From Exosome Biogenesis to Absorption: Key Takeaways for Cancer Research
Abstract
Simple Summary
Abstract
1. Introduction
2. Mechanisms of Exosome Biogenesis
2.1. Endosome Formation
2.2. MVB Formation and Cargo Encapsulation
2.3. Cargo Sorting to MVBs
2.4. Extracellular Release
3. Mechanisms of Exosome Absorption
3.1. Exosome Transport and Targeting to Recipient Cells
3.2. Interaction with Recipient Cells and Internalization
3.3. Fate of Internalized Exosomes
4. Relevance of Exosome Secretion and Uptake in Cancer and Cancer Therapy
4.1. Regulation of Exosome Secretion in Cancer
4.2. The Role of Exosome Targeting in Cancer and Drug Delivery
4.3. Cancer Exosomes as a Therapeutic Target
5. Challenges and Perspectives for Designing Studies on Exosome Biology
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, 6977. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, J.; Ahmadi, M.; Ravanbakhsh, R.; Mojarad, B.; Mahbubfam, S.; Shaban, S.A.; Shadi, K.; Berenjabad, N.J.; Etemadi, T. Tumor-derived extracellular vesicles: The metastatic organotropism drivers. Life Sci. 2022, 289, 120216. [Google Scholar] [CrossRef] [PubMed]
- Crescitelli, R.; Lässer, C.; Lötvall, J. Isolation and characterization of extracellular vesicle subpopulations from tissues. Nat. Protoc. 2021, 16, 1548–1580. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Men, Y.; Yelick, J.; Jin, S.; Tian, Y.; Chiang, M.S.R.; Higashimori, H.; Brown, E.; Jarvis, R.; Yang, Y. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat. Commun. 2019, 10, 4136. [Google Scholar] [CrossRef]
- Nolte-’t Hoen, E.N.; Buschow, S.I.; Anderton, S.M.; Stoorvogel, W.; Wauben, M.H. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood 2009, 113, 1977–1981. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, M.; Gong, A.; Zhang, X.; Wu, X.; Zhu, Y.; Shi, H.; Wu, L.; Zhu, W.; Qian, H.; et al. HucMSC-Exosome Mediated-Wnt4 Signaling Is Required for Cutaneous Wound Healing. Stem Cells 2015, 33, 2158–2168. [Google Scholar] [CrossRef] [PubMed]
- Osaki, M.; Okada, F. Exosomes and Their Role in Cancer Progression. Yonago Acta Med. 2019, 62, 182–190. [Google Scholar] [CrossRef]
- Yang, E.; Wang, X.; Gong, Z.; Yu, M.; Wu, H.; Zhang, D. Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther. 2020, 5, 242. [Google Scholar] [CrossRef]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.F.; Li, W.J.; Hu, K.S.; Gao, J.; Zhai, W.L.; Yang, J.H.; Zhang, S.J. Exosome biogenesis: Machinery, regulation, and therapeutic implications in cancer. Mol. Cancer 2022, 21, 207. [Google Scholar] [CrossRef]
- Kang, T.; Atukorala, I.; Mathivanan, S. Biogenesis of Extracellular Vesicles. Subcell. Biochem. 2021, 97, 19–43. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Kwon, S.H.; Oh, S.; Nacke, M.; Mostov, K.E.; Lipschutz, J.H. Adaptor Protein CD2AP and L-type Lectin LMAN2 Regulate Exosome Cargo Protein Trafficking through the Golgi Complex. J. Biol. Chem. 2016, 291, 25462–25475. [Google Scholar] [CrossRef] [PubMed]
- Di Mattia, T.; Tomasetto, C.; Alpy, F. Faraway, so close! Functions of Endoplasmic reticulum–Endosome contacts. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158490. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- O’Brien, K.; Ughetto, S.; Mahjoum, S.; Nair, A.V.; Breakefield, X.O. Uptake, functionality, and re-release of extracellular vesicle-encapsulated cargo. Cell Rep. 2022, 39, 110651. [Google Scholar] [CrossRef]
- McMahon, H.T.; Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011, 12, 517–533. [Google Scholar] [CrossRef]
- Mayor, S.; Parton, R.G.; Donaldson, J.G. Clathrin-independent pathways of endocytosis. Cold Spring Harb. Perspect. Biol. 2014, 6, 16758. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, O.; Teis, D. The ESCRT machinery. Curr. Biol. 2012, 22, R116–R120. [Google Scholar] [CrossRef]
- Vietri, M.; Radulovic, M.; Stenmark, H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020, 21, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Pfitzner, A.K.; Mercier, V.; Jiang, X.; Moser von Filseck, J.; Baum, B.; Šarić, A.; Roux, A. An ESCRT-III Polymerization Sequence Drives Membrane Deformation and Fission. Cell 2020, 182, 1140–1155.e1118. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Kim, S.B.; Kim, H.R.; Park, M.C.; Cho, S.; Goughnour, P.C.; Han, D.; Yoon, I.; Kim, Y.; Kang, T.; Song, E.; et al. Caspase-8 controls the secretion of inflammatory lysyl-tRNA synthetase in exosomes from cancer cells. J. Cell Biol. 2017, 216, 2201–2216. [Google Scholar] [CrossRef]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219, 4113. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Liu, P.; Gong, D.; Zhou, H.; Li, W.; Zhang, H.; Zheng, W.; Xu, J.; Cheng, H.; et al. Phosphatase Shp2 regulates biogenesis of small extracellular vesicles by dephosphorylating Syntenin. J. Extracell. Vesicles 2021, 10, e12078. [Google Scholar] [CrossRef]
- Hyenne, V.; Labouesse, M.; Goetz, J.G. The Small GTPase Ral orchestrates MVB biogenesis and exosome secretion. Small GTPases 2018, 9, 445–451. [Google Scholar] [CrossRef]
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Kazan, J.M.; Desrochers, G.; Martin, C.E.; Jeong, H.; Kharitidi, D.; Apaja, P.M.; Roldan, A.; St Denis, N.; Gingras, A.C.; Lukacs, G.L.; et al. Endofin is required for HD-PTP and ESCRT-0 interdependent endosomal sorting of ubiquitinated transmembrane cargoes. iScience 2021, 24, 103274. [Google Scholar] [CrossRef] [PubMed]
- Schuldner, M.; Dörsam, B.; Shatnyeva, O.; Reiners, K.S.; Kubarenko, A.; Hansen, H.P.; Finkernagel, F.; Roth, K.; Theurich, S.; Nist, A.; et al. Exosome-dependent immune surveillance at the metastatic niche requires BAG6 and CBP/p300-dependent acetylation of p53. Theranostics 2019, 9, 6047–6062. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Lv, L.; Wei, J.; Lei, X.; Lin, H.; Li, G.; Cao, J.; Xie, J.; Yang, W.; Wu, S.; et al. Vps4A mediates the localization and exosome release of β-catenin to inhibit epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2019, 457, 47–59. [Google Scholar] [CrossRef]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid. Res. 2019, 60, 9–18. [Google Scholar] [CrossRef]
- Dawson, G. Isolation of Lipid Rafts (Detergent-Resistant Microdomains) and Comparison to Extracellular Vesicles (Exosomes). Methods Mol. Biol. 2021, 2187, 99–112. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Leidal, A.M.; Huang, H.H.; Marsh, T.; Solvik, T.; Zhang, D.; Ye, J.; Kai, F.; Goldsmith, J.; Liu, J.Y.; Huang, Y.H.; et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 2020, 22, 187–199. [Google Scholar] [CrossRef]
- Adam-Klages, S.; Adam, D.; Wiegmann, K.; Struve, S.; Kolanus, W.; Schneider-Mergener, J.; Krönke, M. FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell 1996, 86, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.R.; Sanchez-Heras, E.; Tsapara, A.; Sobota, A.; Levine, T.P.; Futter, C.E. Annexin A1 Tethers Membrane Contact Sites that Mediate ER to Endosome Cholesterol Transport. Dev. Cell 2016, 37, 473–483. [Google Scholar] [CrossRef]
- Albacete-Albacete, L.; Navarro-Lérida, I.; López, J.A.; Martín-Padura, I.; Astudillo, A.M.; Ferrarini, A.; Van-Der-Heyden, M.; Balsinde, J.; Orend, G.; Vázquez, J.; et al. ECM deposition is driven by caveolin-1-dependent regulation of exosomal biogenesis and cargo sorting. J. Cell Biol. 2020, 219, 6178. [Google Scholar] [CrossRef]
- Phuyal, S.; Hessvik, N.P.; Skotland, T.; Sandvig, K.; Llorente, A. Regulation of exosome release by glycosphingolipids and flotillins. FEBS J. 2014, 281, 2214–2227. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, L.P.; Wendling, C.; Védie, B.; Kobayashi, T.; Chenard, M.P.; Tomasetto, C.; Drin, G.; Alpy, F. STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J. 2017, 36, 1412–1433. [Google Scholar] [CrossRef] [PubMed]
- Strauss, K.; Goebel, C.; Runz, H.; Möbius, W.; Weiss, S.; Feussner, I.; Simons, M.; Schneider, A. Exosome secretion ameliorates lysosomal storage of cholesterol in Niemann-Pick type C disease. J. Biol. Chem. 2010, 285, 26279–26288. [Google Scholar] [CrossRef]
- Kummer, D.; Steinbacher, T.; Schwietzer, M.F.; Thölmann, S.; Ebnet, K. Tetraspanins: Integrating cell surface receptors to functional microdomains in homeostasis and disease. Med. Microbiol. Immunol. 2020, 209, 397–405. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Bun, P.; Huveneers, S.; van Niel, G.; Pegtel, D.M.; Verweij, F.J. Real-time imaging of multivesicular body-plasma membrane fusion to quantify exosome release from single cells. Nat. Protoc. 2020, 15, 102–121. [Google Scholar] [CrossRef]
- Guix, F.X.; Sannerud, R.; Berditchevski, F.; Arranz, A.M.; Horré, K.; Snellinx, A.; Thathiah, A.; Saido, T.; Saito, T.; Rajesh, S.; et al. Tetraspanin 6: A pivotal protein of the multiple vesicular body determining exosome release and lysosomal degradation of amyloid precursor protein fragments. Mol. Neurodegener. 2017, 12, 25. [Google Scholar] [CrossRef]
- Ghossoub, R.; Chéry, M.; Audebert, S.; Leblanc, R.; Egea-Jimenez, A.L.; Lembo, F.; Mammar, S.; Le Dez, F.; Camoin, L.; Borg, J.P.; et al. Tetraspanin-6 negatively regulates exosome production. Proc. Natl. Acad. Sci. USA 2020, 117, 5913–5922. [Google Scholar] [CrossRef]
- Escola, J.M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef]
- Mazurov, D.; Barbashova, L.; Filatov, A. Tetraspanin protein CD9 interacts with metalloprotease CD10 and enhances its release via exosomes. FEBS J. 2013, 280, 1200–1213. [Google Scholar] [CrossRef]
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.V.; da Rosa Soares, A.; Ramalho, J.; Máximo Carvalho, C.; Cardoso, M.H.; Pintado, P.; Carvalho, A.S.; Beck, H.C.; Matthiesen, R.; Zuzarte, M.; et al. LAMP2A regulates the loading of proteins into exosomes. Sci. Adv. 2022, 8, eabm1140. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, C.; Ma, B.; Xu, M.; Xu, S.; Liu, J.; Tian, Y.; Fu, Y.; Luo, Y. Mutant p53 Drives Cancer Metastasis via RCP-Mediated Hsp90α Secretion. Cell Rep. 2020, 32, 107879. [Google Scholar] [CrossRef]
- Grabuschnig, S.; Bronkhorst, A.J.; Holdenrieder, S.; Rosales Rodriguez, I.; Schliep, K.P.; Schwendenwein, D.; Ungerer, V.; Sensen, C.W. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int. J. Mol. Sci. 2020, 21, 8062. [Google Scholar] [CrossRef]
- Yokoi, A.; Villar-Prados, A.; Oliphint, P.A.; Zhang, J.; Song, X.; De Hoff, P.; Morey, R.; Liu, J.; Roszik, J.; Clise-Dwyer, K.; et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849. [Google Scholar] [CrossRef]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287. [Google Scholar] [CrossRef] [PubMed]
- Rabas, N.; Palmer, S.; Mitchell, L.; Ismail, S.; Gohlke, A.; Riley, J.S.; Tait, S.W.G.; Gammage, P.; Soares, L.L.; Macpherson, I.R.; et al. PINK1 drives production of mtDNA-containing extracellular vesicles to promote invasiveness. J. Cell Biol. 2021, 220, 6049. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef]
- Zhu, J.; Lu, K.; Zhang, N.; Zhao, Y.; Ma, Q.; Shen, J.; Lin, Y.; Xiang, P.; Tang, Y.; Hu, X.; et al. Myocardial reparative functions of exosomes from mesenchymal stem cells are enhanced by hypoxia treatment of the cells via transferring microRNA-210 in an nSMase2-dependent way. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1659–1670. [Google Scholar] [CrossRef]
- Bakirtzi, K.; Man Law, I.K.; Fang, K.; Iliopoulos, D.; Pothoulakis, C. MiR-21 in Substance P-induced exosomes promotes cell proliferation and migration in human colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G802–G810. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Zhang, Q.; Jiang, L. Current Knowledge on Exosome Biogenesis, Cargo-Sorting Mechanism and Therapeutic Implications. Membranes 2022, 12, 498. [Google Scholar] [CrossRef] [PubMed]
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell. Vesicles 2020, 10, e12043. [Google Scholar] [CrossRef]
- Chen, C.; Luo, Y.; He, W.; Zhao, Y.; Kong, Y.; Liu, H.; Zhong, G.; Li, Y.; Li, J.; Huang, J.; et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Investig. 2020, 130, 404–421. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zheng, H.; Luo, Y.; Kong, Y.; An, M.; Li, Y.; He, W.; Gao, B.; Zhao, Y.; Huang, H.; et al. SUMOylation promotes extracellular vesicle-mediated transmission of lncRNA ELNAT1 and lymph node metastasis in bladder cancer. J. Clin. Investig. 2021, 131, 146431. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yu, H.; Han, F.; Lai, X.; Ye, K.; Lei, S.; Mai, M.; Lai, M.; Zhang, H. Tumor-suppressive circRHOBTB3 is excreted out of cells via exosome to sustain colorectal cancer cell fitness. Mol. Cancer 2022, 21, 46. [Google Scholar] [CrossRef]
- Pan, Z.; Zhao, R.; Li, B.; Qi, Y.; Qiu, W.; Guo, Q.; Zhang, S.; Zhao, S.; Xu, H.; Li, M.; et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer 2022, 21, 16. [Google Scholar] [CrossRef]
- Iavello, A.; Frech, V.S.; Gai, C.; Deregibus, M.C.; Quesenberry, P.J.; Camussi, G. Role of Alix in miRNA packaging during extracellular vesicle biogenesis. Int. J. Mol. Med. 2016, 37, 958–966. [Google Scholar] [CrossRef]
- Roden, C.; Gladfelter, A.S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021, 22, 183–195. [Google Scholar] [CrossRef]
- Ryan, V.H.; Dignon, G.L.; Zerze, G.H.; Chabata, C.V.; Silva, R.; Conicella, A.E.; Amaya, J.; Burke, K.A.; Mittal, J.; Fawzi, N.L. Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol. Cell 2018, 69, 465–479.e467. [Google Scholar] [CrossRef]
- Liu, X.M.; Ma, L.; Schekman, R. Selective sorting of microRNAs into exosomes by phase-separated YBX1 condensates. eLife 2021, 10, 71982. [Google Scholar] [CrossRef]
- Wani, S.; Kaul, D. Cancer cells govern miR-2909 exosomal recruitment through its 3’-end post-transcriptional modification. Cell Biochem. Funct. 2018, 36, 106–111. [Google Scholar] [CrossRef]
- Yoon, T.Y.; Munson, M. SNARE complex assembly and disassembly. Curr. Biol. 2018, 28, R397–R401. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Xu, X.; Chen, C.; Sanmillan, M.L.; Cai, T.; Zhou, Y.; Giraudo, C.; Le, A.; Shi, S. The Fas/Fap-1/Cav-1 complex regulates IL-1RA secretion in mesenchymal stem cells to accelerate wound healing. Sci. Transl. Med. 2018, 10, 8524. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Peng, X.; Li, Y.; Zhang, X.; Ma, Y.; Wu, C.; Fan, Q.; Wei, S.; Li, H.; Liu, J. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol. Cancer 2019, 18, 78. [Google Scholar] [CrossRef]
- Hoshino, D.; Kirkbride, K.C.; Costello, K.; Clark, E.S.; Sinha, S.; Grega-Larson, N.; Tyska, M.J.; Weaver, A.M. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013, 5, 1159–1168. [Google Scholar] [CrossRef]
- Sinha, S.; Hoshino, D.; Hong, N.H.; Kirkbride, K.C.; Grega-Larson, N.E.; Seiki, M.; Tyska, M.J.; Weaver, A.M. Cortactin promotes exosome secretion by controlling branched actin dynamics. J. Cell Biol. 2016, 214, 197–213. [Google Scholar] [CrossRef]
- Marat, A.L.; Ioannou, M.S.; McPherson, P.S. Connecdenn 3/DENND1C binds actin linking Rab35 activation to the actin cytoskeleton. Mol. Biol. Cell 2012, 23, 163–175. [Google Scholar] [CrossRef]
- Beghein, E.; Devriese, D.; Van Hoey, E.; Gettemans, J. Cortactin and fascin-1 regulate extracellular vesicle release by controlling endosomal trafficking or invadopodia formation and function. Sci. Rep. 2018, 8, 15606. [Google Scholar] [CrossRef] [PubMed]
- Jongsma, M.L.; Bakker, J.; Cabukusta, B.; Liv, N.; van Elsland, D.; Fermie, J.; Akkermans, J.L.; Kuijl, C.; van der Zanden, S.Y.; Janssen, L.; et al. SKIP-HOPS recruits TBC1D15 for a Rab7-to-Arl8b identity switch to control late endosome transport. EMBO J. 2020, 39, e102301. [Google Scholar] [CrossRef]
- Granger, E.; McNee, G.; Allan, V.; Woodman, P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin. Cell Dev. Biol. 2014, 31, 20–29. [Google Scholar] [CrossRef]
- Jordens, I.; Fernandez-Borja, M.; Marsman, M.; Dusseljee, S.; Janssen, L.; Calafat, J.; Janssen, H.; Wubbolts, R.; Neefjes, J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 2001, 11, 1680–1685. [Google Scholar] [CrossRef]
- Wozniak, A.L.; Adams, A.; King, K.E.; Dunn, W.; Christenson, L.K.; Hung, W.T.; Weinman, S.A. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J. Cell Biol. 2020, 219, 12074. [Google Scholar] [CrossRef]
- Wei, D.; Zhan, W.; Gao, Y.; Huang, L.; Gong, R.; Wang, W.; Zhang, R.; Wu, Y.; Gao, S.; Kang, T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021, 31, 157–177. [Google Scholar] [CrossRef]
- Kinchen, J.M.; Ravichandran, K.S. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 2010, 464, 778–782. [Google Scholar] [CrossRef]
- Fei, X.; Li, Z.; Yang, D.; Kong, X.; Lu, X.; Shen, Y.; Li, X.; Xie, S.; Wang, J.; Zhao, Y.; et al. Neddylation of Coro1a determines the fate of multivesicular bodies and biogenesis of extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12153. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Baixauli, F.; Mittelbrunn, M.; Fernández-Delgado, I.; Torralba, D.; Moreno-Gonzalo, O.; Baldanta, S.; Enrich, C.; Guerra, S.; Sánchez-Madrid, F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat. Commun. 2016, 7, 13588. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef]
- Alnaas, A.A.; Watson-Siriboe, A.; Tran, S.; Negussie, M.; Henderson, J.A.; Osterberg, J.R.; Chon, N.L.; Harrott, B.M.; Oviedo, J.; Lyakhova, T.; et al. Multivalent lipid targeting by the calcium-independent C2A domain of synaptotagmin-like protein 4/granuphilin. J. Biol. Chem. 2021, 296, 100159. [Google Scholar] [CrossRef]
- He, J.; Johnson, J.L.; Monfregola, J.; Ramadass, M.; Pestonjamasp, K.; Napolitano, G.; Zhang, J.; Catz, S.D. Munc13-4 interacts with syntaxin 7 and regulates late endosomal maturation, endosomal signaling, and TLR9-initiated cellular responses. Mol. Biol. Cell 2016, 27, 572–587. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, K.; Qi, L.; Hu, Q.; Shen, Z.; Liu, B.; Deng, J.; Zhang, C.; Zhang, Y. DENN domain-containing protein FAM45A regulates the homeostasis of late/multivesicular endosomes. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 916–929. [Google Scholar] [CrossRef]
- Imai, A.; Ishida, M.; Fukuda, M.; Nashida, T.; Shimomura, H. MADD/DENN/Rab3GEP functions as a guanine nucleotide exchange factor for Rab27 during granule exocytosis of rat parotid acinar cells. Arch. Biochem. Biophys. 2013, 536, 31–37. [Google Scholar] [CrossRef]
- Song, L.; Tang, S.; Han, X.; Jiang, Z.; Dong, L.; Liu, C.; Liang, X.; Dong, J.; Qiu, C.; Wang, Y.; et al. KIBRA controls exosome secretion via inhibiting the proteasomal degradation of Rab27a. Nat. Commun. 2019, 10, 1639. [Google Scholar] [CrossRef]
- Savina, A.; Vidal, M.; Colombo, M.I. The exosome pathway in K562 cells is regulated by Rab11. J. Cell Sci. 2002, 115, 2505–2515. [Google Scholar] [CrossRef]
- Hsu, C.; Morohashi, Y.; Yoshimura, S.; Manrique-Hoyos, N.; Jung, S.; Lauterbach, M.A.; Bakhti, M.; Grønborg, M.; Möbius, W.; Rhee, J.; et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J. Cell Biol. 2010, 189, 223–232. [Google Scholar] [CrossRef]
- Messenger, S.W.; Woo, S.S.; Sun, Z.; Martin, T.F.J. A Ca2+-stimulated exosome release pathway in cancer cells is regulated by Munc13-4. J. Cell Biol. 2018, 217, 2877–2890. [Google Scholar] [CrossRef]
- Savina, A.; Fader, C.M.; Damiani, M.T.; Colombo, M.I. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 2005, 6, 131–143. [Google Scholar] [CrossRef]
- Qin, J.; Xu, Q. Functions and application of exosomes. Acta Pol. Pharm. 2014, 71, 537–543. [Google Scholar]
- Morishita, M.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Pharmacokinetics of Exosomes—An Important Factor for Elucidating the Biological Roles of Exosomes and for the Development of Exosome-Based Therapeutics. J. Pharm. Sci. 2017, 106, 2265–2269. [Google Scholar] [CrossRef]
- Smyth, T.; Kullberg, M.; Malik, N.; Smith-Jones, P.; Graner, M.W.; Anchordoquy, T.J. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J. Control. Release 2015, 199, 145–155. [Google Scholar] [CrossRef]
- Wiklander, O.P.; Nordin, J.Z.; O’Loughlin, A.; Gustafsson, Y.; Corso, G.; Mäger, I.; Vader, P.; Lee, Y.; Sork, H.; Seow, Y.; et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 2015, 4, 26316. [Google Scholar] [CrossRef] [PubMed]
- Horibe, S.; Tanahashi, T.; Kawauchi, S.; Murakami, Y.; Rikitake, Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer 2018, 18, 47. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Vinas, J.L.; Spence, M.; Gutsol, A.; Knoll, W.; Burger, D.; Zimpelmann, J.; Allan, D.S.; Burns, K.D. Receptor-Ligand Interaction Mediates Targeting of Endothelial Colony Forming Cell-derived Exosomes to the Kidney after Ischemic Injury. Sci. Rep. 2018, 8, 16320. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Yue, S.; Stadel, D.; Zoller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef]
- Toda, Y.; Takata, K.; Nakagawa, Y.; Kawakami, H.; Fujioka, S.; Kobayashi, K.; Hattori, Y.; Kitamura, Y.; Akaji, K.; Ashihara, E. Effective internalization of U251-MG-secreted exosomes into cancer cells and characterization of their lipid components. Biochem. Biophys. Res. Commun. 2015, 456, 768–773. [Google Scholar] [CrossRef]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef]
- Sancho-Albero, M.; Navascués, N.; Mendoza, G.; Sebastián, V.; Arruebo, M.; Martín-Duque, P.; Santamaría, J. Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. J. Nanobiotechnol. 2019, 17, 16. [Google Scholar] [CrossRef]
- Hazan-Halevy, I.; Rosenblum, D.; Weinstein, S.; Bairey, O.; Raanani, P.; Peer, D. Cell-specific uptake of mantle cell lymphoma-derived exosomes by malignant and non-malignant B-lymphocytes. Cancer Lett. 2015, 364, 59–69. [Google Scholar] [CrossRef]
- Qiao, L.; Hu, S.; Huang, K.; Su, T.; Li, Z.; Vandergriff, A.; Cores, J.; Dinh, P.U.; Allen, T.; Shen, D.; et al. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics 2020, 10, 3474–3487. [Google Scholar] [CrossRef]
- Saari, H.; Lázaro-Ibáñez, E.; Viitala, T.; Vuorimaa-Laukkanen, E.; Siljander, P.; Yliperttula, M. Microvesicle- and exosome-mediated drug delivery enhances the cytotoxicity of Paclitaxel in autologous prostate cancer cells. J. Control. Release 2015, 220, 727–737. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Prajuabjinda, O.; Soe, Z.Y.; Darkwah, S.; Appiah, M.G.; Kawamoto, E.; Momose, F.; Shiku, H.; Shimaoka, M. Exosomal regulation of lymphocyte homing to the gut. Blood. Adv. 2019, 3, 24877. [Google Scholar] [CrossRef]
- Laulagnier, K.; Javalet, C.; Hemming, F.J.; Chivet, M.; Lachenal, G.; Blot, B.; Chatellard, C.; Sadoul, R. Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons. Cell Mol. Life Sci. 2018, 75, 757–773. [Google Scholar] [CrossRef]
- Logtenberg, M.E.W.; Scheeren, F.A.; Schumacher, T.N. The CD47-SIRPα Immune Checkpoint. Immunity 2020, 52, 742–752. [Google Scholar] [CrossRef]
- Clayton, A.; Harris, C.L.; Court, J.; Mason, M.D.; Morgan, B.P. Antigen-presenting cell exosomes are protected from complement-mediated lysis by expression of CD55 and CD59. Eur. J. Immunol. 2003, 33, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Munich, S.; Sobo-Vujanovic, A.; Buchser, W.J.; Beer-Stolz, D.; Vujanovic, N.L. Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. Oncoimmunology 2012, 1, 1074–1083. [Google Scholar] [CrossRef]
- Tkach, M.; Kowal, J.; Zucchetti, A.E.; Enserink, L.; Jouve, M.; Lankar, D.; Saitakis, M.; Martin-Jaular, L.; Théry, C. Qualitative differences in T-cell activation by dendritic cell-derived extracellular vesicle subtypes. EMBO J. 2017, 36, 3012–3028. [Google Scholar] [CrossRef]
- Chernomordik, L.V.; Kozlov, M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 675–683. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef]
- Prada, I.; Meldolesi, J. Binding and Fusion of Extracellular Vesicles to the Plasma Membrane of Their Cell Targets. Int. J. Mol. Sci. 2016, 17, 1296. [Google Scholar] [CrossRef]
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455. [Google Scholar] [CrossRef]
- Mettlen, M.; Chen, P.H.; Srinivasan, S.; Danuser, G.; Schmid, S.L. Regulation of Clathrin-Mediated Endocytosis. Annu. Rev. Biochem. 2018, 87, 871–896. [Google Scholar] [CrossRef]
- Feng, D.; Zhao, W.L.; Ye, Y.Y.; Bai, X.C.; Liu, R.Q.; Chang, L.F.; Zhou, Q.; Sui, S.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Teissier, E.; Pécheur, E.I. Lipids as modulators of membrane fusion mediated by viral fusion proteins. Eur. Biophys. J. 2007, 36, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Glebov, O.O.; Bright, N.A.; Nichols, B.J. Flotillin-1 defines a clathrin-independent endocytic pathway in mammalian cells. Nat. Cell Biol. 2006, 8, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Koumangoye, R.B.; Sakwe, A.M.; Goodwin, J.S.; Patel, T.; Ochieng, J. Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. PLoS ONE 2011, 6, e24234. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Useros, N.; Naranjo-Gómez, M.; Archer, J.; Hatch, S.C.; Erkizia, I.; Blanco, J.; Borràs, F.E.; Puertas, M.C.; Connor, J.H.; Fernández-Figueras, M.T.; et al. Capture and transfer of HIV-1 particles by mature dendritic cells converges with the exosome-dissemination pathway. Blood 2009, 113, 2732–2741. [Google Scholar] [CrossRef]
- Kerr, M.C.; Teasdale, R.D. Defining macropinocytosis. Traffic 2009, 10, 364–371. [Google Scholar] [CrossRef]
- Lim, J.P.; Gleeson, P.A. Macropinocytosis: An endocytic pathway for internalising large gulps. Immunol. Cell Biol. 2011, 89, 836–843. [Google Scholar] [CrossRef]
- Nakase, I.; Kobayashi, N.B.; Takatani-Nakase, T.; Yoshida, T. Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes. Sci. Rep. 2015, 5, 10300. [Google Scholar] [CrossRef]
- Nanbo, A.; Kawanishi, E.; Yoshida, R.; Yoshiyama, H. Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J. Virol. 2013, 87, 10334–10347. [Google Scholar] [CrossRef] [PubMed]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Mörgelin, M.; Belting, M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 2013, 288, 17713–17724. [Google Scholar] [CrossRef] [PubMed]
- Rudt, S.; Müller, R.H. In vitro phagocytosis assay of nano- and microparticles by chemiluminescence. III. Uptake of differently sized surface-modified particles, and its correlation to particle properties and in vivo distribution. Eur. J. Pharm. Sci. 1993, 1, 31–39. [Google Scholar] [CrossRef]
- Gordon, S. Phagocytosis: An Immunobiologic Process. Immunity 2016, 44, 463–475. [Google Scholar] [CrossRef]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Stolz, D.B.; Sullivan, M.L.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef]
- Yuyama, K.; Sun, H.; Mitsutake, S.; Igarashi, Y. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-β by microglia. J. Biol. Chem. 2012, 287, 10977–10989. [Google Scholar] [CrossRef]
- Chen, C.; Zong, S.; Wang, Z.; Lu, J.; Zhu, D.; Zhang, Y.; Zhang, R.; Cui, Y. Visualization and intracellular dynamic tracking of exosomes and exosomal miRNAs using single molecule localization microscopy. Nanoscale 2018, 10, 5154–5162. [Google Scholar] [CrossRef]
- Tian, T.; Zhu, Y.L.; Hu, F.H.; Wang, Y.Y.; Huang, N.P.; Xiao, Z.D. Dynamics of exosome internalization and trafficking. J. Cell Physiol. 2013, 228, 1487–1495. [Google Scholar] [CrossRef]
- Shelke, G.V.; Yin, Y.; Jang, S.C.; Lässer, C.; Wennmalm, S.; Hoffmann, H.J.; Li, L.; Gho, Y.S.; Nilsson, J.A.; Lötvall, J. Endosomal signalling via exosome surface TGFβ-1. J. Extracell. Vesicles 2019, 8, 1650458. [Google Scholar] [CrossRef]
- Heusermann, W.; Hean, J.; Trojer, D.; Steib, E.; von Bueren, S.; Graff-Meyer, A.; Genoud, C.; Martin, K.; Pizzato, N.; Voshol, J.; et al. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J. Cell Biol. 2016, 213, 173–184. [Google Scholar] [CrossRef]
- Wang, S.; Sun, H.; Tanowitz, M.; Liang, X.H.; Crooke, S.T. Intra-endosomal trafficking mediated by lysobisphosphatidic acid contributes to intracellular release of phosphorothioate-modified antisense oligonucleotides. Nucleic Acids Res. 2017, 45, 5309–5322. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.D.; Donaldson, J.G. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2009, 10, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Lakhal, S.; Wood, M.J. Exosome nanotechnology: An emerging paradigm shift in drug delivery: Exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. Bioessays 2011, 33, 737–741. [Google Scholar] [CrossRef]
- Zhong, L.; Liao, D.; Li, J.; Liu, W.; Wang, J.; Zeng, C.; Wang, X.; Cao, Z.; Zhang, R.; Li, M.; et al. Rab22a-NeoF1 fusion protein promotes osteosarcoma lung metastasis through its secretion into exosomes. Signal Transduct. Target. Ther. 2021, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Imjeti, N.S.; Menck, K.; Egea-Jimenez, A.L.; Lecointre, C.; Lembo, F.; Bouguenina, H.; Badache, A.; Ghossoub, R.; David, G.; Roche, S.; et al. Syntenin mediates SRC function in exosomal cell-to-cell communication. Proc. Natl. Acad. Sci. USA 2017, 114, 12495–12500. [Google Scholar] [CrossRef]
- Datta, A.; Kim, H.; Lal, M.; McGee, L.; Johnson, A.; Moustafa, A.A.; Jones, J.C.; Mondal, D.; Ferrer, M.; Abdel-Mageed, A.B. Manumycin A suppresses exosome biogenesis and secretion via targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in castration-resistant prostate cancer cells. Cancer Lett. 2017, 408, 73–81. [Google Scholar] [CrossRef]
- Wang, F.; Oudaert, I.; Tu, C.; Maes, A.; Van der Vreken, A.; Vlummens, P.; De Bruyne, E.; De Veirman, K.; Wang, Y.; Fan, R.; et al. System Xc(-) inhibition blocks bone marrow-multiple myeloma exosomal crosstalk, thereby countering bortezomib resistance. Cancer Lett. 2022, 535, 215649. [Google Scholar] [CrossRef]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.G.; Harris, C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018, 9, 771. [Google Scholar] [CrossRef]
- Mitani, F.; Lin, J.; Sakamoto, T.; Uehara, R.; Hikita, T.; Yoshida, T.; Setiawan, A.; Arai, M.; Oneyama, C. Asteltoxin inhibits extracellular vesicle production through AMPK/mTOR-mediated activation of lysosome function. Sci. Rep. 2022, 12, 6674. [Google Scholar] [CrossRef] [PubMed]
- Dorayappan, K.D.P.; Wanner, R.; Wallbillich, J.J.; Saini, U.; Zingarelli, R.; Suarez, A.A.; Cohn, D.E.; Selvendiran, K. Hypoxia-induced exosomes contribute to a more aggressive and chemoresistant ovarian cancer phenotype: A novel mechanism linking STAT3/Rab proteins. Oncogene 2018, 37, 3806–3821. [Google Scholar] [CrossRef]
- Verweij, F.J.; Bebelman, M.P.; Jimenez, C.R.; Garcia-Vallejo, J.J.; Janssen, H.; Neefjes, J.; Knol, J.C.; de Goeij-de Haas, R.; Piersma, S.R.; Baglio, S.R.; et al. Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J. Cell Biol. 2018, 217, 1129–1142. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.D.; Zhuang, X.P.; Cai, D.L.; Cao, C.; Gu, Q.S.; Liu, X.N.; Zheng, B.B.; Guan, B.J.; Yu, L.; Li, J.K.; et al. Let-7a regulates EV secretion and mitochondrial oxidative phosphorylation by targeting SNAP23 in colorectal cancer. J. Exp. Clin. Cancer Res. 2021, 40, 31. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.W.; Cao, C.H.; Han, K.; Zhao, Y.X.; Cai, M.Y.; Xiang, Z.C.; Zhang, J.X.; Chen, J.W.; Zhong, L.P.; Huang, Y.; et al. APC-activated long noncoding RNA inhibits colorectal carcinoma pathogenesis through reduction of exosome production. J. Clin. Investig. 2019, 129, 727–743. [Google Scholar] [CrossRef]
- Tan, Z.; Cao, L.; Wu, Y.; Wang, B.; Song, Z.; Yang, J.; Cheng, L.; Yang, X.; Zhou, X.; Dai, Z.; et al. Bisecting GlcNAc modification diminishes the pro-metastatic functions of small extracellular vesicles from breast cancer cells. J. Extracell. Vesicles 2020, 10, e12005. [Google Scholar] [CrossRef]
- Giovannone, A.J.; Reales, E.; Bhattaram, P.; Fraile-Ramos, A.; Weimbs, T. Monoubiquitination of syntaxin 3 leads to retrieval from the basolateral plasma membrane and facilitates cargo recruitment to exosomes. Mol. Biol. Cell 2017, 28, 2843–2853. [Google Scholar] [CrossRef]
- Sun, H.; Meng, Q.; Shi, C.; Yang, H.; Li, X.; Wu, S.; Familiari, G.; Relucenti, M.; Aschner, M.; Wang, X.; et al. Hypoxia-Inducible Exosomes Facilitate Liver-Tropic Premetastatic Niche in Colorectal Cancer. Hepatology 2021, 74, 2633–2651. [Google Scholar] [CrossRef]
- Yang, K.; Fan, M.; Wang, X.; Xu, J.; Wang, Y.; Tu, F.; Gill, P.S.; Ha, T.; Liu, L.; Williams, D.L.; et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ. 2022, 29, 133–146. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Gong, H.; Luo, S.; Cui, Y. The Role of Exosomes and Their Applications in Cancer. Int. J. Mol. Sci. 2021, 22, 2204. [Google Scholar] [CrossRef]
- Baig, M.S.; Roy, A.; Rajpoot, S.; Liu, D.; Savai, R.; Banerjee, S.; Kawada, M.; Faisal, S.M.; Saluja, R.; Saqib, U.; et al. Tumor-derived exosomes in the regulation of macrophage polarization. Inflamm. Res. 2020, 69, 435–451. [Google Scholar] [CrossRef]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef]
- Cheng, Q.; Shi, X.; Han, M.; Smbatyan, G.; Lenz, H.J.; Zhang, Y. Reprogramming Exosomes as Nanoscale Controllers of Cellular Immunity. J. Am. Chem. Soc. 2018, 140, 16413–16417. [Google Scholar] [CrossRef]
- Kooijmans, S.A.; Aleza, C.G.; Roffler, S.R.; van Solinge, W.W.; Vader, P.; Schiffelers, R.M. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. Vesicles 2016, 5, 31053. [Google Scholar] [CrossRef] [PubMed]
- de Gassart, A.; Geminard, C.; Fevrier, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef] [PubMed]
- Tallon, C.; Hollinger, K.R.; Pal, A.; Bell, B.J.; Rais, R.; Tsukamoto, T.; Witwer, K.W.; Haughey, N.J.; Slusher, B.S. Nipping disease in the bud: nSMase2 inhibitors as therapeutics in extracellular vesicle-mediated diseases. Drug Discov. Today 2021, 26, 1656–1668. [Google Scholar] [CrossRef]
- Leblanc, R.; Kashyap, R.; Barral, K.; Egea-Jimenez, A.L.; Kovalskyy, D.; Feracci, M.; Garcia, M.; Derviaux, C.; Betzi, S.; Ghossoub, R.; et al. Pharmacological inhibition of syntenin PDZ2 domain impairs breast cancer cell activities and exosome loading with syndecan and EpCAM cargo. J. Extracell. Vesicles 2020, 10, e12039. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.; Kim, H.; McGee, L.; Johnson, A.E.; Talwar, S.; Marugan, J.; Southall, N.; Hu, X.; Lal, M.; Mondal, D.; et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: A drug repurposing strategy for advanced cancer. Sci. Rep. 2018, 8, 8161. [Google Scholar] [CrossRef]
- Wang, G.; Xie, L.; Li, B.; Sang, W.; Yan, J.; Li, J.; Tian, H.; Li, W.; Zhang, Z.; Tian, Y.; et al. A nanounit strategy reverses immune suppression of exosomal PD-L1 and is associated with enhanced ferroptosis. Nat. Commun. 2021, 12, 5733. [Google Scholar] [CrossRef]
- Xu, X.; Liu, Y.; Li, Y.; Chen, H.; Zhang, Y.; Liu, J.; Deng, S.; Zheng, Y.; Sun, X.; Wang, J.; et al. Selective exosome exclusion of miR-375 by glioma cells promotes glioma progression by activating the CTGF-EGFR pathway. J. Exp. Clin. Cancer Res. 2021, 40, 16. [Google Scholar] [CrossRef]
- Menck, K.; Sönmezer, C.; Worst, T.S.; Schulz, M.; Dihazi, G.H.; Streit, F.; Erdmann, G.; Kling, S.; Boutros, M.; Binder, C.; et al. Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J. Extracell. Vesicles 2017, 6, 1378056. [Google Scholar] [CrossRef] [PubMed]
- Marleau, A.M.; Chen, C.S.; Joyce, J.A.; Tullis, R.H. Exosome removal as a therapeutic adjuvant in cancer. J. Transl. Med. 2012, 10, 134. [Google Scholar] [CrossRef] [PubMed]
- Ruzicka, M.; Xiao, F.; Abujrad, H.; Al-Rewashdy, Y.; Tang, V.A.; Langlois, M.A.; Sorisky, A.; Ooi, T.C.; Burger, D. Effect of hemodialysis on extracellular vesicles and circulating submicron particles. BMC Nephrol. 2019, 20, 294. [Google Scholar] [CrossRef] [PubMed]
- Nishida-Aoki, N.; Tominaga, N.; Takeshita, F.; Sonoda, H.; Yoshioka, Y.; Ochiya, T. Disruption of Circulating Extracellular Vesicles as a Novel Therapeutic Strategy against Cancer Metastasis. Mol. Ther. 2017, 25, 181–191. [Google Scholar] [CrossRef]
- Hayatudin, R.; Fong, Z.; Ming, L.C.; Goh, B.H.; Lee, W.L.; Kifli, N. Overcoming Chemoresistance via Extracellular Vesicle Inhibition. Front. Mol. Biosci. 2021, 8, 629874. [Google Scholar] [CrossRef]
- Kirchhausen, T.; Macia, E.; Pelish, H.E. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzym. 2008, 438, 77–93. [Google Scholar] [CrossRef]
- Dutta, D.; Donaldson, J.G. Search for inhibitors of endocytosis: Intended specificity and unintended consequences. Cell Logist. 2012, 2, 203–208. [Google Scholar] [CrossRef]
- Chiba, M.; Kubota, S.; Sato, K.; Monzen, S. Exosomes released from pancreatic cancer cells enhance angiogenic activities via dynamin-dependent endocytosis in endothelial cells in vitro. Sci. Rep. 2018, 8, 11972. [Google Scholar] [CrossRef]
- Elkin, S.R.; Oswald, N.W.; Reed, D.K.; Mettlen, M.; MacMillan, J.B.; Schmid, S.L. Ikarugamycin: A Natural Product Inhibitor of Clathrin-Mediated Endocytosis. Traffic 2016, 17, 1139–1149. [Google Scholar] [CrossRef]
- Sento, S.; Sasabe, E.; Yamamoto, T. Application of a Persistent Heparin Treatment Inhibits the Malignant Potential of Oral Squamous Carcinoma Cells Induced by Tumor Cell-Derived Exosomes. PLoS ONE 2016, 11, e0148454. [Google Scholar] [CrossRef]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.P.; Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef] [PubMed]
- Bonjoch, L.; Gironella, M.; Iovanna, J.L.; Closa, D. REG3β modifies cell tumor function by impairing extracellular vesicle uptake. Sci. Rep. 2017, 7, 3143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lyden, D. Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat. Protoc. 2019, 14, 1027–1053. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Chiang, C.Y.; Chen, C. Toward characterizing extracellular vesicles at a single-particle level. J. Biomed. Sci. 2019, 26, 9. [Google Scholar] [CrossRef] [PubMed]
- Frolova, L.; Li, I.T.S. Targeting Capabilities of Native and Bioengineered Extracellular Vesicles for Drug Delivery. Bioengineering 2022, 9, 496. [Google Scholar] [CrossRef]
- Hung, M.E.; Leonard, J.N. Stabilization of exosome-targeting peptides via engineered glycosylation. J. Biol. Chem. 2015, 290, 8166–8172. [Google Scholar] [CrossRef]
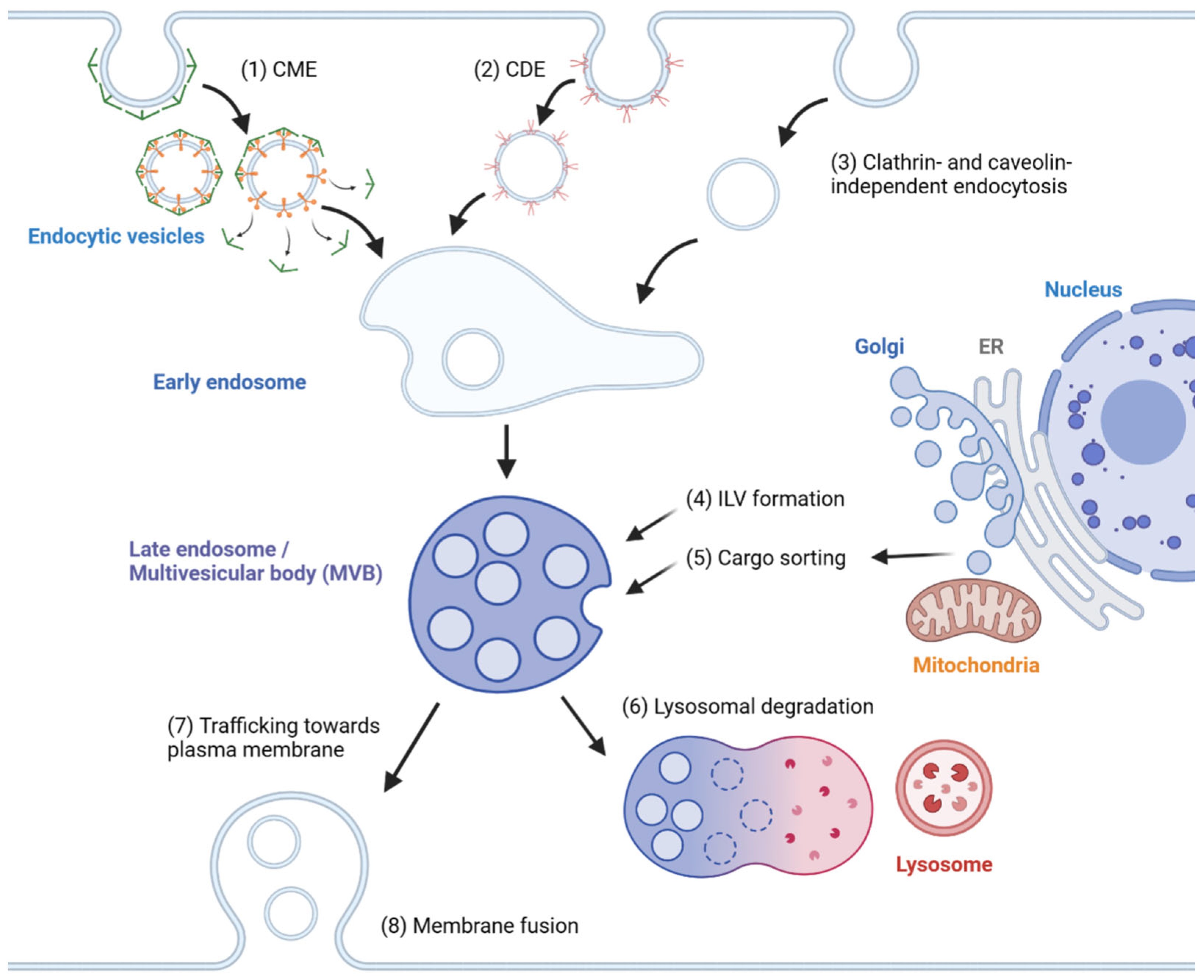
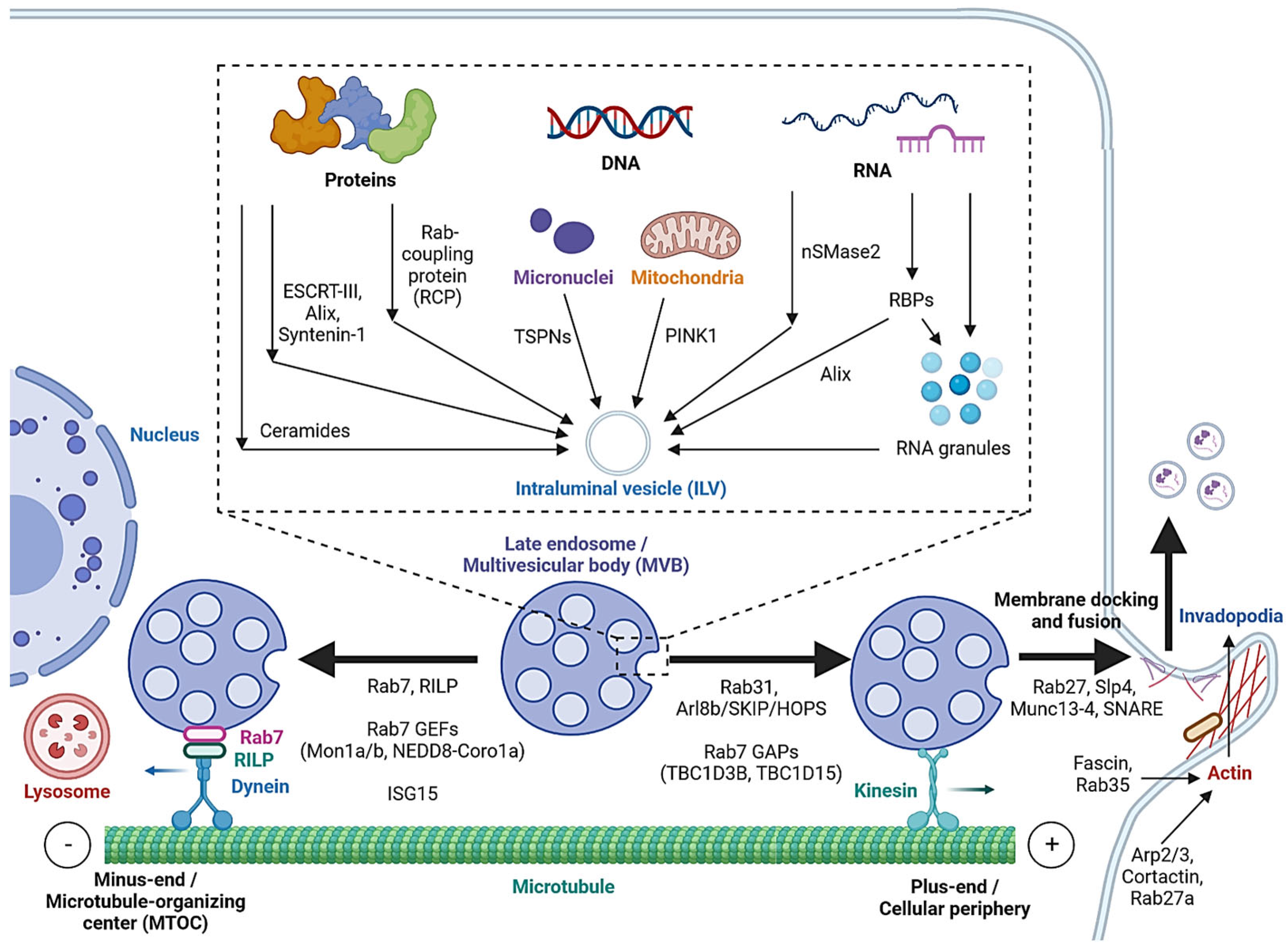
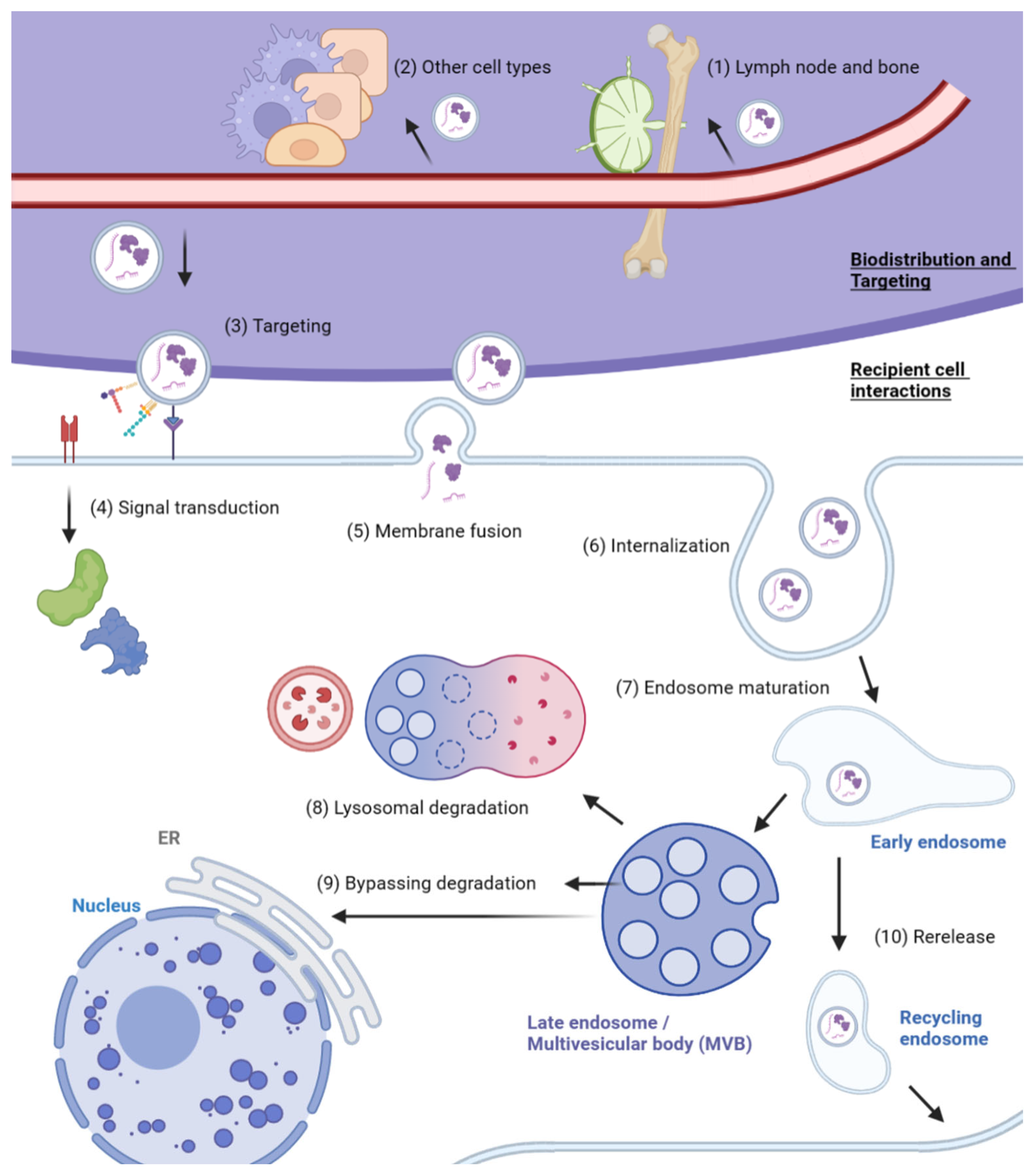

| Steps | Pathways | Mechanisms | References |
|---|---|---|---|
| Exosome biogenesis | |||
| Endosome formation | Clathrin-mediated endocytosis | FCHO, AP2, adaptor proteins, Dynamin, HSC70, auxilin | [20] |
| Caveolin-dependent endocytosis | Caveolin-1 | [21] | |
| Clathrin- and caveolin-independent endocytosis | CLIC/GEEC; ARF6; Actin | [20,21] | |
| MVB formation | ESCRT-dependent pathway | ESCRT-0/I/II/III, VPS4 | [22,23,24] |
| Non-canonical ESCRT-dependent pathways | Syndecan-Syntenin-Alix, heparinase; HD-PTP | [25,28,29,30,31] | |
| ESCRT-independent pathway | LC3, nSMase2, ceramide; cholesterol, caveolin-1, flotillins; TSPNs (CD63, TSPN6, CD9, CD81) | [34,35,36,37,38] [39,40,41,42,43] [45,46,47,48,49,50,51] | |
| Cargo sorting | Protein-sorting pathways | ESCRT or ESCRT-independent pathways; Rab coupling protein | [25,26,27,32,33,53,54] |
| DNA-sorting pathways | TSPN interaction with micronuclei; PINK1 | [56,58] | |
| RNA-sorting pathways | nSMase2, SP/NK1R, RBPs, ESCRT-II, Alix; LLPS, 3′ end adenylation or uridylation | [5,59,60,61,62,63,64,65,66,67,68] [69,70,71,72] | |
| Extracellular release | SNARE-mediated membrane fusion | Fas/Fap-1/caveolin-1, lncRNA HOTAIR | [74,75] |
| Actin reorganization | Cortactin, Arp2/3, Rab27a, Coronin1b; Rab25, Fascin | [76,77,78,79] | |
| Microtubule-mediated transport | Rab7, RILP; Rab31, Arl8b/SKIP/HOPS, Rab7 GAPs (TBC1D2B, TBC1D15), Rab7 GEFs (Mon1a/b, NEDD8-Coro1a); ISG15 | [80,81,82] [83,84,85,86] [87] | |
| Rab-regulated trafficking and fusion | Rab27a/b, Rab27 GEFs (MADD, FAM45A), KIBRA, Slp4, Munc13-4; Rab11; Rab35, Ca2+ | [88,89,90,91,92,93] [94,95,96,97] | |
| Steps | Factors/Processes | Mechanisms | References |
|---|---|---|---|
| Exosome absorption | |||
| Exosome biodistribution and organ-specific accumulation | Retention in lymph nodes and bone | Exosome size | [14] |
| Route of administration | Intratumoral, intravenous, etc. | [99,100,101] | |
| Cell type-specific rate of exosome uptake | Cell type-dependent factors | [102] | |
| Targeting to recipient cell | Receptor–ligand interaction | RVG, SDF-1; integrins (αvβ5, α6β4, α6β1, α4β7), TSPNs (TSPAN8, CD63), CD47 | [103,104,105] [112,113,114,115,116] |
| Lipid targeting | Phosphatidylethanolamine, sphingomyelin | [106,107] | |
| Cell-type-specific exosome uptake | Recognition moieties from the donor cell type | [108,109,110,111] | |
| Recipient cell interactions | Signal transduction via receptors | MHC-peptide complex, TNF receptors | [118,119] |
| Membrane fusion | Rabs, SNARE; membrane rigidity and lipid content | [107,122] | |
| Clathrin-mediated endocytosis | Dynamin-2; transferrin | [117,124,125] | |
| Lipid raft endocytosis | Cholesterol, sphingolipid, flotillin | [126,127,128,129] | |
| Macropinocytosis | Rac1 GTPase, actin, cholesterol, EGFR | [130,131,132] | |
| Caveolin-dependent endocytosis | Caveolin-1 | [133,134] | |
| Phagocytosis | PI3K, phosphatidylserine | [125,135,136,137,138] | |
| Internalized exosomes | Lysosomal degradation | Trafficking to lysosomes (e.g., Rab7) | [80,139] |
| Bypassing lysosomal degradation | Passive diffusion; cargo activation by acidification; RNA transfer to ER; fusion with endosomal membrane (e.g., LBPA) | [108,140,141,142,143] | |
| Re-release | Sorting to recycling endosomes | [144,145] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lau, N.C.H.; Yam, J.W.P. From Exosome Biogenesis to Absorption: Key Takeaways for Cancer Research. Cancers 2023, 15, 1992. https://doi.org/10.3390/cancers15071992
Lau NCH, Yam JWP. From Exosome Biogenesis to Absorption: Key Takeaways for Cancer Research. Cancers. 2023; 15(7):1992. https://doi.org/10.3390/cancers15071992
Chicago/Turabian StyleLau, Nicolas Cheuk Hang, and Judy Wai Ping Yam. 2023. "From Exosome Biogenesis to Absorption: Key Takeaways for Cancer Research" Cancers 15, no. 7: 1992. https://doi.org/10.3390/cancers15071992
APA StyleLau, N. C. H., & Yam, J. W. P. (2023). From Exosome Biogenesis to Absorption: Key Takeaways for Cancer Research. Cancers, 15(7), 1992. https://doi.org/10.3390/cancers15071992







