 3559 variants assayed, 2035 failed to identify any heterozygous (het) carriers with the assay. These variants were considered to have failed performance, since the assay should have at least identified the original cousin pair carriers. For 562 of the variants, although some het carriers were identified, the assay did not identify the original cousin pair as carriers, so these variants were also considered to have failed the assay. For 78 of the variants, the only carriers identified in the assay were the original cousin pair; these variant assays were considered to have performed appropriately, but they did not identify any additional carriers of the candidate variants. This finding was not unexpected given the rare nature of the variants, but these variants did not provide any additional evidence for segregation and were not further considered here. For the remaining 884 of the variant assays, all original cousin pair carriers were identified, as well as additional het carriers. These 884 candidate variants were considered the best rare candidate lethal prostate cancer predisposition variants. These variants were examined further in the 1195 prostate cancer cases assayed, to determine if segregation to other related prostate cancer cases was observed.
All het carriers for each of these 884 rare candidate variants were analyzed by identifying all genealogical relationships among the het carriers using the UPDB linked genealogy. This analysis identified many more clusters than the original 51 pedigrees analyzed since all genetic relationships among all cases were considered; 1959 total clusters (pedigrees) that included 3 or more related het carriers for at least 1 of these 884 variants were identified. Although none of these pedigrees overlapped completely, a case could belong to more than one.
These 884 candidate variants were all originally selected for their low frequency (<0.005) based on review of GnomAD. However, some of the variants were observed at much higher frequency than 0.005 in the assay of 1195 prostate cancer cases. It is possible that these variants might represent truly rare variants that were observed in much higher frequencies in the assay of prostate cancer cases because they are, in fact, strongly associated with prostate cancer risk. However, these results might also be due to inaccuracies in reporting of frequencies for what were, in fact, more common variants, or due to assay performance failure. We could not confirm the lack of a data problem for these more commonly observed variants, so we elected to exclude consideration of any of the 1959 variant-segregating pedigrees for any variant for which more than 10% (n > 110 carriers) het carriers were observed in the assay of 1195 prostate cancer cases.
This left 1070 pedigrees of interest that included at least 3 related het carriers of 1 of the candidate variants. Because some of these clusters of het carriers might represent random clusters rather than high-risk prostate cancer pedigrees, we additionally excluded those variant-sharing pedigrees that did not exhibit a significant excess of prostate cancer cases (p < 0.05) among the descendants of the common founding ancestor of all the related carriers. This left 814 high-risk prostate cancer pedigrees, including 559 different rare candidate variants in 398 genes that showed evidence of segregation of the candidate variant with prostate cancer. The list of these 559 rare candidate predisposition variants is provided in Supplemental Table S1, which includes the UK Biobank association test results for those variants with data, the diagnosis and interpretation data from ClinVar, and identifies those variants associated with a prostate cancer pathway in Ingenuity.
Only 1 of these 559 candidate variants was significantly validated for risk association with prostate cancer (LRBA). This LRBA variant (rs62346982) is classified in ClinVar with “conflicting interpretations of pathogenicity”. The LRBA variant was observed to segregate with prostate cancer in a total of five high-risk prostate cancer pedigrees. Figure 1 shows segregation of the rare LRBA variant in the largest LRBA-segregating pedigree identified. The founder of this pedigree was born in the late 1700s in Vermont and has almost 32,000 descendants in UPDB, with a total of 230 prostate cancer cases observed and 174.9 expected (p = 4.0 × 10−5) among all descendants (additional UPDB data not shown); variant-carrying cases are shown. A multiple marriage in the third generation is noted with a triangle on the two horizontal marriage lines, variant carriers are noted by “+”, and the decade of prostate cancer diagnosis is noted below each case. It must be noted that complete prostate cancer case diagnosis data for Utah are only available from 1973, which explains why cases are only observed in the bottom, most recent few generations of the high-risk pedigrees shown.
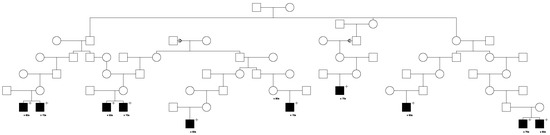
Figure 1.
One of the high-risk prostate cancer pedigrees segregating the rare LRBA candidate prostate cancer predisposition variant.
3.3. Protein Structure Prediction
Lipopolysaccharide-responsive and beige-like anchor protein (LRBA), the single candidate predisposition variant identified in a cousin pair and also confirmed with significant association to prostate cancer in the UK Biobank case/control analysis, is involved in coupling signal transduction and vesicle trafficking to enable polarized secretion and/or membrane deposition of immune effector molecules (this function was assigned by similarity). It is involved in phagophore growth during mitophagy by regulating ATG9A trafficking to mitochondria. A mutation of this protein has been associated with CVID8, an autosomal recessive immunologic disorder associated with defective B-cell differentiation.
The variant considered here corresponds to rs62346982. PolyPhen-2 predictor predicts this variant as benign with a score of 0.167 (sensitivity 0.92 and specificity 0.87) [29]. The overall predicted sequences do not show any significant changes upon the amino acid replacement T461P in the model considered here, but careful inspection of the region close to the replacement (440–480) shows that the mutated structure loses a helical structure two residues after the replacement (see Figure 2 and Figure 3). The water accessibility does not change upon mutation, keeping this region buried.

Figure 2.
Secondary structure comparison in the region of the T461P replacement.
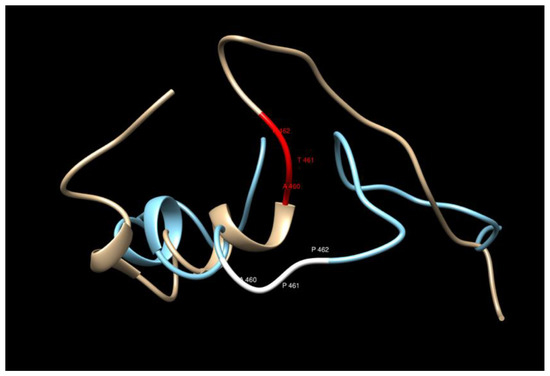
Figure 3.
The 3D structure comparison in the region of the T461P replacement. The bronze/red structure corresponds to the WT and the blue/white one to the MT.
3.4. Consideration of Other Likely Candidate Predisposition Variants Identified
In addition to the significant evidence identifying the rare LRBA variant as a strong candidate prostate cancer predisposition variant, this study identified a large set of other rare candidate predisposition variants, many of which are worthy of further consideration. We have used various criteria to select a subset of these LPrCa candidate predisposition variants for presentation in more detail below. These criteria include: (i) known cancer pathogenic variants, (ii) variants in genes recognized to be implicated in breast cancer (BROCA genes), and (iii) noncoding variants with suggestive Regulome DB scores.
- (i)
- Cancer pathogenic variants
Only one candidate variant that was classified as “pathogenic/likely pathogenic” in ClinVar was identified, in MUTYH (seq_1_45797228_C-T_MUTYH). This variant was identified in 29/1195 of the assayed cases and segregated in 3 high-risk prostate cancer pedigrees including 6, 3, and 3 sampled related cases, respectively. Figure 4 below shows the largest of these three pedigrees, with full shading for prostate cancer cases; the six assayed het carriers are identified with “+”. One inferred case carrier (not sampled) is also shown; decade of age at diagnosis of prostate cancer is shown below each case. The founder of this pedigree was born in New York in the early 1800s, with a total of almost 16,000 descendants included in UPDB; only the descending lines to the 6 identified het carriers are shown. In the UPDB, a total of 114 prostate cancer cases were observed among all descendants, with 68.3 cases expected (p = 2.7 × 10−7). The founding male had two marriages, indicated with a small triangle on the top horizontal marriage lines.
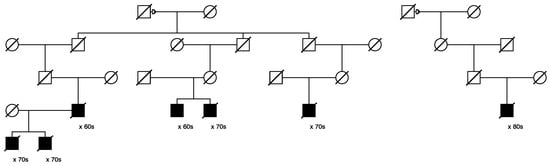
Figure 4.
High-risk prostate cancer pedigree segregating the rare pathogenic MUTYH variant.
Eleven candidate variants found to segregate in high-risk pedigrees were classified with “conflicting interpretations of pathogenicity” in ClinVar; these included variants in DUOX2, LRBA (discussed previously), MYO3A, EVC, CP, FBN1, MYH11, KCND3, ACNT2, ADAM9, and PTCH1. The largest pedigree observed for the “conflicting” variant identified for DUOX2 (n = 14 case carriers) is shown in Figure 5, and the largest pedigree observed for LRBA (n = 10) was shown in Figure 1. PTCH1 was also selected as a BROCA gene, and a pedigree is shown later. A variant in MSH6 was the only variant of 14 classified in ClinVar as of “uncertain significance” that also reported a cancer-related clinical diagnosis; it was also selected as a BROCA variant and is shown later.
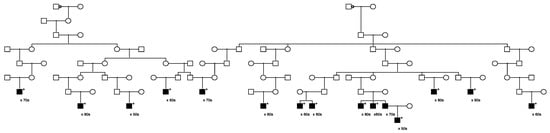
Figure 5.
High-risk prostate cancer pedigree segregating a rare DUOX2 variant.
Figure 5 shows the segregation of the rare DUOX2 variant in a high-risk prostate cancer pedigree. The founder of this pedigree was born in the mid-1700s in North Carolina and has almost 153,000 descendants in UPDB; 897 prostate cancer cases were observed among all descendants, with 811.5 expected (p = 0.0016). The male founder of this pedigree had two spouses, shown with a triangle on the top horizontal marriage lines. All 14 prostate cancer case variant carriers are shown with “+”, and 1 inferred case (father of an affected carrier who was not sampled) is also shown.
- (ii)
- Variants in recognized BROCA genes
Seven of the rare candidate predisposition variants found to segregate in prostate cancer cases who were members of high-risk prostate cancer pedigrees were in known BROCA genes, including the MUTYH variant already discussed, as well as variants in APC (ClinVar: conflicting interpretations of pathogenicity), BRCA1 (benign), MSH6 (uncertain significance), PTCH1 (conflicting), SDHC (not classified), and SLX4 (benign/likely benign). The pedigrees segregating the rare variants in BRCA1, MSH6, and PTCH1 are shown in Figure 6, Figure 7 and Figure 8.
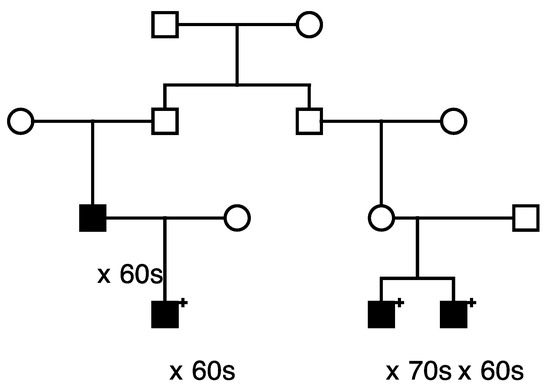
Figure 6.
High-risk prostate cancer pedigree segregating a rare BRCA1 variant.
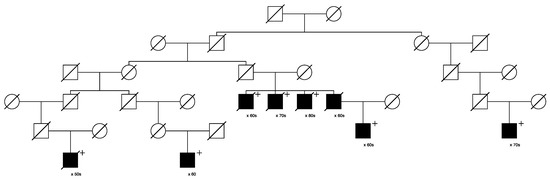
Figure 7.
High-risk prostate cancer pedigree segregating a rare MSH6 variant.
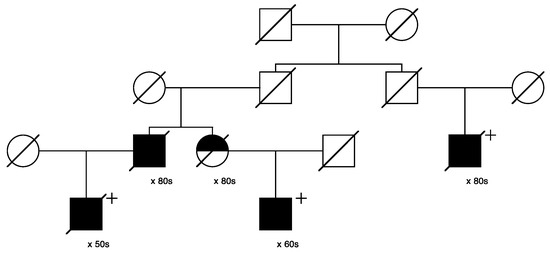
Figure 8.
High-risk prostate cancer pedigree segregating a rare PTCH1 variant.
Figure 6 shows the single high-risk prostate cancer pedigree segregating a BRCA1 variant (seq_17_41228587_T-G_BRCA1). This variant has been classified as “benign” in ClinVar, based on the breast cancer phenotype. The founder of this pedigree was born in the early 1800s in Canada and has a total of over 2300 descendants in the UPDB; a total of 25 prostate cancer cases were observed among all descendants, with 12.1 expected (p = 7.6 × 10−4). The three identified variant carriers are shown with “+”; also shown is the unsampled pedigree member father of one of the affected pair cases who was also diagnosed with prostate cancer. This pedigree also has a significant excess of breast cancer (18 observed, 11.3 expected, p = 0.04). There are no identified breast cancer-affected descendants of the first son of the founder pair shown in Figure 6, but the second son of the founder has eight descendants diagnosed with breast cancer (UPDB breast cancer cases not shown).
Figure 7 shows the single high-risk pedigree segregating a rare MSH6 variant (seq_2_48026979_A-C_MSH6). This variant is classified as “uncertain” in ClinVar. The founder of this pedigree was born in the early 1800s in Massachusetts and has almost 23,000 descendants in UPDB; 166 prostate cancer cases were observed among all descendants, with 113.0 expected (p = 1.8 × 10−6); 129 breast cancers were observed with 109.8 expected (p = 0.04); 23 ovarian cancers were observed with 13.9 expected (p = 0.016). Neither colorectal nor endometrial cancers were observed in excess among the descendants in this pedigree. Prostate cancer case variant carriers are shown with “+”; one unsampled father of a carrier was also diagnosed with prostate cancer as shown. Two of the prostate cancer case carriers had an additional cancer diagnosis in UPDB (one kidney and one chronic lymphocytic leukemia); data are not shown to protect privacy.
Figure 8 shows the single high-risk pedigree segregating a rare PTCH1 variant (seq_9_98240378_C-T_PTCH1). This variant is classified as “conflicting” in ClinVar. The founder of this pedigree was born in the mid-1800s with no recorded birthplace and has almost 4600 descendants in UPDB; a total of 33 prostate cancers were observed among all descendants, with 20.0 expected (p = 0.0047). This pedigree also exhibits a significant excess of breast cancer: a total of 37 breast cancers were observed in the entire descending pedigree, with 23.1 expected (p = 0.0045). One of these breast cancer cases was the mother of a carrier, shown with half shading.
- (iii)
- Noncoding variants with suggestive Regulome DB scores
Regulome DB (RGDB) scores ranging from “2a” to “4” were observed among the rare candidate variants. The two variants with the most evidence for a regulatory role based on RGDB score were in PAX6 and ZWILCH, and both were scored “2a”. Two pedigrees with the PAX6 variant were observed with three het carriers each, and one pedigree with the ZWILCH variant also included three het carriers; these pedigrees are not shown.
Multiple variants with an RGDB score of “2b” were identified, but pedigrees are not shown here. The founder of the largest pedigree, with 11 het carriers of an APCDD1L-DT variant with score “2b”, was born in the late 1700s in Massachusetts and has over 36,000 total descendants, with 268 prostate cancer cases observed among all descendants, and 167.8 expected (p = 6.6 × 10−13).
4. Discussion
It is estimated that 10–20% of prostate cancer cases occur in a familial context [1,30]. Genetic predisposition to prostate cancer development has been associated with both rare variants in moderate- to high-penetrance genes, as well as with common genetic alterations in low-risk genes (e.g., [7,8]). Almost 200 common variants have been identified in large case–control cohorts, but other evidence for their direct association with prostate cancer is lacking, and only a small fraction of familial prostate cancer is associated with known rare predisposition variants.
High-risk pedigree studies remain a powerful mechanism for identification of predisposition genes and variants [31,32,33,34]. This has proven true for prostate cancer [35,36], although such high-risk prostate cancer pedigrees remain infrequently presented. Here, we have taken advantage of unique Utah resources, combined with an unusual and powerful study design that includes sampled affected cousin pairs, to generate, and begin to evaluate, a strong list of candidate predisposition variants for prostate cancer.
Extensive linked genealogic and disease registries existing in Utah have been used to identify and study thousands of Utah high-risk pedigrees [37]. We have previously used this same sequencing approach in affected cousins belonging to high-risk pedigrees to identify multiple candidate predisposition variants for several different phenotypes, including GOLM1 for melanoma [38], ERF for bladder cancer [36], FANCM for colorectal cancer [39], MEGF for osteoporosis [40], HOXC4 for Chiari Malformations [41], and multiple candidates for Alzheimer’s [42] and exceptional longevity [43], among others.
Using the combined genealogy and cancer registry resource in the UPDB, the strongest evidence for an inherited contribution to prostate cancer has been shown for the subset of lethal prostate cancer cases [44]. The present analysis of available germline DNA for 51 pairs of cousins who each died of prostate cancer and belonged to a pedigree with a significant excess of prostate cancer among the descendants has identified thousands of rare, shared coding and noncoding variants in those cousins, each of which represents a candidate predisposition variant for prostate cancer which can be explored further. Since validation of segregation and risk association was based on the prostate cancer phenotype, rather than on the rarer (and remaining unknown for most cases) phenotype of lethal prostate cancer, conclusions may therefore be limited to prostate cancer rather than lethal prostate cancer, pending further study.
One outstanding candidate, in LRBA, was the only candidate predisposition variant to be further validated in the independent UK Biobank population for association with prostate cancer risk; this variant was found to segregate in five independent Utah high-risk pedigrees. While protein prediction modeling for this variant did not show any significant changes, the mutated structure does lose a helical structure two residues after the replacement (Figure 2 and Figure 3). The COACH results did not find any binding site in the region considered for this protein, nor is there any ligand information from experimental studies; therefore, it is not clear how to relate the additional helix structure with possible increase of loss of function of the protein [45,46]. Further study of the specific Utah high-risk pedigrees identified here, as well as analysis in other independent populations of prostate cancer cases, can further clarify whether, and how, this variant is specifically associated with increased risk for prostate cancer or lethal prostate cancer.
While not all the thousands of rare, shared candidate prostate cancer predisposition variants identified in this study have been reviewed in detail due to limitations of time and space, several specific subsets of variants were presented in more detail. Subsets considered included known cancer pathogenic variants, BROCA genes, and noncoding variants with suggestive Regulome DB scores. The identification of multiple strong candidate variants in these subsets, some of which represented more than one subset (e.g., PTCH1 or MSH6) demonstrates the power of this approach and serves as validation of this approach to identify both known and new predisposition variants.
The strengths of this study include the large number of affected cousin pairs analyzed, leading to many rare, shared candidate predisposition variants being identified. Strength also comes from the analysis of the phenotype of lethal prostate cancer in the proband pairs, which was confirmed both by a linked Utah death certificate for prostate cancer and validation of the cancer diagnosis within the linked Utah Cancer Registry. The available stored germline DNA from thousands of Utah prostate cancer cases, many of whom were related to the affected cousin pairs sequenced, allowed further validation of candidate variants by the confirmation of segregation of the variants. Limitations of this study include the censoring of some death, cancer, and genealogy data, which may have occurred through errors of reporting or record linking, or through lack of records. Also of note is that the founding population of Utah is largely northern European [47], and while immigration has been significant, these results may only specifically apply to this limited population and will require confirmation in other independent and more diverse populations. Confirmation of these results for the lethal prostate cancer phenotype will have to be made when more data for this more extreme phenotype are available in other independent populations.
5. Conclusions
Analysis of high-risk cancer pedigrees identified in this powerful Utah resource previously provided the identification of BRCA1 and BRCA2 [48,49], and CDKN2A [50], which remain the most common cancer predisposition genes to be identified. Large-scale case and pedigree studies strongly suggest that other common cancer predisposition genes may not remain to be identified and that, rather, most familial cancer predispositions might be the result of many varied, rare predisposition genes and variants [34]. Whether or not this is the case, studies such as this one, which analyze related affected individuals within a large number of high-risk pedigrees, have shown the strong potential to identify many candidate predisposition genes and variants for many different phenotypes. Such studies should be pursued, and the candidate predisposition variants identified are worthy of further exploration.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15072085/s1, Table S1: LPRCA_Shared_Genes.
Author Contributions
Conceptualization, L.A.C.-A.; data curation, J.S., J.C.F. and C.C.T.; formal analysis, L.A.C.-A., J.S., J.C.F. and C.C.T.; funding acquisition, L.A.C.-A. and N.A.; investigation, K.A.-B.; methodology, L.A.C.-A., J.S., J.C.F. and C.C.T.; project administration, L.A.C.-A. and K.A.-B.; resources, L.A.C.-A. and N.A.; software, J.S., J.C.F. and C.C.T.; validation, L.A.C.-A., C.C.T. and K.A.-B.; visualization, J.C.F.; writing—original draft, L.A.C.-A., J.S., J.C.F. and C.C.T.; writing—review and editing, L.A.C.-A., J.S., J.C.F., C.C.T., K.A.-B. and N.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the U.S. Department of Defense Prostate Cancer Research Program of the Office of the Congressionally Directed Medical Research Programs, grant number DOD PC170413, Lethal Prostate Cancer Gene Identification, awarded to Lisa Cannon-Albright. This research was conducted using the UK Biobank resource under application number 43460. This work was partially supported by the Utah Center for Clinical and Translational Science funded by NCATS award 1ULTR002538. Computer resources were provided by the University of Utah Center for High-Performance Computing, which has been partially funded by the NIH Shared Instrumentation Grant 1S10OD02164401A1. This research was supported by the Utah Cancer Registry, which is funded by the National Cancer Institute’s SEER Program, contract no. HHSN261201800016I, the U.S. Center for Disease Control and Prevention’s National Program of Cancer Registries, cooperative agreement no. NU58DP0063200-01, with additional support from the University of Utah and Huntsman Cancer Foundation. Partial support for all datasets within the Utah Population Database is provided by the University of Utah, Huntsman Cancer Institute, and the Huntsman Cancer Institute Cancer Center Support grant P30 CA42014 from the National Cancer Institute. L.A.C.-A. acknowledges partial support from the Huntsman Cancer Institute Cancer Center Support grant P30 CA42014 from the National Cancer Institute.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of the University of Utah School of Medicine (protocol code 4192 approved March 2023).
Informed Consent Statement
This project was approved by the University of Utah Institutional Review Board and has approval from the Resource for Genetic Epidemiology, which has oversight for the UPDB. Informed consent was obtained from each subject.
Data Availability Statement
All study subjects have relatives within the study set, which may allow identification from genetic data. Use of UPDB relationship data requires separate project application and approval. Interested researchers can contact the authors, and data will be made available to those who obtain appropriate approvals.
Conflicts of Interest
L.A.C.-A., J.S., J.C.F., C.C.T. and K.A.-B. declare no potential conflict of interest. N.A. (lifetime disclosures): consultancy to Astellas, Astra Zeneca, Aveo, Bayer, Bristol Myers Squibb, Calithera, Clovis, Eisai, Eli Lilly, EMD Serono, Exelixis, Foundation Medicine, Genentech, Gilead, Janssen, Merck, MEI Pharma, Nektar, Novartis, Pfizer, Pharmacyclics, and Seattle Genetics. Research funding to Neeraj Agarwal’s institution: Arnivas, Astellas, Astra Zeneca, Bavarian Nordic, Bayer, Bristol Myers Squibb, Calithera, Celldex, Clovis, Crispr, Eisai, Eli Lilly, EMD Serono, Exelixis, Genentech, Gilead, Glaxo Smith Kline, Immunomedics, Janssen, Lava, Medivation, Merck, Nektar, Neoleukin, New Link Genetics, Novartis, Oric, Pfizer, Prometheus, Rexahn, Roche, Sanofi, Seattle Genetics, Takeda, and Tracon.
References
- Cannon, L.; Bishop, D.T.; Skolnick, M.H.; Hunt, S.; Lyon, J.L.; Smart, C. Genetic epidemiology of prostate cancer in the Utah Mormon genealogy. Cancer Surv. 1982, 1, 48–69. [Google Scholar]
- Pritchard, C.C.; Mateo, J.; Walsh, M.F.; De Sarkar, N.; Abida, W.; Beltran, H.; Garofalo, A.; Gulati, R.; Carreira, S.; Eeles, R.; et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. NEJM 2016, 375, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Rantapero, T.; Wahlfors, T.; Kähler, A.; Hultman, C.; Lindberg, J.; Tammela, T.L.; Nykter, M.; Schleutker, J.; Wiklund, F. Inherited DNA repair gene mutations in men with lethal prostate cancer. Genes 2020, 11, 314. [Google Scholar] [CrossRef] [PubMed]
- Albright, F.S.; Stephenson, R.A.; Agarwal, N.; Teerlink, C.C.; Lowrance, W.T.; Farnham, J.M.; Cannon-Albright, L.A. Prostate cancer risk prediction based on complete prostate cancer family history. Prostate 2015, 75, 390–398. [Google Scholar] [CrossRef]
- Albright, F.S.; Stephenson, R.A.; Agarwal, N.; Cannon-Albright, L.A. Relative Risks for lethal prostate cancer based on complete family history of prostate cancer death. Prostate 2017, 77, 41–48. [Google Scholar] [CrossRef]
- Brandt, A.; Sundquist, J.; Hemminki, K. Risk for incident and fatal prostate cancer in men with a family history of any incident and fatal cancer. Ann. Oncol. 2012, 23, 251–256. [Google Scholar] [CrossRef]
- Schumacher, F.R.; Al Olama, A.A.; Berndt, S.I.; Benlloch, S.; Ahmed, M.; Saunders, E.J.; Dadaev, T.; Leongamornlert, D.; Anokian, E.; Cieza-Borrella, C.; et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018, 50, 928–936. [Google Scholar] [CrossRef]
- Castro, E.; Eeles, R. The role of BRCA1 and BRCA2 in prostate cancer. Asian J. Androl. 2012, 14, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lange, E.M.; Lu, L.; Zheng, S.L.; Wang, Z.; Thibodeau, S.N.; Cannon-Albright, L.A.; Teerlink, C.C.; Camp, N.J.; Johnson, A.M.; et al. International Consortium for Prostate Cancer Genetics. HOXB13 is a susceptibility gene for prostate cancer: Results from the International Consortium for Prostate Cancer Genetics (ICPCG). Hum. Genet. 2013, 132, 5–14. [Google Scholar] [CrossRef]
- Das, S.; Salami, S.S.; Spratt, D.E.; Kaffenberger, S.D.; Jacobs, M.F.; Morgan, T.M. Bringing prostate cancer germline genetics into clinical practice. J. Urol. 2019, 202, 223–230. [Google Scholar] [CrossRef]
- Hernandez, R.; Facelli, J.C. Understanding protein structural changes for oncogenic missense variants. Heliyon 2021, 7, e06013. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Abraham, G.; Qiu, Y.; Inouye, M. FlashPCA2: Principal component analysis of Biobank-scale genotype datasets. Bioinformatics 2017, 33, 2776–2778. [Google Scholar] [CrossRef]
- The Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Deelen, P.; Bonder, M.J.; van der Velde, K.J.; Westra, H.; Winder, E.; Hendriksen, D.; Franke, L.; Swertz, M.A. Genotype harmonizer: Automatic strand alignment and format conversion for genotype data integration. BMC Res. Notes 2014, 7, 901. [Google Scholar] [CrossRef] [PubMed]
- Loh, P.R.; Palamara, P.F.; Price, A.L. Fast and accurate long-range phasing in a UK Biobank cohort. Nat. Genet. 2016, 48, 811–816. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef]
- Ziv, E.; Dean, E.; Hu, D.; Martino, A.; Serie, D.; Curtin, K.; Campa, D.; Aftab, B.; Bracci, P.; Buda, G.; et al. Genome-wide association study identifies variants at 16p13 associated with survival in multiple myeloma patients. Nat. Commun. 2015, 6, 7539. [Google Scholar] [CrossRef] [PubMed]
- Huyghe, J.R.; Blen, S.A.; Harrison, T.A.; Kang, H.M.; Chen, S.; Schmit, S.L.; Conti, D.V.; Qu, C.; Jeon, J.; Edlund, C.K.; et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 2019, 51, 76–87. [Google Scholar] [CrossRef]
- Lopez-Herrera, G.; Tampella, G.; Pan-Hammarstrom, Q.; Herholz, P.; Trujillo-Vargas, C.M.; Phadwal, K.; Simon, A.K.; Moutschen, M.; Etzioni, A.; Mory, A.; et al. Deleterious mutations in LRBA are associated with a syndrome of immune deficiency and autoimmunity. Am. J. Hum. Genet. 2012, 90, 986–1001. [Google Scholar] [CrossRef]
- Gebauer, D.; Li, J.; Jogl, G.; Shen, Y.; Myszka, D.G.; Tong, L. Crystal structure of the PH-BEACH domains of human LRBA/BGL. Biochemistry 2004, 43, 14873–14880. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins Struct. Funct. Bioinform. 2009, 77, 100–113. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protocols 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Carter, B.S.; Bova, G.S.; Beaty, T.H.; Steinberg, G.D.; Childs, B.; Issacs, W.B.; Walsh, P.C. Hereditary prostate cancer: Epidemiologic and clinical features. J. Urol. 1993, 150, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nat. Cell. Biol. 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Wijsman, E.M. The role of large pedigrees in an era of high-throughput sequencing. Qual. Life Res. 2012, 131, 1555–1563. [Google Scholar] [CrossRef]
- Ott, J.; Wang, J.; Leal, S.M. Genetic linkage analysis in the age of whole-genome sequencing. Nat. Rev. Genet. 2015, 16, 275–284. [Google Scholar] [CrossRef]
- Terwilliger, J.D.; Goring, H.H. Gene mapping in the 20th and 21st centuries: Statistical methods, data analysis, and experimental design. Hum. Biol. 2009, 81, 663–728. [Google Scholar] [CrossRef] [PubMed]
- Ewing, C.M.; Ray, A.M.; Lange, E.M.; Zuhlke, K.A.; Robbins, C.M.; Tembe, W.D.; Wiley, K.E.; Isaacs, S.D.; Johng, D.; Wang, Y.; et al. Germline mutations in HOXB13 and prostate cancer risk. N. Engl. J. Med. 2012, 366, 141–149. [Google Scholar] [CrossRef]
- Cannon-Albright, L.A.; Teerlink, C.C.; Stevens, J.; Huang, F.W.; Sipeky, C.; Schleutker, J.; Hernandez, R.; Facelli, J.; Agarwal, N.; Trump, D.L. A Rare Variant in ERF (rs144812092) Predisposes to Prostate and Bladder Cancers in an Extended Pedigree. Cancers 2021, 13, 2399. [Google Scholar] [CrossRef]
- Cannon-Albright, L.A. Utah family-based analysis: Past, present and future. Hum. Hered. 2008, 65, 209–220. [Google Scholar] [CrossRef]
- Teerlink, C.C.; Huff, C.; Stevens, J.; Yu, Y.; Holmen, S.L.; Silvis, M.R.; Trombetti, K.; Zhao, H.; Grossman, D.; Farnham, J.M.; et al. A non-synonymous variant in GOLM1 in cutaneous malignant melanoma. JNCI J. Natl. Cancer Inst. 2018, 110, 1380–1385. [Google Scholar]
- Cannon-Albright, L.A.; Teerlink, C.C.; Stevens, J.; Snow, A.K.; Thompson, B.A.; Bell, R.; Nguyen, K.N.; Sargent, N.R.; Kohlmann, W.; Neklason, D.W.; et al. FANCM c5791CT stopgain mutation (rs144567652) is a familial colorectal cancer risk factor. Mol. Genet. Genom. Med. 2020, 8, e1532. [Google Scholar]
- Teerlink, C.C.; Jurynec, M.J.; Hernande, R.; Stevens, J.; Hughes, D.C.; Brunker, C.P.; Rowe, K.; Grunwald, D.J.; Facelli, J.C.; Cannon-Albright, L.A. A role for the MEGF6 gene in predisposition to osteoporosis. Ann. Hum. Genet. 2021, 85, 58–72. [Google Scholar] [CrossRef]
- Brockmeyer, D.L.; Cheshier, S.H.; Stevens, J.; Facelli, J.C.; Rowe, K.; Heiss, J.D.; Musolf, A.; Viskochil, D.H.; Allen-Brady, K.L.; Cannon-Albright, L.A. A likely HOXC4 predisposition variant for Chiari malformations. J. Neurosurg. 2022, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Teerlink, C.C.; Miller, J.B.; Vance, E.L.; Staley, L.A.; Stevens, J.; Tavana, J.P.; Cloward, M.E.; Page, M.L.; Dayton, L.; Alzheimer’s Disease Genetics Consortium; et al. Analysis of high-risk pedigrees identifies 11 candidate variants for Alzheimer’s disease. Alzheimer’s Dement 2022, 18, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.B.; Ward, E.; Staley, L.A.; Stevens, J.; Teerlink, C.C.; Tavana, J.P.; Cloward, M.; Page, M.; Dayton, L.; Alzheimer’s Disease Genetics Consortium; et al. Identification and genomic analysis of pedigrees with exceptional longevity identifies candidate rare variants. Neurobiol. Dis. 2020, 143, 104972. [Google Scholar] [CrossRef] [PubMed]
- Nelson, Q.; Agarwal, N.; Stephenson, R.; Cannon-Albright, L.A. A population-based analysis of clustering identifies a strong genetic contribution to lethal prostate cancer. Front. Genet. 2013, 4, 152. [Google Scholar] [CrossRef]
- Yang, J.; Roy, A.; Zhang, Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef]
- Yang, J.; Roy, A.; Zhang, Y. BioLiP: A semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 2013, 41, D1096–D1103. [Google Scholar] [CrossRef] [PubMed]
- Cannon-Albright, L.A.; Farnham, J.M.; Thomas, A.; Camp, N.J. Identification and study of Utah pseudo-isolate populations-prospects for gene identification. Am. J. Med. Genet. A 2005, 137, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W.; et al. A Strong Candidate for the Breast and Ovarian Cancer susceptibility Gene BRCA 1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef]
- Tavtigian, S.V.; Simard, J.; Rommens, J.; Couch, F.; Shattuck-Eidens, D.; Neuhausen, S.; Merajver, S.; Thorlacius, S.; Offit, K.; Stoppa-Lyonnet, D.; et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat. Genet. 1996, 12, 333–337. [Google Scholar] [CrossRef]
- Kamb, A.; Shattuck-Eidens, D.; Eeles, R.; Liu, Q.; Gris, N.A.; Ding, W.; Hussey, C.; Tran, T.; Miki, Y.; Weaver-Feldhaus, J.; et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat. Genet. 1994, 8, 23–26. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).