Cervical Fluids Are a Source of Protein Biomarkers for Early, Non-Invasive Endometrial Cancer Diagnosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Recruitment and Sample Collection
- M1 was obtained with the Rovers Cervex Brush® (Rovers Medical Devices, Oss, The Netherlands). This cervical brush is used to obtain samples for cervical liquid cytology. It has a shape designed to obtain a good representation of endocervical and exocervical material.
- M2 was obtained with the Wooden cervical scrape or Ayres spatula (Goodwood medical care, Dalian, China). It is generally used to obtain the exocervical representation of the pap-smears. It can also be used to obtain a vaginal sample but it was not used for this purpose in the present study.
- M3 was obtained with the endocervical swab HC2 DNA collection device Digene (QIAGEN, Hilden, Germany), used to get an endocervical mucus sample. It is generally used to perform the hybrid capture test to rule out human papillomavirus infections of the lower genital tract. It has little bristles that are introduced in the endocervix to collect the mucus.
- M4 was obtained with a cotton swab (Deltalab, Barcelona, Spain), usually used to take superficial samples to perform bacterial cultures of any location. It is blunt and only impregnates with the secretions of the endocervix without scratching the tissue.
- M5 was obtained using an endocervical brush (Bexen medical, Mondragón, Spain). It is the tool used to obtain an endocervical representation in pap-smears.
- Pipelle biopsies were obtained with the Cornier Pipelle (Eurogine Ref. 03040200, Spain) following the standard procedure. The device was introduced through the cervical canal into the uterine cavity, and the pipelle biopsy was obtained by applying negative pressure. This device requires cervical permeability to access the uterine cavity. Once collected, PBS 1X was added in a 1:1 volume.
2.2. Sample Preparation
2.3. DDA Analysis on a Tims-TOF Pro Mass Spectrometer
2.4. DDA Data Processing
2.5. PRM Analysis
2.6. Data Analysis
3. Results
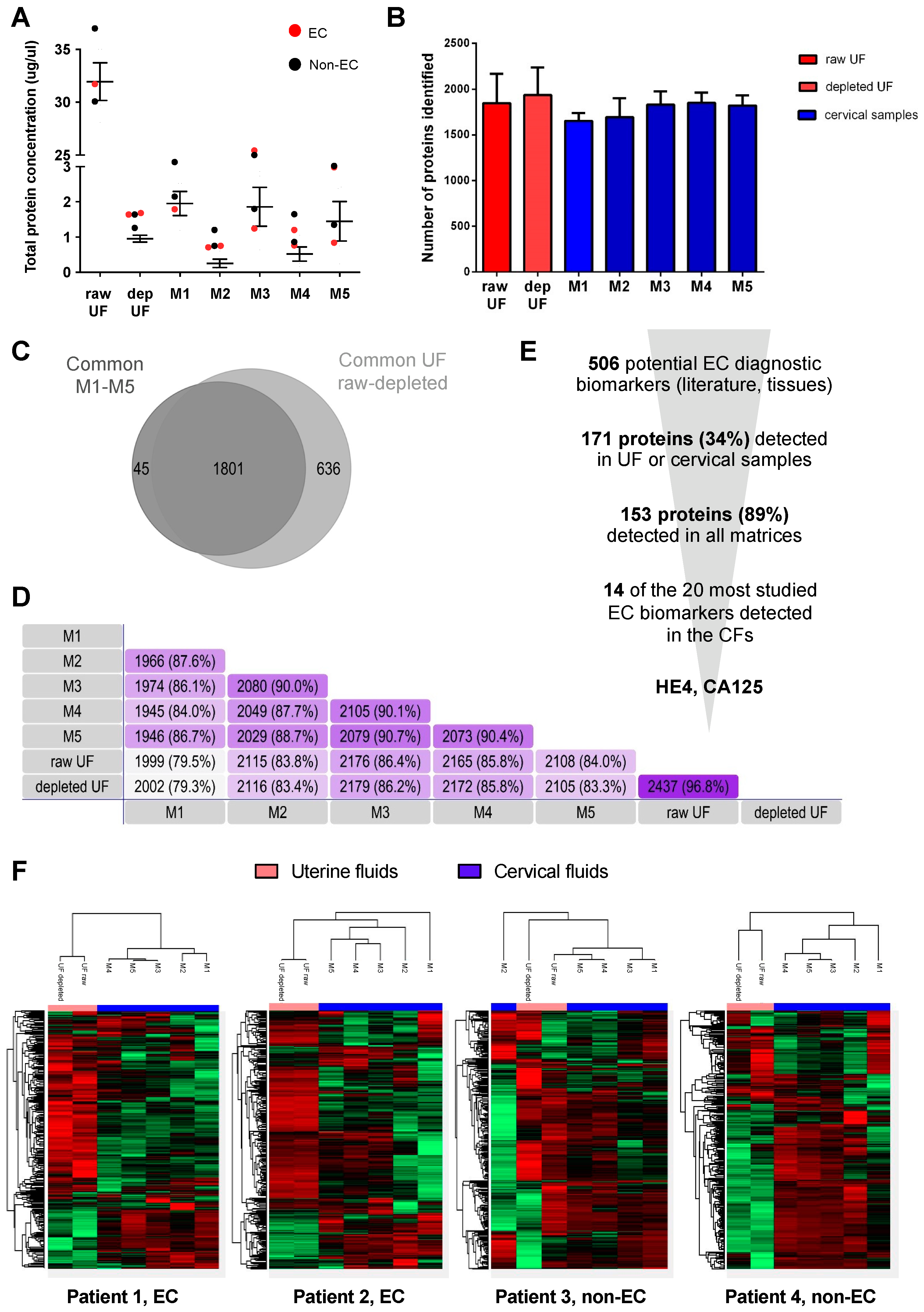
3.1. Proteomic Characterization of Cervical Fluids
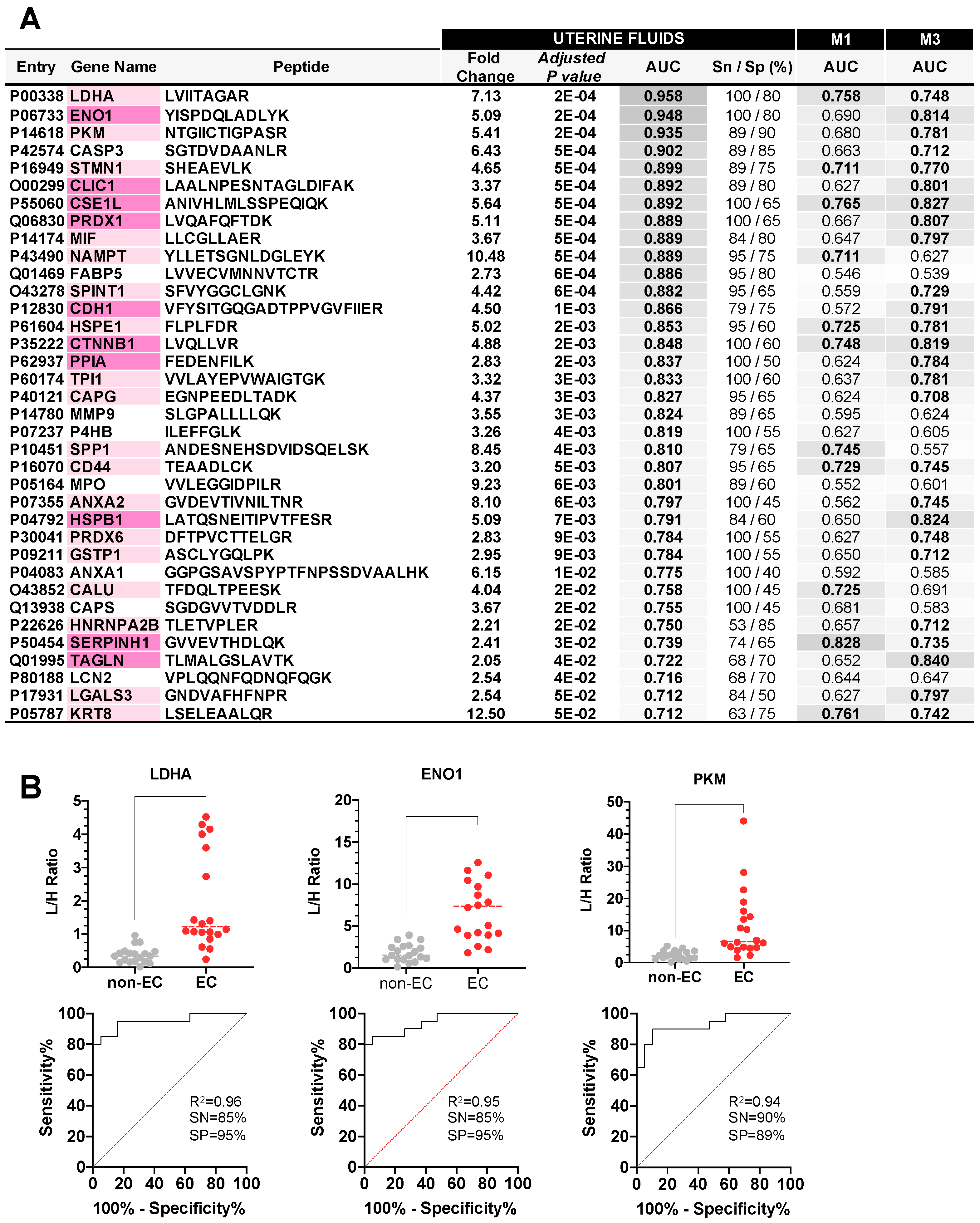
3.2. Measurement of EC-Related Protein Biomarkers in CF
3.3. Selection of M1 and M3 Cervical Samples
3.4. Verification Study of EC Biomarker Candidates in Raw UF and CF
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Clarke, M.A.; Long, B.J.; Del Mar Morillo, A.; Arbyn, M.; Bakkum-Gamez, J.N.; Wentzensen, N. Association of Endometrial Cancer Risk with Postmenopausal Bleeding in Women a Systematic Review and Meta-Analysis. JAMA Intern. Med. 2018, 178, 1201–1208. [Google Scholar] [CrossRef]
- van Hanegem, N.; Prins, M.M.C.; Bongers, M.Y.; Opmeer, B.C.; Sahota, D.S.; Mol, B.W.J.; Timmermans, A. The Accuracy of Endometrial Sampling in Women with Postmenopausal Bleeding: A Systematic Review and Meta-Analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 197, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Raffone, A.; Troisi, J.; Boccia, D.; Travaglino, A.; Capuano, G.; Insabato, L.; Mollo, A.; Guida, M.; Zullo, F. Metabolomics in Endometrial Cancer Diagnosis: A Systematic Review. Acta Obstet. Gynecol. Scand. 2020, 99, 1135–1146. [Google Scholar] [CrossRef]
- Troisi, J.; Mollo, A.; Lombardi, M.; Scala, G.; Richards, S.M.; Symes, S.J.K.; Travaglino, A.; Neola, D.; de Laurentiis, U.; Insabato, L.; et al. The Metabolomic Approach for the Screening of Endometrial Cancer: Validation from a Large Cohort of Women Scheduled for Gynecological Surgery. Biomolecules 2022, 12, 1229. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, E.; Lopez-Gil, C.; Campoy, I.; Vallve, J.; Coll, E.; Cabrera, S.; Ramon Y Cajal, S.; Matias-Guiu, X.; van Oostrum, J.; Reventos, J.; et al. Advances in Endometrial Cancer Protein Biomarkers for Use in the Clinic. Expert Rev. Proteom. 2018, 15, 81–99. [Google Scholar] [CrossRef] [PubMed]
- Barr, C.E.; Njoku, K.; Jones, E.R.; Crosbie, E.J. Serum CA125 and HE4 as Biomarkers for the Detection of Endometrial Cancer and Associated High-Risk Features. Diagnostics 2022, 12, 2834. [Google Scholar] [CrossRef]
- Knific, T.; Osredkar, J.; Smrkolj, Š.; Tonin, I.; Vouk, K.; Blejec, A.; Frković Grazio, S.; Rižner, T.L. Novel Algorithm Including CA-125, HE4 and Body Mass Index in the Diagnosis of Endometrial Cancer. Gynecol. Oncol. 2017, 147, 126–132. [Google Scholar] [CrossRef]
- Dong, C.; Liu, P.; Li, C. Value of HE4 Combined with Cancer Antigen 125 in the Diagnosis of Endometrial Cancer. Pak. J. Med. Sci. 2017, 33, 1013. [Google Scholar] [CrossRef]
- Li, L.; Zhu, Y.; Zhong, Y.; Su, T.; Fan, X.; Xi, Q.; Li, M.; Fu, J.; Tan, H.; Liu, S. Human Epididymis Protein 4 in Endometrial Cancer: A Meta-Analysis. Clin. Chim. Acta 2018, 482, 215–223. [Google Scholar] [CrossRef]
- Colas, E.; Perez, C.; Cabrera, S.; Pedrola, N.; Monge, M.; Castellvi, J.; Eyzaguirre, F.; Gregorio, J.; Ruiz, A.; Llaurado, M.; et al. Molecular Markers of Endometrial Carcinoma Detected in Uterine Aspirates. Int. J. Cancer 2011, 129, 2435–2444. [Google Scholar] [CrossRef]
- Perez-Sanchez, C.; Colas, E.; Cabrera, S.; Falcon, O.; Sanchez-del-Río, A.; García, E.; Fernández-de-Castillo, L.; Muruzabal, J.C.; Alvarez, E.; Fiol, G.; et al. Molecular Diagnosis of Endometrial Cancer from Uterine Aspirates. Int. J. Cancer 2013, 133, 2383–2391. [Google Scholar] [CrossRef]
- Martinez-Garcia, E.; Lesur, A.; Devis, L.; Campos, A.; Cabrera, S.; van Oostrum, J.; Matias-Guiu, X.; Gil-Moreno, A.; Reventos, J.; Colas, E.; et al. Development of a Sequential Workflow Based on LC-PRM for the Verification of Endometrial Cancer Protein Biomarkers in Uterine Aspirate Samples. Oncotarget 2016, 7, 53102–53115. [Google Scholar] [CrossRef]
- Martinez-Garcia, E.; Lesur, A.; Devis, L.; Cabrera, S.; Matias-Guiu, X.; Hirschfeld, M.; Asberger, J.; van Oostrum, J.; Casares de Cal, M.d.L.Á.; Gómez-Tato, A.; et al. Targeted Proteomics Identifies Proteomic Signatures in Liquid Biopsies of the Endometrium to Diagnose Endometrial Cancer and Assist in the Prediction of the Optimal Surgical Treatment. Clin. Cancer Res. 2017, 23, 6458–6467. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-Wide Label-Free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Costas, L.; Frias-Gomez, J.; Guardiola, M.; Benavente, Y.; Pineda, M.; Pavón, M.Á.; Martínez, J.M.; Climent, M.; Barahona, M.; Canet, J.; et al. New Perspectives on Screening and Early Detection of Endometrial Cancer. Int. J. Cancer 2019, 145, 3194–3206. [Google Scholar] [CrossRef]
- Frias-Gomez, J.; Benavente, Y.; Ponce, J.; Brunet, J.; Ibáñez, R.; Peremiquel-Trillas, P.; Baixeras, N.; Zanca, A.; Piulats, J.M.; Aytés, Á.; et al. Sensitivity of Cervico-Vaginal Cytology in Endometrial Carcinoma: A Systematic Review and Meta-Analysis. Cancer Cytopathol. 2020, 128, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Bakkum-Gamez, J.N.; Wentzensen, N.; Maurer, M.J.; Hawthorne, K.M.; Voss, J.S.; Kroneman, T.N.; Famuyide, A.O.; Clayton, A.C.; Halling, K.C.; Kerr, S.E.; et al. Detection of Endometrial Cancer via Molecular Analysis of DNA Collected with Vaginal Tampons. Gynecol. Oncol. 2015, 137, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Reijnen, C.; van der Putten, L.J.M.; Bulten, J.; Snijders, M.P.L.M.; Küsters-Vandevelde, H.V.N.; Sweegers, S.; Vos, M.C.; van der Wurff, A.A.M.; Ligtenberg, M.J.L.; Massuger, L.F.A.G.; et al. Mutational Analysis of Cervical Cytology Improves Diagnosis of Endometrial Cancer: A Prospective Multicentre Cohort Study. Int. J. Cancer 2020, 146, 2628–2635. [Google Scholar] [CrossRef]
- Liew, P.-L.; Huang, R.-L.; Wu, T.-I.; Liao, C.-C.; Chen, C.-W.; Su, P.-H.; Wang, H.-C.; Weng, Y.-C.; Lai, H.-C. Combined Genetic Mutations and DNA-Methylated Genes as Biomarkers for Endometrial Cancer Detection from Cervical Scrapings. Clin. Epigenetics 2019, 11, 1–10. [Google Scholar] [CrossRef]
- Sangtani, A.; Wang, C.; Weaver, A.; Hoppman, N.L.; Kerr, S.E.; Abyzov, A.; Shridhar, V.; Staub, J.; Kocher, J.-P.A.; Voss, J.S.; et al. Combining Copy Number, Methylation Markers, and Mutations as a Panel for Endometrial Cancer Detection via Intravaginal Tampon Collection. Gynecol. Oncol. 2020, 156, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Herzog, C.; Marín, F.; Jones, A.; Evans, I.; Reisel, D.; Redl, E.; Schreiberhuber, L.; Paytubi, S.; Pelegrina, B.; Carmona, A.; et al. A Simple Cervicovaginal Epigenetic Test for Screening and Rapid Triage of Women With Suspected Endometrial Cancer: Validation in Several Cohort and Case/Control Sets. J. Clin. Oncol. 2022, 40, 3828–3838. [Google Scholar] [CrossRef]
- Njoku, K.; Chiasserini, D.; Geary, B.; Pierce, A.; Jones, E.R.; Whetton, A.D.; Crosbie, E.J. Comprehensive Library Generation for Identification and Quantification of Endometrial Cancer Protein Biomarkers in Cervico-Vaginal Fluid. Cancers 2021, 13, 3804. [Google Scholar] [CrossRef]



| Method Optimization | Verification Phase | ||||
|---|---|---|---|---|---|
| EC | non-EC | EC | non-EC | ||
| (n = 2) | (n = 2) | (n = 22) | (n = 19) | ||
| Age (years) | |||||
| Mean | 69 | 50 | 70 | 60 | |
| Minimum | 63 | 48 | 49 | 23 | |
| Maximum | 75 | 51 | 93 | 88 | |
| Uterine condition | |||||
| Premenopausal | - | 1 | 1 | 3 | |
| Postmenopausal | 2 | 1 | 21 | 16 | |
| Benign gynecological condition | |||||
| Atrophic endometrium | 2 | 13 | |||
| Normal endometrium | - | 3 | |||
| Endometrial polyp | - | 2 | |||
| Simple hyperplasia, no atypia | - | 1 | |||
| Histological type | |||||
| Endometrioid | 2 | 18 | |||
| Serous | - | 3 | |||
| Others (carcinosarcoma) | - | 1 | |||
| Histological grade | |||||
| Low-grade | 1 | 14 | |||
| High-grade | 1 | 8 | |||
| FIGO stage | |||||
| IA | 1 | 9 | |||
| IB | 1 | 5 | |||
| II | - | 4 | |||
| IIIC2 | - | 3 | |||
| IVB | - | 1 | |||
| Miometrial invasion | |||||
| <50% | 1 | 13 | |||
| >50% | 1 | 9 | |||
| Lymphovascular invasion | |||||
| Yes | - | 7 | |||
| No | 2 | 15 | |||
| Molecular classification | |||||
| POLEmut | - | - | |||
| MMRd | 2 | 9 | |||
| NSMP | - | 5 | |||
| p53mut | - | 2 | |||
| NA | - | 6 | |||
| Samples collected | |||||
| Pipelle biopsy (PB) | PB | PB | |||
| Cervical samples | M1, M2, M3, M4, M5 | M1, M3 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Garcia, E.; Coll-de la Rubia, E.; Lesur, A.; Dittmar, G.; Gil-Moreno, A.; Cabrera, S.; Colas, E. Cervical Fluids Are a Source of Protein Biomarkers for Early, Non-Invasive Endometrial Cancer Diagnosis. Cancers 2023, 15, 911. https://doi.org/10.3390/cancers15030911
Martinez-Garcia E, Coll-de la Rubia E, Lesur A, Dittmar G, Gil-Moreno A, Cabrera S, Colas E. Cervical Fluids Are a Source of Protein Biomarkers for Early, Non-Invasive Endometrial Cancer Diagnosis. Cancers. 2023; 15(3):911. https://doi.org/10.3390/cancers15030911
Chicago/Turabian StyleMartinez-Garcia, Elena, Eva Coll-de la Rubia, Antoine Lesur, Gunnar Dittmar, Antonio Gil-Moreno, Silvia Cabrera, and Eva Colas. 2023. "Cervical Fluids Are a Source of Protein Biomarkers for Early, Non-Invasive Endometrial Cancer Diagnosis" Cancers 15, no. 3: 911. https://doi.org/10.3390/cancers15030911
APA StyleMartinez-Garcia, E., Coll-de la Rubia, E., Lesur, A., Dittmar, G., Gil-Moreno, A., Cabrera, S., & Colas, E. (2023). Cervical Fluids Are a Source of Protein Biomarkers for Early, Non-Invasive Endometrial Cancer Diagnosis. Cancers, 15(3), 911. https://doi.org/10.3390/cancers15030911








