Aberrant Methylation of the Imprinted C19MC and MIR371-3 Clusters in Patients with Non-Small Cell Lung Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Specimens
2.2. DNA Sample
2.3. Illumina 450 K Methylation Assay
2.4. Methylome Data Processing
2.5. Integrated Analysis of the Target mRNA–miRNA Regulatory Network
2.6. Correlation Analysis in Lung Primary Tumors from TCGA Datasets
2.7. Survival Analysis to Assess the Prognostic Value of Validated Target Genes of the C19MC and MIR371-3 miRNA Clusters in NSCLC Patients
3. Results
3.1. DNA Methylation Pattern of the C19MC and MIR371-3 miRNA Clusters in Lung Cancer
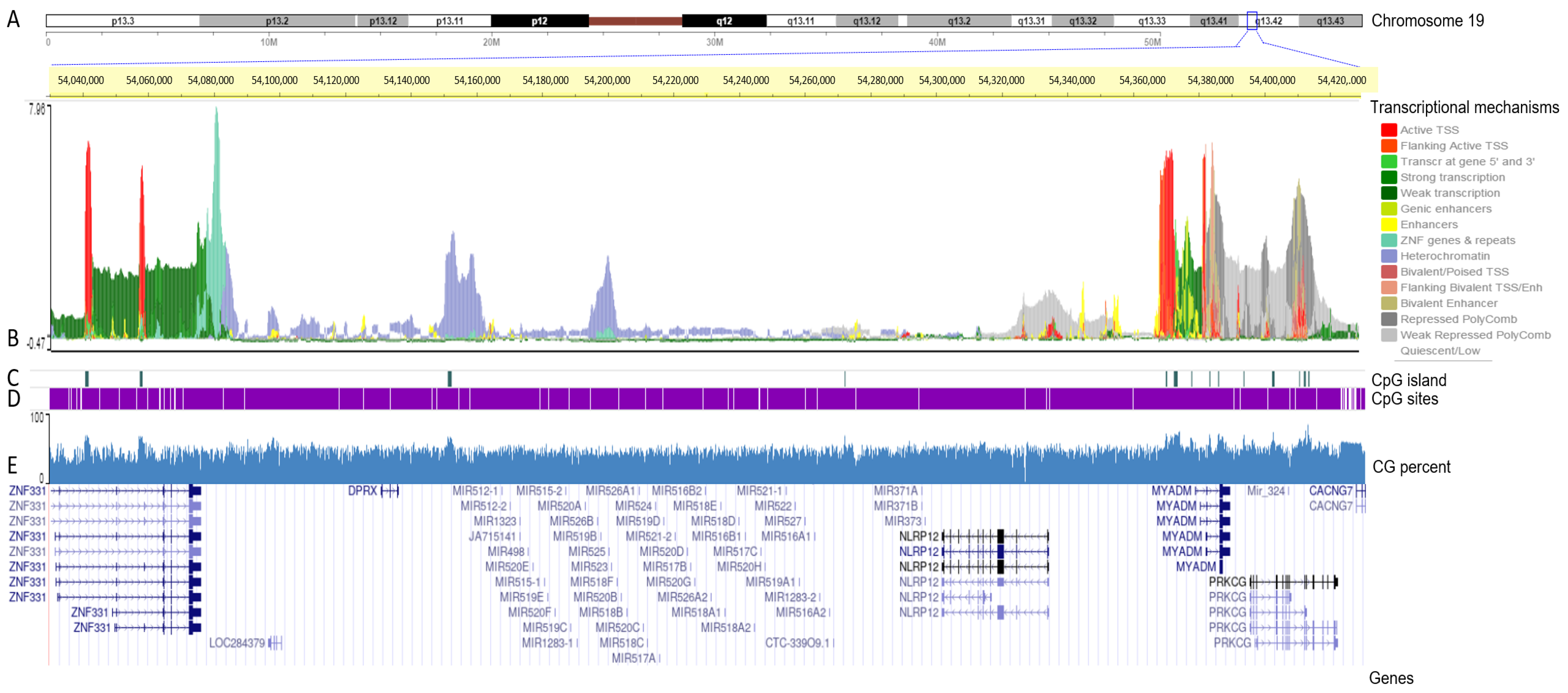
3.2. Transcriptional Mapping of the C19MC and MIR371-3 miRNA Clusters
3.3. Methylation Profile of the C19MC and MIR371-3 miRNA Clusters by Histological Subtypes
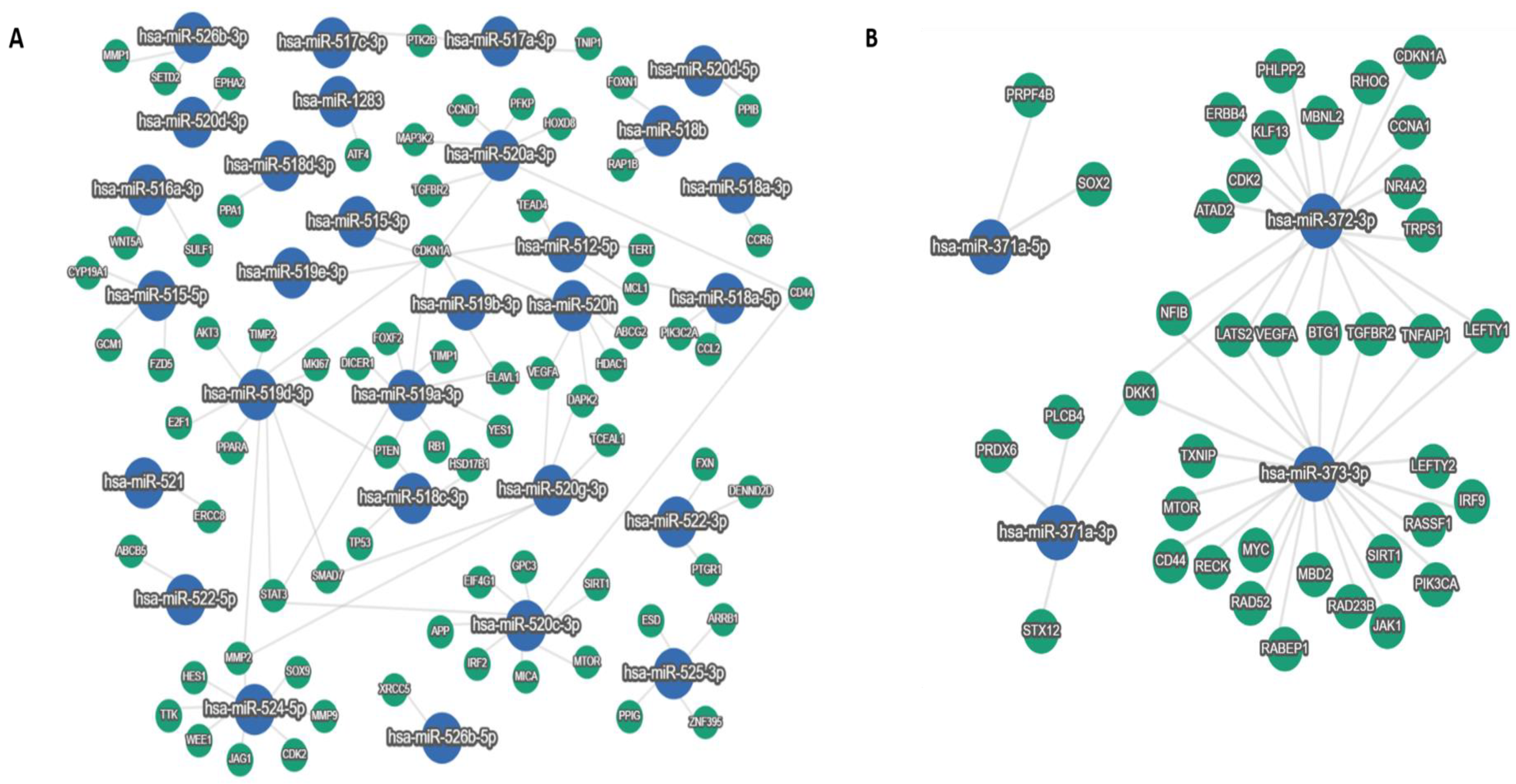
3.4. Experimentally Validated miRNA–Target Interactions for the C19MC and MIR371-2 miRNA Clusters
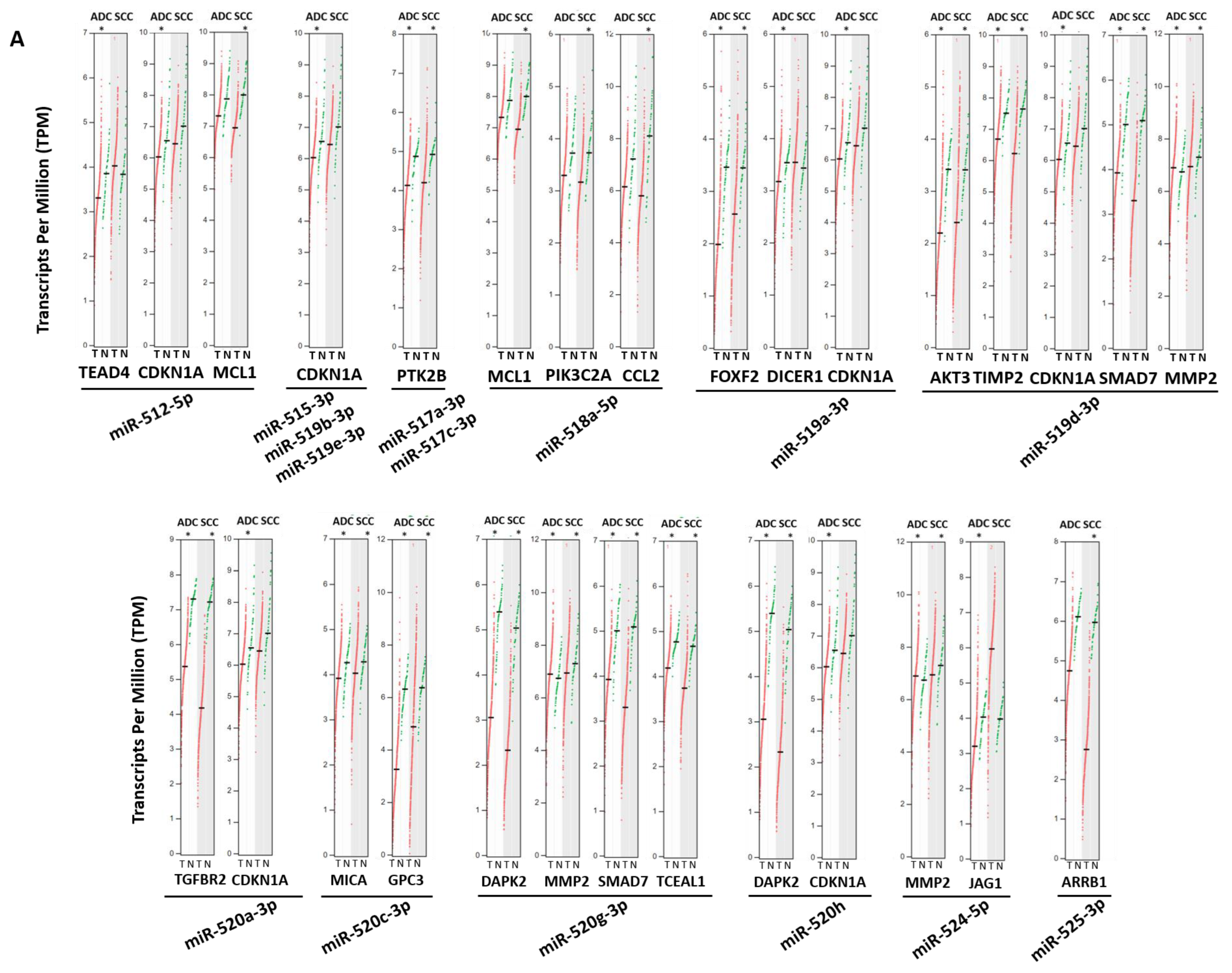
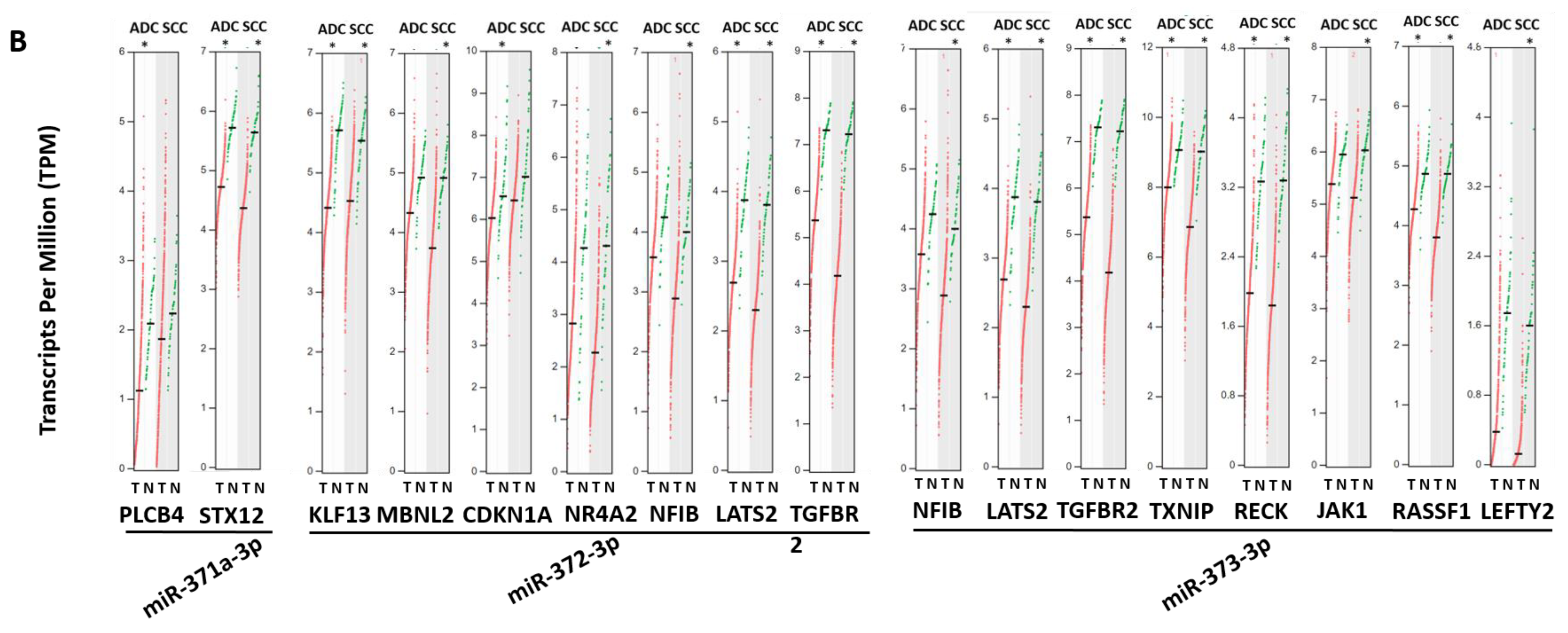
3.5. Aberrant Expression of Validated Target Genes for Both miRNA Clusters in Lung Cancer Patients
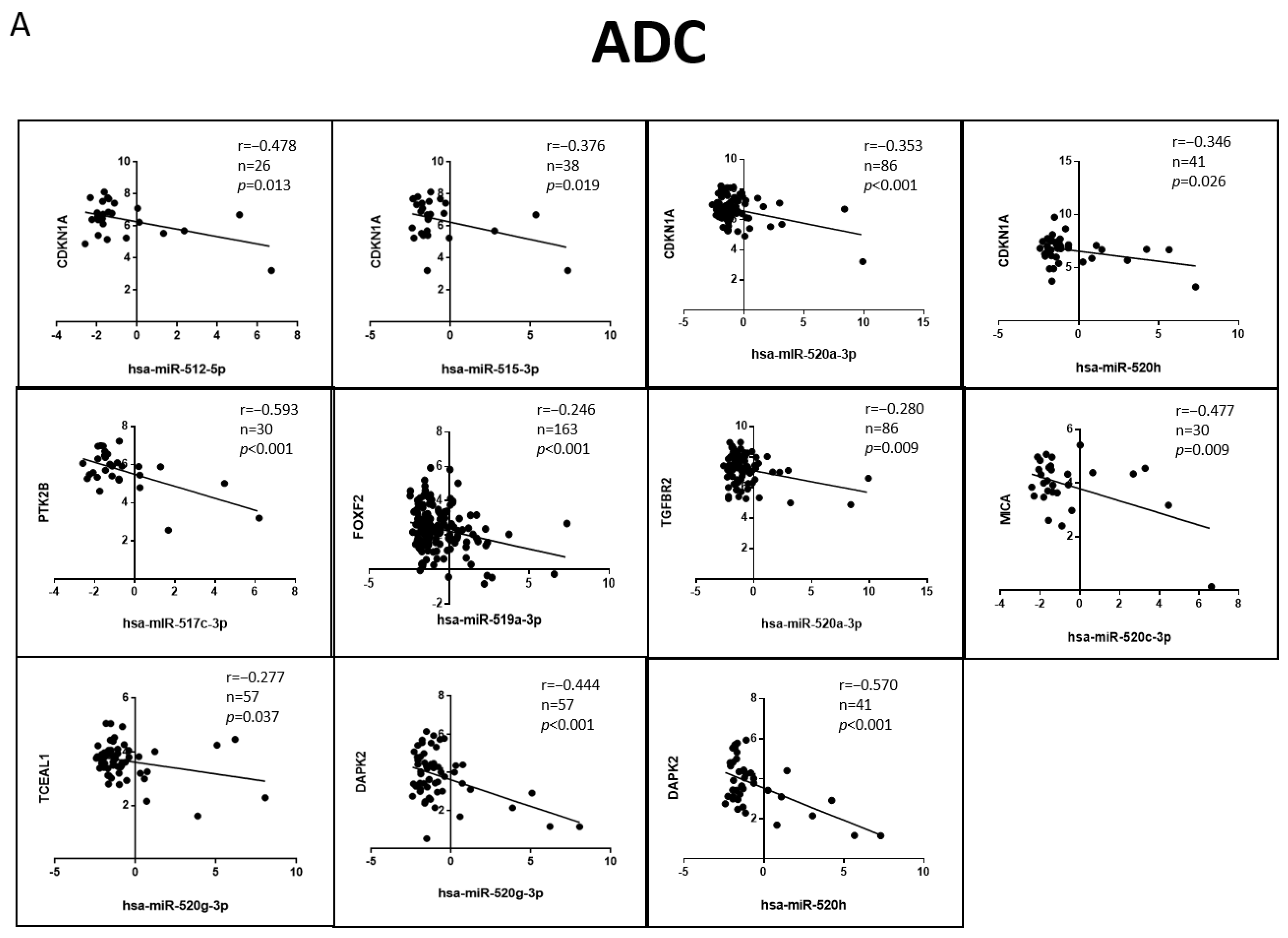
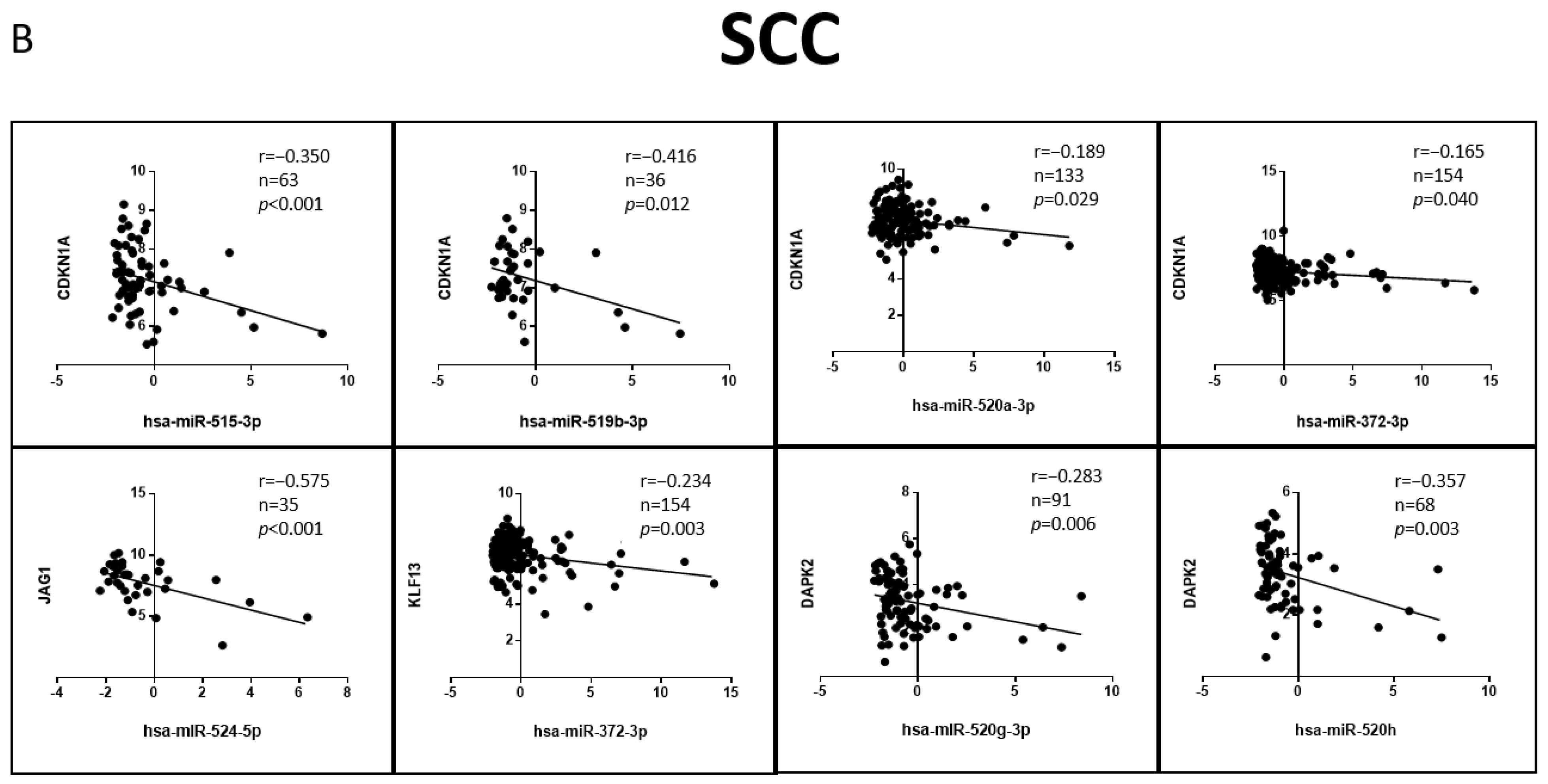
3.6. Correlation of the Validated Target mRNA–miRNA Expression in Lung Primary Tumors
3.7. Prognostic Role of the Target Gene Network of the C19MC and MIR371-3 Clusters in Lung Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Young, R.P.; Hopkins, R.J.; Christmas, T.; Black, P.N.; Metcalf, P.; Gamble, G.D. COPD Prevalence Is Increased in Lung Cancer, Independent of Age, Sex and Smoking History. Eur. Respir. J. 2009, 34, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Young, R.P.; Hopkins, R.J. COPD and Lung Cancer Linked at a Molecular Genetic Level. Chest 2011, 140, 266–267. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Heymach, J.V.; Lippman, S.M. Lung Cancer. N. Engl. J. Med. 2008, 359, 1367–1380. [Google Scholar] [CrossRef]
- Calabrese, F.; Lunardi, F.; Pezzuto, F.; Fortarezza, F.; Vuljan, S.E.; Marquette, C.; Hofman, P. Are There New Biomarkers in Tissue and Liquid Biopsies for the Early Detection of Non-Small Cell Lung Cancer? J. Clin. Med. 2019, 8, 414. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genomics 2010, 11, 537. [Google Scholar] [CrossRef]
- Keller, A.; Fehlmann, T.; Ludwig, N.; Kahraman, M.; Laufer, T.; Backes, C.; Vogelmeier, C.; Diener, C.; Biertz, F.; Herr, C.; et al. Genome-Wide MicroRNA Expression Profiles in COPD: Early Predictors for Cancer Development. Genom. Proteom. Bioinform. 2018, 16, 162–171. [Google Scholar] [CrossRef]
- Zhang, Y.; Sui, J.; Shen, X.; Li, C.; Yao, W.; Hong, W.; Peng, H.; Pu, Y.; Yin, L.; Liang, G. Differential Expression Profiles of MicroRNAs as Potential Biomarkers for the Early Diagnosis of Lung Cancer. Oncol. Rep. 2017, 37, 3543–3553. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Shoorei, H.; Branicki, W.; Taheri, M. Non-Coding RNA Profile in Lung Cancer. Exp. Mol. Pathol. 2020, 114, 104411. [Google Scholar] [CrossRef]
- Zarredar, H.; Ansarin, K.; Baradaran, B.; Shekari, N.; Eyvazi, S.; Safari, F.; Farajnia, S. Critical MicroRNAs in Lung Cancer: Recent Advances and Potential Applications. Anticancer Agents Med. Chem. 2018, 18, 1991–2005. [Google Scholar] [CrossRef]
- Quintanal-Villalonga, Á.; Molina-Pinelo, S. Epigenetics of Lung Cancer: A Translational Perspective. Cell. Oncol. 2019, 42, 739–756. [Google Scholar] [CrossRef] [PubMed]
- Matouk, I.J.; Halle, D.; Gilon, M.; Hochberg, A. The Non-Coding RNAs of the H19-IGF2 Imprinted Loci: A Focus on Biological Roles and Therapeutic Potential in Lung Cancer. J. Transl. Med. 2015, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Qi, P.; Ma, Z. Biology of MiR-17-92 Cluster and Its Progress in Lung Cancer. Int. J. Med. Sci. 2018, 15, 1443. [Google Scholar] [CrossRef] [PubMed]
- Enterina, J.R.; Enfield, K.S.S.; Anderson, C.; Marshall, E.A.; Ng, K.W.; Lam, W.L. DLK1-DIO3 Imprinted Locus Deregulation in Development, Respiratory Disease, and Cancer. Expert Rev. Respir. Med. 2017, 11, 749–761. [Google Scholar] [CrossRef]
- Molina-Pinelo, S.; Salinas, A.; Moreno-Mata, N.; Ferrer, I.; Suarez, R.; Andrés-León, E.; Rodríguez-Paredes, M.; Gutekunst, J.; Jantus-Lewintre, E.; Camps, C.; et al. Impact of DLK1-DIO3 Imprinted Cluster Hypomethylation in Smoker Patients with Lung Cancer. Oncotarget 2018, 9, 4395. [Google Scholar] [CrossRef]
- Merkerova, M.D.; Remesova, H.; Krejcik, Z.; Loudova, N.; Hrustincova, A.; Szikszai, K.; Cermak, J.; Jonasova, A.; Belickova, M. Relationship between Altered MiRNA Expression and DNA Methylation of the DLK1-DIO3 Region in Azacitidine-Treated Patients with Myelodysplastic Syndromes and Acute Myeloid Leukemia with Myelodysplasia-Related Changes. Cells 2018, 7, 138. [Google Scholar] [CrossRef] [PubMed]
- Honda, S.; Chatterjee, A.; Leichter, A.L.; Miyagi, H.; Minato, M.; Fujiyoshi, S.; Ara, M.; Kitagawa, N.; Tanaka, M.; Tanaka, Y.; et al. A MicroRNA Cluster in the DLK1-DIO3 Imprinted Region on Chromosome 14q32.2 Is Dysregulated in Metastatic Hepatoblastomas. Front. Oncol. 2020, 10, 513601. [Google Scholar] [CrossRef]
- McCarthy, E.C.; Dwyer, R.M. Emerging Evidence of the Functional Impact of the MiR379/MiR656 Cluster (C14MC) in Breast Cancer. Biomedicines 2021, 9, 827. [Google Scholar] [CrossRef]
- Flor, I.; Bullerdiek, J. The Dark Side of a Success Story: MicroRNAs of the C19MC Cluster in Human Tumours. J. Pathol. 2012, 227, 270–274. [Google Scholar] [CrossRef]
- Fonseca, Á.Y.G.; Santos, J.G.; Aristizábal-Pachón, A.F. An Overview of C19MC Cluster Subgroup 3 and Cancer. MicroRNA 2021, 10, 154–163. [Google Scholar] [CrossRef]
- Shah, J.A.; Khattak, S.; Rauf, M.A.; Cai, Y.; Jin, J. Potential Biomarkers of MiR-371-373 Gene Cluster in Tumorigenesis. Life 2021, 11, 984. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Yang, C.S.; Rana, T.M. Evolutionary Emergence of MicroRNAs in Human Embryonic Stem Cells. PLoS ONE 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Hromadnikova, I.; Kotlabova, K.; Doucha, J.; Dlouha, K.; Krofta, L. Absolute and Relative Quantification of Placenta-Specific Micrornas in Maternal Circulation with Placental Insufficiency-Related Complications. J. Mol. Diagn. 2012, 14, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, S.; Ding, J.; Zhao, Y.; Liang, L.; Liu, T.; Zhan, R.; He, X. Multiple MicroRNAs Modulate P21Cip1/Waf1 Expression by Directly Targeting Its 3’ Untranslated Region. Oncogene 2010, 29, 2302–2308. [Google Scholar] [CrossRef]
- Fornari, F.; Milazzo, M.; Chieco, P.; Negrini, M.; Marasco, E.; Capranico, G.; Mantovani, V.; Marinello, J.; Sabbioni, S.; Callegari, E.; et al. In Hepatocellular Carcinoma MiR-519d Is up-Regulated by P53 and DNA Hypomethylation and Targets CDKN1A/P21, PTEN, AKT3 and TIMP2. J. Pathol. 2012, 227, 275–285. [Google Scholar] [CrossRef]
- Zhou, A.D.; Diao, L.T.; Xu, H.; Xiao, Z.D.; Li, J.H.; Zhou, H.; Qu, L.H. β-Catenin/LEF1 Transactivates the MicroRNA-371-373 Cluster That Modulates the Wnt/β-Catenin-Signaling Pathway. Oncogene 2012, 31, 2968–2978. [Google Scholar] [CrossRef]
- Bullerdiek, J.; Flor, I. Exosome-Delivered MicroRNAs of “Chromosome 19 MicroRNA Cluster” as Immunomodulators in Pregnancy and Tumorigenesis. Mol. Cytogenet. 2012, 5, 27. [Google Scholar] [CrossRef]
- Li, Y.; Liang, M.; Zhang, Y.; Yuan, B.; Gao, W.; Shi, Z.; Bai, J. MiR-93, MiR-373, and MiR-17-5p Negatively Regulate the Expression of TBP2 in Lung Cancer. Front. Oncol. 2020, 10, 526. [Google Scholar] [CrossRef]
- Schönberger, S.; Mohseni, M.M.; Ellinger, J.; Tran, G.V.Q.; Becker, M.; Claviez, A.; Classen, C.F.; Hermes, B.; Driever, P.H.; Jorch, N.; et al. MicroRNA-Profiling of MiR-371~373- and MiR-302/367-Clusters in Serum and Cerebrospinal Fluid Identify Patients with Intracranial Germ Cell Tumors. J. Cancer Res. Clin. Oncol. 2023, 149, 791–802. [Google Scholar] [CrossRef]
- Xue, Q.; Yang, Y.; Yang, L.; Yan, X.; Shen, Z.; Liu, J.; Xue, J.; Zhao, W.; Liu, X. MiR-371b-5p-Engineered Exosomes Enhances Tumor Inhibitory Effect. Front. Cell Dev. Biol. 2021, 9, 750171. [Google Scholar] [CrossRef]
- Assenov, Y.; Müller, F.; Lutsik, P.; Walter, J.; Lengauer, T.; Bock, C. Comprehensive Analysis of DNA Methylation Data with RnBeads. Nat. Methods 2014, 11, 1138–1140. [Google Scholar] [CrossRef] [PubMed]
- Maksimovic, J.; Gordon, L.; Oshlack, A. SWAN: Subset-Quantile within Array Normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol. 2012, 13, R44. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Diyagama, D.; Neilson, J.; van Laar, R.; Dobrovic, A.; Holloway, A.; Smyth, G.K. Empirical Array Quality Weights in the Analysis of Microarray Data. BMC Bioinform. 2006, 7, 261. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, D.; Lowdon, R.F.; Costello, J.F.; Wang, T. MethylC Track: Visual Integration of Single-Base Resolution DNA Methylation Data on the WashU EpiGenome Browser. Bioinformatics 2014, 30, 2206–2207. [Google Scholar] [CrossRef] [PubMed]
- Kern, F.; Aparicio-Puerta, E.; Li, Y.; Fehlmann, T.; Kehl, T.; Wagner, V.; Ray, K.; Ludwig, N.; Lenhof, H.P.; Meese, E.; et al. MiRTargetLink 2.0-Interactive MiRNA Target Gene and Target Pathway Networks. Nucleic Acids Res. 2021, 49, W409–W416. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Lin, Y.C.D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. MiRTarBase Update 2022: An Informative Resource for Experimentally Validated MiRNA-Target Interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making Genome-Scale Phylogenetics Accessible to All. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for Representation and Analysis of Molecular Networks Involving Diseases and Drugs. Nucleic Acids Res. 2010, 38, D355–D360. [Google Scholar] [CrossRef]
- Li, R.; Qu, H.; Wang, S.; Chater, J.M.; Wang, X.; Cui, Y.; Yu, L.; Zhou, R.; Jia, Q.; Traband, R.; et al. CancerMIRNome: An Interactive Analysis and Visualization Database for MiRNome Profiles of Human Cancer. Nucleic Acids Res. 2022, 50, D1139–D1146. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Hoang, P.H.; Landi, M.T. DNA Methylation in Lung Cancer: Mechanisms and Associations with Histological Subtypes, Molecular Alterations, and Major Epidemiological Factors. Cancers 2022, 14, 961. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Soejima, K.; Arai, E.; Hamamoto, J.; Yasuda, H.; Arai, D.; Ishioka, K.; Ohgino, K.; Naoki, K.; Kohno, T.; et al. Prognostic Implication of PTPRH Hypomethylation in Non-Small Cell Lung Cancer. Oncol. Rep. 2015, 34, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Swiatnicki, M.R.; Rennhack, J.P.; Ortiz, M.M.O.; Hollern, D.P.; Perry, A.V.; Kubiak, R.; Riveria Riveria, S.M.; OReilly, S.; Andrechek, E.R. Elevated Phosphorylation of EGFR in NSCLC Due to Mutations in PTPRH. PLoS Genet. 2022, 18, e1010362. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Cheung, W.K.C.; Chen, S.; Lu, G.; Wang, Z.; Xie, D.; Li, K.; Lin, M.C.M.; Kung, H.F. Computational Identification and Characterization of Primate-Specific MicroRNAs in Human Genome. Comput. Biol. Chem. 2010, 34, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Suh, M.R.; Lee, Y.; Kim, J.Y.; Kim, S.K.; Moon, S.H.; Lee, J.Y.; Cha, K.Y.; Chung, H.M.; Yoon, H.S.; Moon, S.Y.; et al. Human Embryonic Stem Cells Express a Unique Set of MicroRNAs. Dev. Biol. 2004, 270, 488–498. [Google Scholar] [CrossRef]
- Vaira, V.; Elli, F.; Forno, I.; Guarnieri, V.; Verdelli, C.; Ferrero, S.; Scillitani, A.; Vicentini, L.; Cetani, F.; Mantovani, G.; et al. The MicroRNA Cluster C19MC Is Deregulated in Parathyroid Tumours. J. Mol. Endocrinol. 2012, 49, 115–124. [Google Scholar] [CrossRef]
- Ivey, K.N.; Srivastava, D. MicroRNAs as Regulators of Differentiation and Cell Fate Decisions. Cell Stem Cell 2010, 7, 36–41. [Google Scholar] [CrossRef]
- Rippe, V.; Dittberner, L.; Lorenz, V.N.; Drieschner, N.; Nimzyk, R.; Sendt, W.; Junker, K.; Belge, G.; Bullerdiek, J. The Two Stem Cell MicroRNA Gene Clusters C19MC and MiR-371-3 Are Activated by Specific Chromosomal Rearrangements in a Subgroup of Thyroid Adenomas. PLoS ONE 2010, 5, e9485. [Google Scholar] [CrossRef]
- Vilela-Salgueiro, B.; Barros-Silva, D.; Lobo, J.; Costa, A.L.; Guimarães, R.; Cantante, M.; Lopes, P.; Braga, I.; Oliveira, J.; Henrique, R.; et al. Germ Cell Tumour Subtypes Display Differential Expression of MicroRNA371a-3p. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed]
- Leão, R.; Albersen, M.; Looijenga, L.H.J.; Tandstad, T.; Kollmannsberger, C.; Murray, M.J.; Culine, S.; Coleman, N.; Belge, G.; Hamilton, R.J.; et al. Circulating MicroRNAs, the Next-Generation Serum Biomarkers in Testicular Germ Cell Tumours: A Systematic Review. Eur. Urol. 2021, 80, 456–466. [Google Scholar] [CrossRef]
- Zhao, J.J.; Yang, J.; Lin, J.; Yao, N.; Zhu, Y.; Zheng, J.; Xu, J.; Cheng, J.Q.; Lin, J.Y.; Ma, X. Identification of MiRNAs Associated with Tumorigenesis of Retinoblastoma by MiRNA Microarray Analysis. Child’s Nerv. Syst. 2009, 25, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Gumireddy, K.; Schrier, M.; le Sage, C.; Nagel, R.; Nair, S.; Egan, D.A.; Li, A.; Huang, G.; Klein-Szanto, A.J.; et al. The MicroRNAs MiR-373 and MiR-520c Promote Tumour Invasion and Metastasis. Nat. Cell Biol. 2008, 10, 202–210. [Google Scholar] [CrossRef]
- Cho, W.J.; Shin, J.M.; Kim, J.S.; Lee, M.R.; Hong, K.S.; Lee, J.H.; Koo, K.H.; Park, J.W.; Kim, K.S. MiR-372 Regulates Cell Cycle and Apoptosis of Ags Human Gastric Cancer Cell Line through Direct Regulation of LATS2. Mol. Cells 2009, 28, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Feber, A.; Xi, L.; Luketich, J.D.; Pennathur, A.; Landreneau, R.J.; Wu, M.; Swanson, S.J.; Godfrey, T.E.; Litle, V.R. MicroRNA Expression Profiles of Esophageal Cancer. J. Thorac. Cardiovasc. Surg. 2008, 135, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A MicroRNA Expression Signature of Human Solid Tumors Defines Cancer Gene Targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Fan, X.; Tao, S.; Li, Q.; Deng, B.; Tan, Q.Y.; Jin, H. The MiR-23a/27a/24-2 Cluster Promotes Postoperative Progression of Early-Stage Non-Small Cell Lung Cancer. Mol. Ther. Oncolytics 2021, 24, 205–217. [Google Scholar] [CrossRef]
- Valdmanis, P.N.; Roy-Chaudhuri, B.; Kim, H.K.; Sayles, L.C.; Zheng, Y.; Chuang, C.H.; Caswell, D.R.; Chu, K.; Zhang, Y.; Winslow, M.M.; et al. Upregulation of the MicroRNA Cluster at the Dlk1-Dio3 Locus in Lung Adenocarcinoma. Oncogene 2015, 34, 94–103. [Google Scholar] [CrossRef]
- González-Vallinas, M.; Rodríguez-Paredes, M.; Albrecht, M.; Sticht, C.; Stichel, D.; Gutekunst, J.; Pitea, A.; Sass, S.; Sánchez-Rivera, F.J.; Lorenzo-Bermejo, J.; et al. Epigenetically Regulated Chromosome 14q32 MiRNA Cluster Induces Metastasis and Predicts Poor Prognosis in Lung Adenocarcinoma Patients. Mol. Cancer Res. 2018, 16, 390–402. [Google Scholar] [CrossRef]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A Polycistronic MicroRNA Cluster, MiR-17-92, Is Overexpressed in Human Lung Cancers and Enhances Cell Proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Jia, X.; Zhou, J.; Sun, Q.; Ma, Z. The MiR-17-92 Gene Cluster Is a Blood-Based Marker for Cancer Detection in Non-Small-Cell Lung Cancer. Am. J. Med. Sci. 2020, 360, 248–260. [Google Scholar] [CrossRef]
- Nguyen, P.N.N.; Huang, C.J.; Sugii, S.; Cheong, S.K.; Choo, K.B. Selective Activation of MiRNAs of the Primate-Specific Chromosome 19 MiRNA Cluster (C19MC) in Cancer and Stem Cells and Possible Contribution to Regulation of Apoptosis. J. Biomed. Sci. 2017, 24, 20. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Aksoy, M.; Shi, J.; Houbaviy, H.B. Evolution of the MiR-290-295/MiR-371-373 Cluster Family Seed Repertoire. PLoS ONE 2014, 9, e8519. [Google Scholar] [CrossRef]
- Bunz, F.; Dutriaux, A.; Lengauer, C.; Waldman, T.; Zhou, S.; Brown, J.P.; Sedivy, J.M.; Kinzler, K.W.; Vogelstein, B. Requirement for P53 and P21 to Sustain G2 Arrest after DNA Damage. Science 1998, 282, 1497–1501. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Chen, X.; Chen, L.; Ma, Y.; Zhou, L.; Qi, Q.; Liu, Y.; Zhang, S.; Luo, J.; Zhou, X. Upregulation of the MiR-212/132 Cluster Suppresses Proliferation of Human Lung Cancer Cells. Oncol. Rep. 2015, 33, 705–712. [Google Scholar] [CrossRef]
- Li, C.; Lyu, J.; Meng, Q.H. MiR-93 Promotes Tumorigenesis and Metastasis of Non-Small Cell Lung Cancer Cells by Activating the PI3K/Akt Pathway via Inhibition of LKB1/ PTEN/ CDKN1A. J. Cancer 2017, 8, 870–879. [Google Scholar] [CrossRef]
- Wang, L.Q.; Yu, P.; Li, B.; Guo, Y.H.; Liang, Z.R.; Zheng, L.L.; Yang, J.H.; Xu, H.; Liu, S.; Zheng, L.S.; et al. MiR-372 and MiR-373 Enhance the Stemness of Colorectal Cancer Cells by Repressing Differentiation Signaling Pathways. Mol. Oncol. 2018, 12, 1949–1964. [Google Scholar] [CrossRef]
- Chen, C.H.; Wang, W.J.; Kuo, J.C.; Tsai, H.C.; Lin, J.R.; Chang, Z.F.; Chen, R.H. Bidirectional Signals Transduced by DAPK-ERK Interaction Promote the Apoptotic Effect of DAPK. EMBO J. 2005, 24, 294–304. [Google Scholar] [CrossRef]
- Jin, M.; Li, G.; Liu, W.; Wu, X.; Zhu, J.; Zhao, D.; Zeng, Z.; Xiong, M.; Song, Y.; He, X.; et al. Cigarette Smoking Induces Aberrant N6-Methyladenosine of DAPK2 to Promote Non-Small Cell Lung Cancer Progression by Activating NF-ΚB Pathway. Cancer Lett. 2021, 518, 214–229. [Google Scholar] [CrossRef]
- Schlegel, C.R.; Fonseca, A.V.; Stöcker, S.; Georgiou, M.L.; Misterek, M.B.; Munro, C.E.; Carmo, C.R.; Seckl, M.J.; Costa-Pereira, A.P. DAPK2 Is a Novel Modulator of TRAIL-Induced Apoptosis. Cell Death Differ. 2014, 21, 1780–1791. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Zhang, J.; Yu, X.L.; Zheng, G.F.; Zhao, F.; Jia, X.J. Death-Associated Protein Kinase Promoter Methylation Correlates with Clinicopathological and Prognostic Features in Nonsmall Cell Lung Cancer Patients: A Cohort Study. J. Cancer Res. Ther. 2018, 14, S65–S71. [Google Scholar] [CrossRef] [PubMed]
- Su, C.M.; Wang, M.Y.; Hong, C.C.; Chen, H.A.; Su, Y.H.; Wu, C.H.; Huang, M.T.; Chang, Y.W.; Jiang, S.S.; Sung, S.Y.; et al. MiR-520h Is Crucial for DAPK2 Regulation and Breast Cancer Progression. Oncogene 2016, 35, 1134–1142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, L.; Sun, Y.; Xiang, J.; Zhou, D.; Wang, L.; Xu, H.; Yang, X.; Du, N.; Zhang, M.; et al. MicroRNA-520g Promotes Epithelial Ovarian Cancer Progression and Chemoresistance via DAPK2 Repression. Oncotarget 2016, 7, 26516–26534. [Google Scholar] [CrossRef]
- Li, T.; Wang, H.; Xu, J.; Li, C.; Zhang, Y.; Wang, G.; Liu, Y.; Cai, S.; Fang, W.; Li, J.; et al. TGFBR2 Mutation Predicts Resistance to Immune Checkpoint Inhibitors in Patients with Non-Small Cell Lung Cancer. Ther. Adv. Med. Oncol. 2021, 13, 17588359211038477. [Google Scholar] [CrossRef]
- Kong, P.Z.; Li, G.M.; Tian, Y.; Song, B.; Shi, R.Y. Decreased Expression of FOXF2 as New Predictor of Poor Prognosis in Stage I Non-Small Cell Lung Cancer. Oncotarget 2016, 7, 55601–55610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, Y.Z.; Dai, D.Q. Aberrant DNA Methylation-Mediated FOXF2 Dysregulation Is a Prognostic Risk Factor for Gastric Cancer. Front. Mol. Biosci. 2021, 8, 645470. [Google Scholar] [CrossRef] [PubMed]
- Neel, J.-C.; Humbert, L.; Lebrun, J.-J. The Dual Role of TGFβ in Human Cancer: From Tumor Suppression to Cancer Metastasis. ISRN Mol. Biol. 2012, 2012, 381428. [Google Scholar] [CrossRef]
- Lo Sardo, F.; Pulito, C.; Sacconi, A.; Korita, E.; Sudol, M.; Strano, S.; Blandino, G. YAP/TAZ and EZH2 Synergize to Impair Tumor Suppressor Activity of TGFBR2 in Non-Small Cell Lung Cancer. Cancer Lett. 2021, 500, 51–63. [Google Scholar] [CrossRef]
- Feng, S.; Jacobsen, S.E.; Reik, W. Epigenetic Reprogramming in Plant and Animal Development. Science 2010, 330, 622–627. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, C.; Wu, C.; Cui, W.; Wang, L. DNA Methyltransferases in Cancer: Biology, Paradox, Aberrations, and Targeted Therapy. Cancers 2020, 12, 2123. [Google Scholar] [CrossRef] [PubMed]
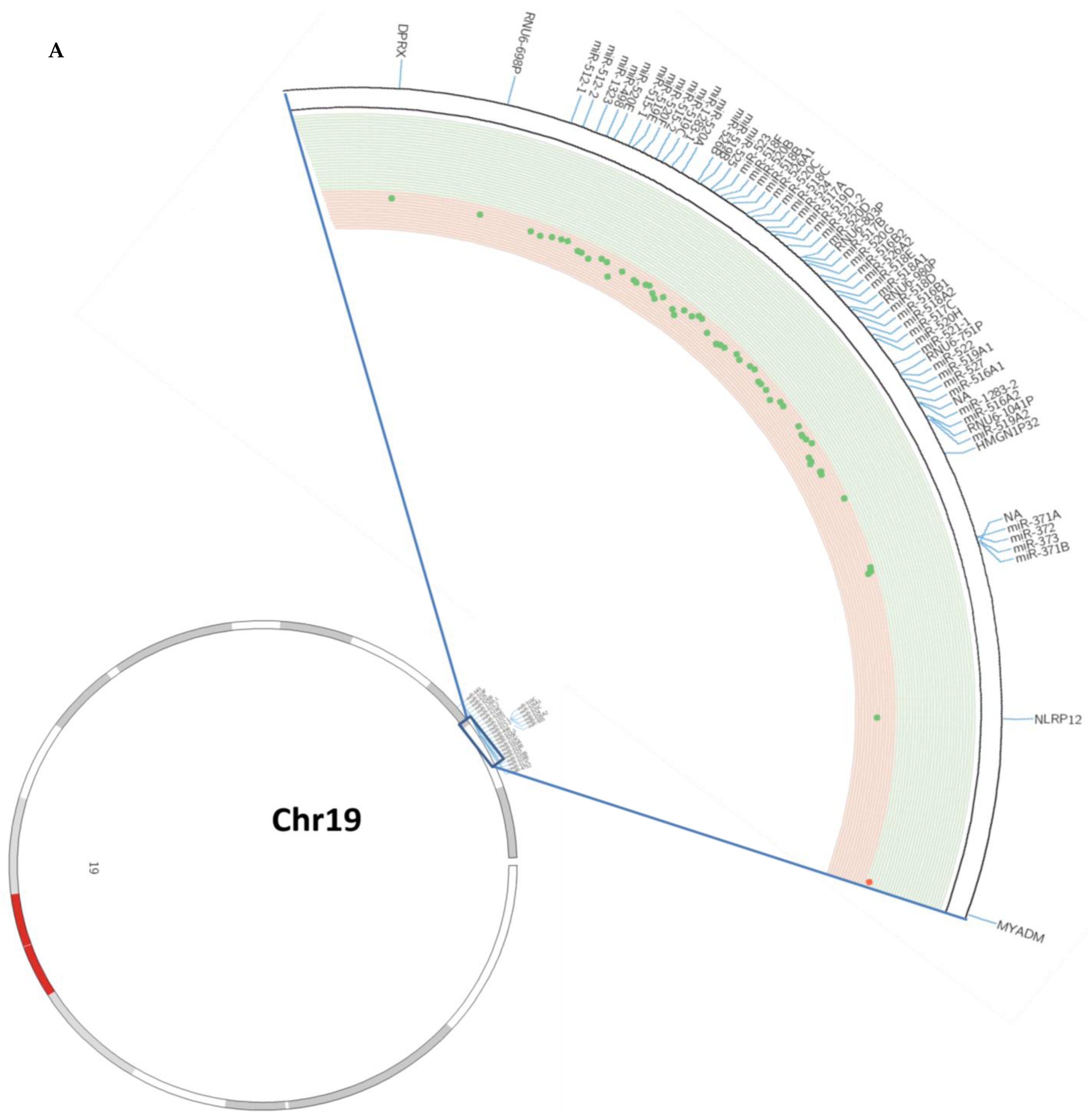
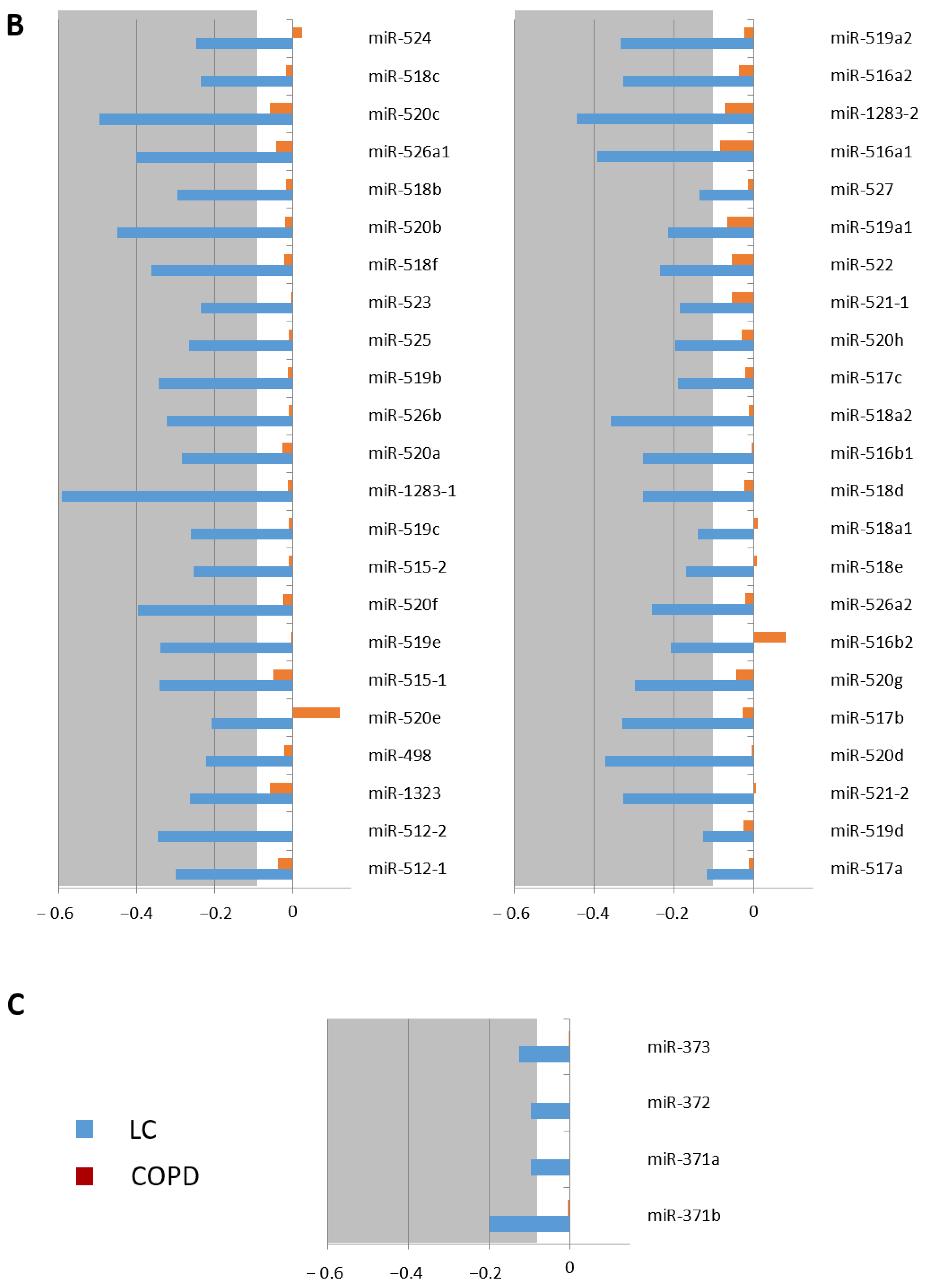


| Genes | Relative Methylation Changes (log2) | Adjusted p-Value | |
|---|---|---|---|
| C19MC | miR-512-1 | −0.307 | 6.3946 × 10−14 |
| miR-512-2 | −0.341 | 7.2597 × 10−9 | |
| miR-1323 | −0.245 | 1.0347 × 10−6 | |
| miR-498 | −0.241 | 2.7704 × 10−5 | |
| miR-520e | −0.214 | 5.7827 × 10−5 | |
| miR-515-1 | −0.346 | 2.0661 × 10−12 | |
| miR-519e | −0.340 | 3.1486 × 10−11 | |
| miR-520f | −0.402 | 4.644 × 10−13 | |
| miR-515-2 | −0.258 | 2.686 × 10−9 | |
| miR-519c | −0.264 | 2.9066 × 10−10 | |
| miR-1283-1 | −0.601 | 4.8535 × 10−17 | |
| miR-520a | −0.279 | 4.1063 × 10−11 | |
| miR-526b | −0.329 | 5.2657 × 10−14 | |
| miR-519b | −0.348 | 1.5969 × 10−15 | |
| miR-525 | −0.360 | 1.2282 × 10−10 | |
| miR-523 | −0.267 | 1.1234 × 10−11 | |
| miR-518f | −0.368 | 7.9986 × 10−15 | |
| miR-520b | −0.441 | 4.5322 × 10−16 | |
| miR-518b | −0.290 | 1.7394 × 10−9 | |
| miR-526a1 | −0.474 | 3.4512 × 10−14 | |
| miR-520c | −0.590 | 4.7234 × 10−18 | |
| miR-518c | −0.271 | 6.5541 × 10−9 | |
| miR-524 | −0.261 | 1.5518 × 10−12 | |
| miR-517a | −0.129 | 1.6843 × 10−5 | |
| miR-519d | −0.140 | 3.7082 × 10−5 | |
| miR-521-2 | −0.343 | 1.1838 × 10−14 | |
| miR-520d | −0.378 | 2.9354 × 10−13 | |
| miR-517b | −0.327 | 2.6624 × 10−12 | |
| miR-520g | −0.293 | 9.6092 × 10−11 | |
| miR-516b2 | −0.215 | 1.1738 × 10−8 | |
| miR-526a2 | −0.303 | 9.5413 × 10−10 | |
| miR-518e | −0.168 | 6.8329 × 10−7 | |
| miR-518a1 | −0.139 | 7.0465 × 10−5 | |
| miR-518d | −0.412 | 2.144 × 10−12 | |
| miR-516b1 | −0.269 | 9.8287 × 10−10 | |
| miR-518a2 | −0.364 | 2.9253 × 10−14 | |
| miR-517c | −0.199 | 1.476 × 10−10 | |
| miR-520h | −0.211 | 2.5861 × 10−10 | |
| miR-521-1 | −0.183 | 8.9968 × 10−5 | |
| miR-522 | −0.235 | 5.605 × 10−10 | |
| miR-519a1 | −0.217 | 9.8489 × 10−11 | |
| miR-527 | −0.130 | 7.4475 × 10−9 | |
| miR-516a1 | −0.400 | 2.5393 × 10−9 | |
| miR-1283-2 | −0.453 | 2.9539 × 10−13 | |
| miR-516a2 | −0.340 | 3.3131 × 10−16 | |
| miR-519a2 | −0.344 | 1.2518 × 10−14 | |
| MIR371-3 | miR-371b | −0.209 | 1.4784 × 10−8 |
| miR-371a | −0.106 | 7.6971 × 10−7 | |
| miR-372 | −0.106 | 7.6971 × 10−7 | |
| miR-373 | −0.137 | 1.183 × 10−7 |
| Gene | Gene Description | miRNA |
|---|---|---|
| BTG1 | B-Cell Translocation Gene 1 | miRNA-372-3p, miRNA-373-3p |
| CD44 | Cluster of Differentiation 44 | miRNA-520a-3p, miRNA-520c-3p, miRNA-373-3p |
| CDK2 | Cyclin Dependent Kinase 2 | miRNA-524-5p, miRNA-372-3p |
| CDKN1A | Cyclin Dependent Kinase Inhibitor 1A | miRNA-512-5p, miRNA-515-3p, miRNA-519a-3p, miRNA-519b-3p, miRNA-519d-3p, miRNA-519e-3p, miRNA-520a-3p, miRNA-520h, miRNA-373-3p |
| DAPK2 | Death Associated Protein Kinase 2 | miRNA-520h, miRNA-520g-3p |
| DKK1 | Dickkopf WNT Signaling Pathway Inhibitor 1 | miRNA-371a-3p, miRNA-372-3p, miRNA-373-3p |
| ELAVL1 | ELAV (Embryonic Lethal, Abnormal Vision, Drosophila)-Like RNA Binding Protein 1 | miRNA-519a-3p, miRNA-519b-3p |
| LATS2 | Large Tumor Suppressor Kinase 2 | miRNA-372-3p, miRNA-373-3p |
| LEFTY1 | Left-Right Determination Factor 1 | miRNA-372-3p, miRNA-373-3p |
| MCL1 | Myeloid Cell Leukemia Sequence 1 | miRNA-512-5p, miRNA-518a-5p |
| MMP2 | Matrix Metallopeptidase 2 | miRNA-519d-3p, miRNA-520g-3p, miRNA-524-5p |
| MTOR | Mechanistic Target of Rapamycin Kinase | miRNA-520c-3p, miRNA-373-3p |
| NFIB | Nuclear Factor I B | miRNA-372-3p, miRNA-373-3p |
| PTEN | Phosphatase and Tensin Homolog | miRNA-518c-3p, miRNA-519a-3p, miRNA-519d-3p |
| PTK2B | Protein Tyrosine Kinase 2 Beta | miRNA-517a-3p, miRNA-517c-3p |
| SIRT1 | Sirtuin 1 | miRNA-520c-3p, miRNA-373-3p |
| SAMD7 | Sterile Alpha Motif Domain Containing 7 | miRNA-519d-3p, miRNA-520g-3p |
| STAT3 | Signal Transducer and Activator of Transcription 3 | miRNA-519a-3p, miRNA-519g-3p, miRNA-520c-3p |
| TGFBR2 | Transforming Growth Factor Beta Receptor 2 | miRNA-520a-3p, miRNA-372-3p, miRNA-373-3p |
| TNFAIP1 | Tumor Necrosis Factor Alpha-Induced Protein 1 | miRNA-372-3p, miRNA-373-3p |
| VEGFA | Vascular Endothelial Growth Factor A | miRNA-520g-3p, miRNA-520h, miRNA-372-3p, miRNA-373-3p |
| Symbol | Description |
|---|---|
| ARRB1 | Arrestin Beta 1 |
| AKT3 | AKT Serine/Threonine Kinase 3 |
| CCL2 | C-C Motif Chemokine Ligand 2 |
| CDKN1A | Cyclin Dependent Kinase Inhibitor 1A |
| DAPK2 | Death Associated Protein Kinase 2 |
| DICER1 | Double-Stranded RNA-Specific Endoribonuclease |
| FOXF2 | Forkhead Box F2 |
| GPC3 | Glypican 3 |
| JAG1 | Jagged Canonical Notch Ligand 1 |
| JAK1 | Janus Kinase 1 |
| KLF13 | Kruppel Like Factor 13 |
| LEFTY2 | Left-Right Determination Factor 2 |
| LATS2 | Large Tumor Suppressor Kinase 2 |
| MBNL2 | Muscleblind-Like Protein 2 |
| MCL1 | Myeloid Cell Leukemia Sequence 1 |
| MICA | Major Histocompatibility Complex Class I Chain-Related Protein A |
| MMP2 | Matrix Metallopeptidase 2 |
| NFIB | Mechanistic Target of Rapamycin Kinase (MTOR), Nuclear Factor I B |
| NR4A2 | Nuclear Receptor Subfamily 4 Group A Member 2 |
| PIK3C2A | Phosphatidylinositol-4-Phosphate 3-Kinase Catalytic Subunit Type 2 Alpha |
| PLCB4 | Phospholipase C Beta 4 |
| PTK2B | Protein Tyrosine Kinase 2 Beta |
| TCEAL1 | Transcription Elongation Factor A Like 1 |
| TEAD4 | TEA Domain Transcription Factor 4 |
| TGFBR2 | Transforming Growth Factor Beta Receptor 2 |
| TIMP2 | Tissue Inhibitor of Metalloproteinases 2 |
| TXNIP | Thioredoxin Interacting Protein |
| RASSF1 | Ras Association Domain Family Member 1 |
| RECK | Reversion Inducing Cysteine Rich Protein with Kazal Motifs |
| SMAD7 | SMAD (small Mothers Against Decapentaplegic) family member 7 |
| STX12 | Syntaxin 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boyero, L.; Noguera-Uclés, J.F.; Castillo-Peña, A.; Salinas, A.; Sánchez-Gastaldo, A.; Alonso, M.; Benedetti, J.C.; Bernabé-Caro, R.; Paz-Ares, L.; Molina-Pinelo, S. Aberrant Methylation of the Imprinted C19MC and MIR371-3 Clusters in Patients with Non-Small Cell Lung Cancer. Cancers 2023, 15, 1466. https://doi.org/10.3390/cancers15051466
Boyero L, Noguera-Uclés JF, Castillo-Peña A, Salinas A, Sánchez-Gastaldo A, Alonso M, Benedetti JC, Bernabé-Caro R, Paz-Ares L, Molina-Pinelo S. Aberrant Methylation of the Imprinted C19MC and MIR371-3 Clusters in Patients with Non-Small Cell Lung Cancer. Cancers. 2023; 15(5):1466. https://doi.org/10.3390/cancers15051466
Chicago/Turabian StyleBoyero, Laura, José Francisco Noguera-Uclés, Alejandro Castillo-Peña, Ana Salinas, Amparo Sánchez-Gastaldo, Miriam Alonso, Johana Cristina Benedetti, Reyes Bernabé-Caro, Luis Paz-Ares, and Sonia Molina-Pinelo. 2023. "Aberrant Methylation of the Imprinted C19MC and MIR371-3 Clusters in Patients with Non-Small Cell Lung Cancer" Cancers 15, no. 5: 1466. https://doi.org/10.3390/cancers15051466
APA StyleBoyero, L., Noguera-Uclés, J. F., Castillo-Peña, A., Salinas, A., Sánchez-Gastaldo, A., Alonso, M., Benedetti, J. C., Bernabé-Caro, R., Paz-Ares, L., & Molina-Pinelo, S. (2023). Aberrant Methylation of the Imprinted C19MC and MIR371-3 Clusters in Patients with Non-Small Cell Lung Cancer. Cancers, 15(5), 1466. https://doi.org/10.3390/cancers15051466






